Abstract
The evolutionarily conserved spindle checkpoint is a key mechanism ensuring high-fidelity chromosome transmission. The checkpoint monitors attachment between kinetochores and mitotic spindles and the tension between sister kinetochores. In the absence of proper attachment or tension, the spindle checkpoint mediates cell cycle arrest prior to anaphase. Saccharomyces cerevisiae Mad1p is required for the spindle checkpoint and for chromosome transmission fidelity. Moreover, Mad1p associates with the nuclear pore complex (NPC) and is enriched at kinetochores upon checkpoint activation. Using partial mad1 deletion alleles we determined that the C-terminal half of Mad1p is necessary and sufficient for checkpoint activation in response to microtubule depolymerizing agents, high-fidelity transmission of a reporter chromosome fragment, and in vivo association with centromeres, but not for robust NPC association. Thus, spindle checkpoint activation and chromosome transmission fidelity correlate and these Mad1p functions likely involve kinetochore association but not robust NPC association. These studies are the basis for elucidating the role of protein complexes containing Mad1p in the spindle checkpoint pathway and in maintaining genome stability in S. cerevisiae and other systems.
A critical step in the cell cycle is the proper attachment of chromosomes to the mitotic spindle during metaphase. The evolutionarily conserved spindle checkpoint monitors the interaction between the kinetochores, complexes of proteins bound to the centromeres of chromosomes, and the microtubules that form the mitotic spindle (reviewed in Musacchio and Hardwick 2002). In metaphase, sister kinetochores attach to microtubules emanating from opposite spindle pole bodies during alignment of the chromosomes. Tension is generated when the pulling forces exerted by the spindle are opposed by cohesin proteins that hold sister chromatids together. In addition to deficiencies in attachment of kinetochores to spindle microtubules, the spindle checkpoint is sensitive to loss of tension (Musacchio and Hardwick 2002). When absence of kinetochore attachment or tension is detected, the spindle checkpoint halts the cell cycle prior to anaphase. Failure to arrest can result in aneuploidy and cell death and in vertebrates may be a cause of cancer (reviewed in Draviam et al. 2004).
The mitotic arrest deficient (mad) and budding uninhibited by benzimidazole (bub) yeast mutants, corresponding to the MAD1-MAD3 and BUB1-BUB3 genes, were identified by genetic screens in Saccharomyces cerevisiae on the basis of the failure of the mutants to arrest under checkpoint-inducing conditions (Hoyt et al. 1991; Li and Murray 1991). MPS1, an additional checkpoint gene, was originally identified in a screen for mutants of spindle pole body duplication (Weiss and Winey 1996). Orthologs of the MADs, BUBs, and MPS1 exist in other eukaryotes, including humans (reviewed in Skibbens and Hieter 1998). In contrast to budding yeast, where the MAD and BUB genes are not essential, mutation of BUB1 in Drosophila melanogaster or MAD2 or BUB3 in Mus musculus is lethal (Basu et al. 1999; Dobles et al. 2000; Kalitsis et al. 2000) while RNAi against MAD1 or MAD2 orthologs in Caenorhabditis elegans produces embryonic lethality, sterility, and reduced brood size (Kitagawa and Rose 1999). In budding yeast, mutations or deletions in MAD1, MAD2, BUB1, or BUB3 resulted in defects in chromosome transmission fidelity (CTF), while mad3Δ strains did not exhibit a ctf phenotype (Li and Murray 1991; Pangilinan and Spencer 1996; Warren et al. 2002). Mouse cells mutant for MAD2 or BUB3 exhibit phenotypes consistent with chromosome segregation defects (Dobles et al. 2000; Kalitsis et al. 2000). These results indicate that chromosome loss in cells with mutations of checkpoint genes is not restricted to budding yeast.
Current models propose that enrichment of spindle checkpoint proteins at improperly attached kinetochores leads to the production of a diffusible signal that inhibits the anaphase-promoting complex/cyclosome (APC/C; reviewed in Yu 2002). Consistent with this model, budding yeast Mad1p, Mad2p, Bub1p, and Bub3p proteins are enriched at kinetochores (Kerscher et al. 2003; Kitagawa et al. 2003; Gillett et al. 2004) as are the vertebrate homologs of these proteins under conditions that activate the spindle checkpoint (Musacchio and Hardwick 2002). However, it is not clear if there is an absolute requirement for enrichment of checkpoint proteins at the kinetochore for spindle checkpoint activation to occur (Martin-Lluesma et al. 2002). The APC/C mediates the degradation of proteins whose decay is required for anaphase onset [e.g., Pds1p (Cohen-Fix et al. 1996)]. Several different combinations of checkpoint proteins have been proposed to negatively regulate APC activity, although the nature of the in vivo inhibitory complex has yet to be fully elucidated (Yu 2002).
In budding yeast and humans, Mad1p and Mad2p localize to the nuclear pore complex (NPC) (Campbell et al. 2001; Iouk et al. 2002). Xenopus laevis and Schizosaccharomyces pombe Mad1p and Mad2p, as well has human Mps1p, are localized to the nuclear periphery (Chen et al. 1998; Ikui et al. 2002; Liu et al. 2003) and may also reside at the NPC. The NPC is a proteinacious portal through which proteins and RNA traffic between the nucleoplasm and the cytoplasm (Fabre and Hurt 1997). Mad1p has a role in NPC function in budding yeast, as mad1Δ mutants are defective in nuclear import of a protein substrate (Iouk et al. 2002).
Studies to date indicate that Mad1p is required for the spindle checkpoint and for high-fidelity chromosome transmission and that Mad1p localizes to kinetochores upon checkpoint activation and is a component of the NPC. The S. cerevisiae system provided several advantages to investigate the interrelationship between these Mad1p properties. As MAD1 is not essential, we constructed strains in which the MAD1 locus was replaced by MAD1 truncations or MAD1 truncations fused to GFP that represent the only Mad1p in the cell. Using these strains in combination with growth assays, chromosome transmission fidelity experiments, chromatin immunoprecipitation (ChIP), and live cell imaging studies, we established that the C-terminal half of Mad1p is necessary and sufficient for checkpoint function, high-fidelity chromosome transmission, and centromere enrichment, but not for robust NPC association. A genetic strategy indicated that loss of the C-terminal half of Mad1p is lethal when combined with mutations in genes involved in kinetochore function, chromosome cohesion, and spindle function. These results indicate that the processes of spindle checkpoint activation and chromosome transmission are mediated by an activity carried out by the C-terminal half of Mad1p and that these processes likely involve centromere association.
MATERIALS AND METHODS
Yeast media and strains:
Yeast media were made as previously described (Burke et al. 2000). For CTF experiments, synthetic deficient (SD) medium containing 6 μg/ml of adenine was used (Warren et al. 2002). Media used for synthetic genetic analysis (SGA) were as previously described (Tong et al. 2004). All yeast strains used in this study are described in Table 1. These strains were constructed by a PCR strategy such that the mad1 alleles are terminated by the ADH1 terminator and either marked with natR or fused to GFP and marked with HIS5. The alleles were integrated into the genome at the MAD1 locus and were verified by PCR and sequencing.
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| Y3047 | MATα mfa1::ΔMFA1-HIS3 can1Δ::natR his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 |
| YPH278 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB1113 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ leu2Δ1 mad1Δ::kanMX + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2562 | MATα can1Δ::MFA1pr-HIS3 mad1Δ::mad1 3C/natMX his3Δ leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 |
| YMB2566 | MATα can1Δ::MFA1pr-HIS3 mad1Δ::mad1 1-2C/natMX his3Δ leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 |
| YMB2586 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 mad1Δ::mad1 3C/natMX + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2588 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 mad1Δ::mad1 1C/natMX + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2589 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 mad1Δ::mad1 1-2C/natMX + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2623 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 mad1Δ::MAD1 1-2-3C/natMX + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2634 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 mad1Δ::MAD1 1-2-3C-GFP/HIS5 + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2639 | MATα can1Δ::MFA1pr-HIS3 mad1Δ::natMX his3Δ leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 |
| YMB2670 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 mad1Δ::mad1 1-2C-GFP/HIS5 + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2687 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 mad1Δ::mad1 3C-GFP/HIS5 + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2784 | MATα ura3-52 ade2-101 his3Δ200 leu2Δ1 mad1Δ::mad1 1C-GFP/HIS5 + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2817 |
MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ leu2Δ1 mad1Δ::mad1 2C-GFP/HIS5 + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2856 |
MATaura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 mad1Δ::mad1 2-3C-GFP/HIS5 + CFIII (CEN3.L.YPH278) URA3 SUP11 |
| YMB2877 | Matα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 |
Assays for benomyl sensitivity and cell cycle arrest in the presence of nocodazole:
Strains grown to logarithmic phase were serially diluted (fivefold) and 3 μl of each dilution was spotted on medium containing benomyl (12 μg/ml) or DMSO as a control. Plates were incubated at 25° for 3 days. Nocodazole (M1404, Sigma Aldrich) was added to a final concentration of 15 μg/ml to early logarithmic phase cultures that were then incubated for 4 hr at 30°. Following incubation, the cells were fixed with paraformaldehyde and stained with 4′,6-diamidino-2-phenylindole (DAPI; 10 μg/ml; D9542, Sigma, St. Louis). Cell cycle arrest was scored by fluorescence microscopy using an Axioskop 2 (Carl Zeiss MicroImaging, Thornwood, NY) microscope fitted with a DAPI filter set (Chroma Technology, Brattleboro, VT). Each experiment was repeated three times in which 200 cells were examined.
Western blotting:
Protein extracts from the Mad1-GFP strains were made from early logarithmic phase cultures grown in YEPD at 30° as described previously (Lamb et al. 1994). Following separation with SDS-PAGE and transfer to PVDF membrane, antibodies against GFP (1:5000) (1814460; Roche, Indianapolis) or anti-Pgk1p (1:10,000) (A-6457; Molecular Probes, Eugene, OR) and a mouse secondary antibody (1:20,000) (NA931V; Amersham Biosciences, Piscataway, NJ) were used to detect Mad1-GFP and Pgk1 proteins using a SuperSignal West Femto maximum sensitivity substrate kit (34095; Pierce, Rockford, IL).
Chromosome transmission fidelity assays:
This assay was performed as previously described (Hieter et al. 1985; Spencer et al. 1990). A minimum of 4000 colonies were analyzed for each strain (solid red colonies were not counted). Colonies that were completely red were not included in the analyses. Chromosome loss rates are shown as the number of at least half-red/half-white colonies per 1000 non-solid-red colonies (Warren et al. 2002).
Synthetic genetic analysis:
For SGA, we followed the methodology as previously described (Tong et al. 2004), using a VersArray colony picker (Bio-Rad, Hercules, CA) for strain manipulations in 96-colony arrays. Strains with mad1 1-2C (YMB2566) or mad1 3C (YMB2562) marked with natR were mated to 26 strains that are a subset of strains with deletions (deleted with kanMX4) in genes previously shown to genetically interact with MAD1 [MAD1 synthetic partners (MSPs) (Lee and Spencer 2004)]. Controls included mating of the 26 mspΔ strains to a “wild-type” strain in which the CAN1 locus was replaced with natR (Y3047) and to a mad1Δ::natR strain (YMB2639). The growth of the kanMX4- and natR-resistant strains was quantitated after 1 day of growth. The plates were scanned to determine the pixel density of each colony using Scion (Bethesda, MD) Image software and the pixel densities were assigned a scaled score (0–5) with 5 representing maximum growth. All crosses were done at least three times and the average scaled score was determined for all strains analyzed. The average scaled scores were compared using Student's t-test at a 95% confidence interval to determine which mspΔ strains, when combined with the MAD1 alleles, gave rise to growth that was similar in size to the mad1ΔmspΔ or MAD1mspΔ strains.
Fluorescence microscopy:
Early logarithmic phase cells grown at 30° in YEPD were resuspended in 1× PBS with DAPI (1 μg/ml) and incubated for 30 min. Cells were examined using a Delta Vision system (Applied Precision, Issaquah, WA) consisting of an Olympus IX70 inverted microscope (Olympus America, Melville, NY) and a Photometrics (Huntington Beach, CA) CH350 12-bit camera with a KAF1400 chip. Filter sets used included a FITC filter set (Ex 490/20; Em 528/38; Polychroic mirror) and a DAPI filter set (Ex 360/40; Em 457/50; Polychroic mirror) (Chroma Technology).
Chromatin immunoprecipitation:
ChIP assays were done using minor modifications of protocols described previously (Crotti and Basrai 2004; Gillett et al. 2004). Briefly, chromatin was extracted from either logarithmically grown cultures or cultures treated with nocodazole (final concentration of 25 μg/ml) for 3 hr at 25°. Following a 1-hr crosslinking with formaldehyde and termination by addition of glycine, cells were lysed by vortexing in the presence of glass beads. For immunoprecipitation (IP), we used anti-GFP antibodies (632459; Clontech, Palo Alto, CA). Chromatin from total, IP, and mock (no antibody) samples was analyzed using PCR and primers specific for CEN IV or ACT1. The PCR products from total, IP, and mock samples were quantitated using Image Quant TL (Amersham Biosciences). Dilutions from the total were used to create a calibration curve and used to normalize the values obtained for IP and mock. The concentrations of the IP and mock chromatin samples were 10-fold greater than the total and values were adjusted accordingly. Each IP was repeated at least two times and similar results were obtained.
RESULTS
The C terminus of Mad1p is necessary and sufficient for the spindle checkpoint:
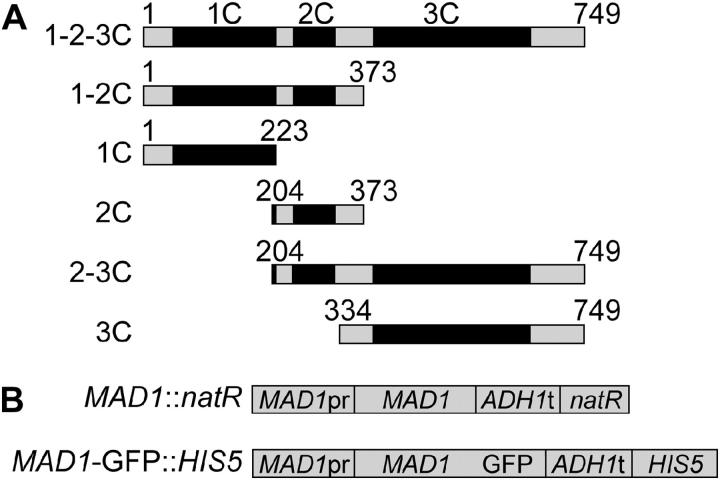
To determine the regions of Mad1p that function in the spindle checkpoint, chromosome transmission fidelity, and CEN enrichment, we undertook molecular dissection of the MAD1 gene. The S. cerevisiae Mad1p protein consists of three predicted coiled coil domains. Alleles of MAD1 were constructed that express different combinations of these domains (Figure 1A). The N-terminal half of Mad1p contains the first and second coiled coil domains (Mad1 1-2C) and the C-terminal half contains the third coiled coil domain (Mad1 3C). The genomic copy of MAD1 was replaced with MAD1 alleles that were fused to GFP (marked with HIS5) or marked with natR (Figure 1B).
Figure 1.—
Schematic of full-length Mad1p and proteins expressed by MAD1 alleles. (A) Mad1p contains three predicted coiled coil domains (“C”) shown as solid boxes. 1-2-3C denotes full-length protein containing all three coiled coil domains, and likewise 1-2C, 1C, 2C, 2-3C, or 3C designations are based on which coiled coil domains are present in the protein. The numbers indicate the amino acids of Mad1p. (B) Strains were made with the MAD1 alleles marked with natR and made with the MAD1 alleles fused to GFP and marked with HIS5.
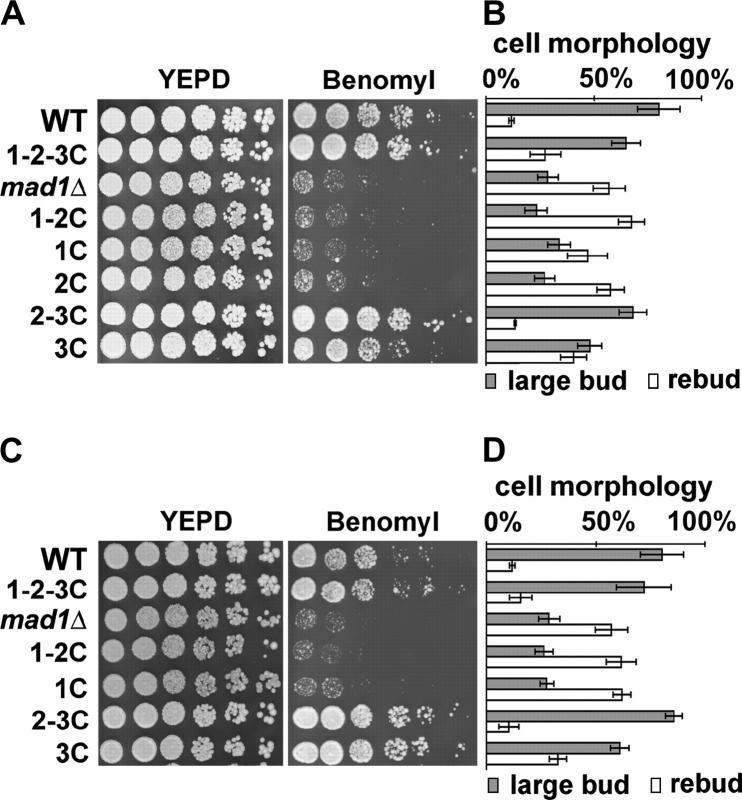
We first examined the strains for checkpoint competence by monitoring their ability to grow on medium containing benomyl. Benomyl is a microtubule-depolymerizing drug that activates the spindle checkpoint (Hoyt et al. 1991; Li and Murray 1991). Failure to activate the checkpoint results in cell death and is visualized as growth inhibition on benomyl-containing medium. As shown in Figure 2A, the growth of a strain expressing full-length Mad1 (1-2-3C-GFP) was similar to that of wild type, while the growth of the mad1Δ strain was inhibited (Figure 2A). The growth of strains expressing the first and second (1-2C-GFP) coiled coil domain, the first coiled coil domain (1C-GFP), or the second coiled coil domain (2C-GFP) was reduced to a level similar to that of mad1Δ (Figure 2A). In contrast, the growth of a strain expressing the second and third coiled coil domain (2-3C-GFP) or the third coiled coil domain (3C-GFP) was similar to that of wild type (Figure 2A). The strains expressing natR-marked MAD1 alleles exhibited characteristics similar to the MAD1-GFP-fusion strains (Figure 2C). These observations indicate that the C-terminal half of Mad1p is necessary and sufficient for benomyl tolerance.
Figure 2.—
The C-terminal half of Mad1p confers checkpoint competence. Benomyl sensitivity and nocodazole arrest assays of strains containing MAD1 alleles are shown. For benomyl sensitivity (A), fivefold serial dilutions of logarithmic-phase cells were spotted on YEPD plates containing either DMSO (control) or benomyl (12 μg/ml) and incubated at 25° for 3 days. (B) Early logarithmic-phase cells were treated with nocodazole (15 μg/ml) for 4 hr at 30° and stained with DAPI. The graph shows the percentages of large-bud arrested cells (shaded bars) and cells that had rebudded without separating nuclear DNA (open bars) for each strain. The percentages of large-bud arrested cells for each strain were: wild-type cells, 81 ± 10%; Mad1 1-2-3C-GFP, 65 ± 7%; mad1Δ, 29 ± 5%; Mad1 1-2C-GFP, 23 ± 5%; Mad1 1C-GFP, 34 ± 5%; Mad1 2C-GFP, 27 ± 5%; Mad1 2-3C-GFP, 68 ± 6%; and Mad1 3C-GFP, 48 ± 6%. The percentage of cells exhibiting neither large-bud arrest nor rebudding, which ranged from 6.0 to 19.0%, is not shown. The experiment was repeated three times and 200 cells of each strain were analyzed. The strains used were: WT (wild type, YPH278), 1-2-3C-GFP (YMB2634), mad1Δ (YMB1113), 1-2C-GFP (YMB2670), 1C-GFP (YMB2784), 2C-GFP (YMB2817), 2-3C-GFP (YMB2856), and 3C-GFP (YMB2687). (C and D) The benomyl sensitivity and nocodazole arrest results for the MAD1 natR-marked strains, respectively. The analysis was carried out as in A and B. The percentages of large-bud arrested cells for each strain were: MAD1 1-2-3C, 72 ± 13%; MAD1 1-2C, 27 ± 4.3%; MAD1 1C, 28 ± 3.3%; MAD1 2-3C, 86 ± 4%; and MAD1 3C, 61 ± 4%. The strains used were: YMB2623 (MAD1 1-2-3C), YMB2589 (MAD1 1-2C), YMB2588 (MAD1 1C), YMB2877 (MAD1 2-3C), and YMB2586 (MAD1 3C). The WT and mad1Δ strains served as controls for all the nocodazole arrest experiments and thus the same data for these strains are plotted in B and D.
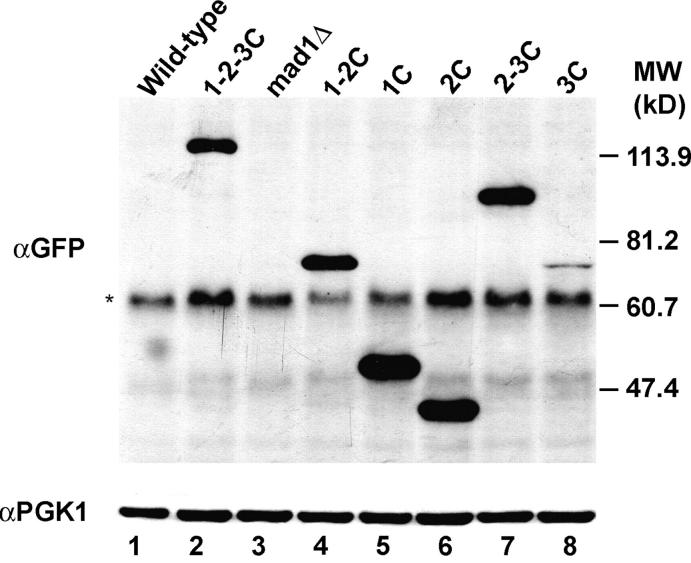
To confirm that the phenotype of strains grown on medium containing benomyl correlated with the ability or failure to arrest the cell cycle, the response of cells grown in liquid medium containing nocodazole, another microtubule depolymerizing agent, was examined. When treated with nocodazole, spindle checkpoint-competent cells arrest as large budded cells with one DAPI-staining mass, while checkpoint-defective cells fail to arrest and rebud with one nuclear mass. As shown in Figure 2B, the majority of wild-type cells (81 ± 10%) arrested as large budded cells with one nuclear mass. Mad1 1-2-3C-GFP-expressing cells also efficiently arrested, while most of the mad1Δ cells failed to arrest. Strains expressing Mad1 1-2C-GFP, Mad1 1C-GFP, or Mad1 2C-GFP failed to arrest efficiently. Mad1 2-3C-GFP and Mad1 3C-GFP strains arrested predominantly as large budded cells with one nuclear mass. Western blotting confirmed the expression of the Mad1-GFP fusion proteins and also revealed that Mad1 3C-GFP accumulates to lower levels relative to the other fusion proteins (Figure 3). The natR-marked MAD1 strains had phenotypes similar to the Mad1-GFP fusion strains (Figure 2, C and D). Taken together, these results indicate that the C-terminal half of Mad1p is both necessary and sufficient for checkpoint activity while the N-terminal half is dispensable for this function.
Figure 3.—
MAD1-GFP fusion alleles are expressed. Western blot analysis is shown of protein samples from wild-type, mad1Δ, and MAD1-GFP/HIS5 strains that were grown to logarithmic phase at 30°. Anti-GFP antibodies (αGFP) were used to detect the MAD1-GFP proteins and anti-Pgk1p antibodies (αPGK1) served as a loading control. The molecular weights corresponding to the marker lane are denoted on the right. The asterisk indicates a nonspecific band.
The C-terminal half of Mad1p does not localize robustly to the NPC:
We localized the Mad1-GFP fusion proteins to determine if the C-terminal half of Mad1p plays a role in NPC localization. As expected, strains expressing Mad1 1-2-3C-GFP exhibited a distinctive pattern of punctate fluorescence at the nuclear periphery (Figure 4a). Our previous genetic, biochemical, and cell biological studies established that this pattern of fluorescence correlates with NPC localization (Iouk et al. 2002). Mad1 1-2C-GFP exhibited a pattern of fluorescence very similar to Mad1 1-2-3C-GFP and was the only Mad1-GFP truncation protein that exhibited a fluorescence pattern indicative of robust NPC association (Figure 4b). Cells expressing Mad1 1C-GFP showed nucleoplasmic accumulation and occasional (∼10% of cells) punctate fluorescence at the nuclear periphery (Figure 4c and data not shown). The punctate fluorescence may indicate weak association with the NPC. The second coiled coil domain alone, Mad1 2C-GFP, showed uniform fluorescence throughout the cells with no distinct localization pattern (Figure 4d). Mad1 2-3C-GFP exhibited fluorescence in the nucleus and, in some cases (∼30%), punctate fluorescence at the nuclear periphery, potentially indicating weak association with the NPC (Figure 4e, large and small arrows, respectively). Mad1 3C-GFP accumulated in the nucleus with no punctate fluorescence indicative of NPC association (Figure 4f). These data indicate that the N-terminal half of Mad1p is necessary for localization similar to wild type and that the C-terminal half of Mad1p does not play a major role in NPC targeting.
Figure 4.—
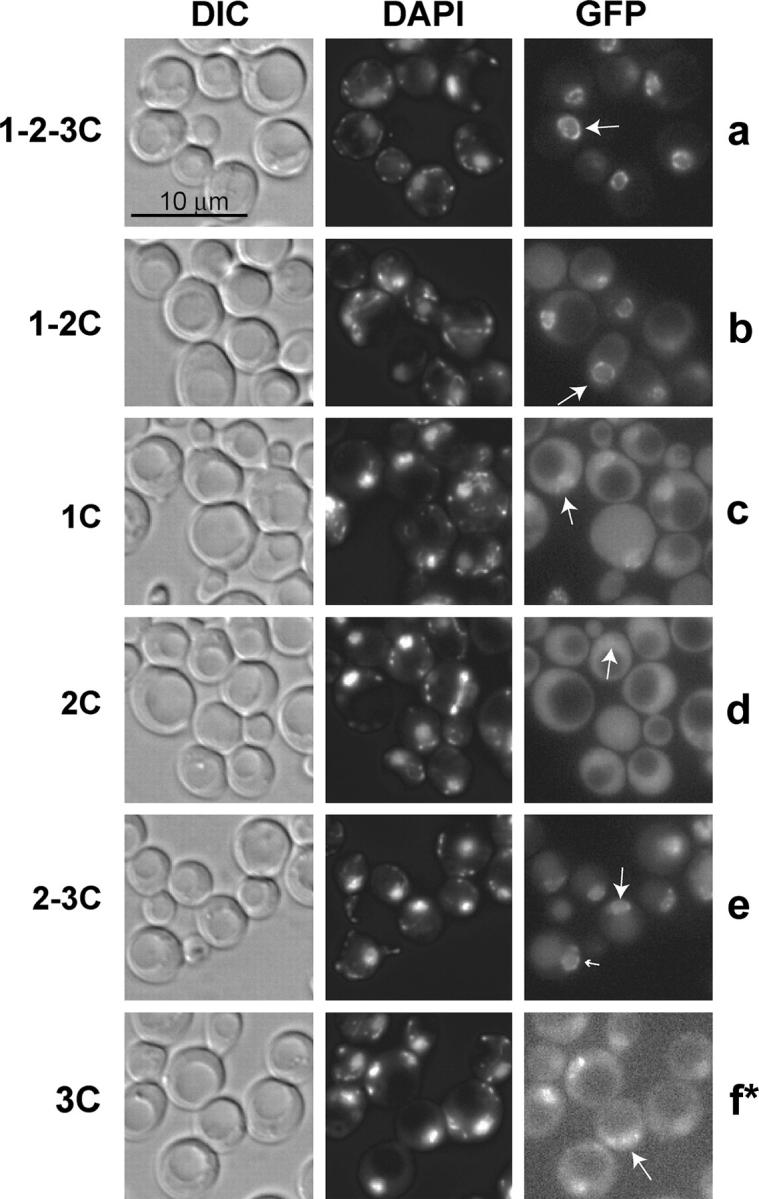
The C-terminal half of Mad1p does not localize robustly to the NPC. Strains containing MAD1-GFP/HIS5 fusions were grown to logarithmic phase at 30°, stained with DAPI, and analyzed by fluorescence microscopy (a–f). Arrows indicate the phenotype observed in >60% of the cells. Bar, 10 μm. At least 100 cells of each strain were examined. The asterisk in f indicates that the signal intensity was adjusted to facilitate visualization of the low level of Mad1 3C-GFP, while a–e are normalized. The strains used were: 1-2-3C-GFP (YMB2634), 1-2C-GFP (YMB2670), 1C-GFP (YMB2784), 2C-GFP (YMB2817), 2-3C-GFP (YMB2856), and 3C-GFP (YMB2687).
The C-terminal half of Mad1p mediates CTF:
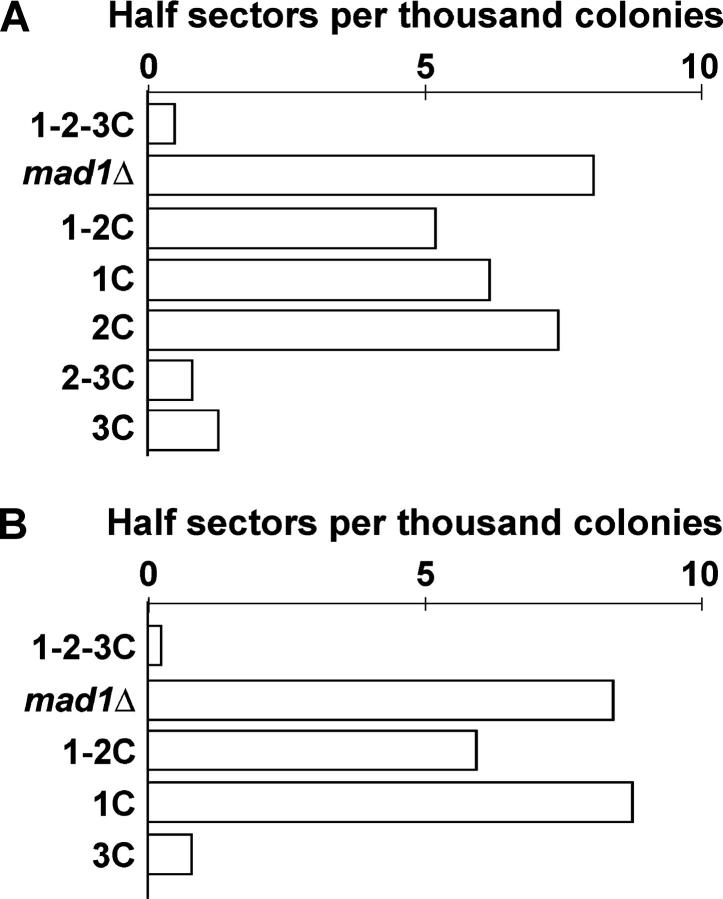
Having determined that checkpoint function, but not robust NPC association, resides in the C-terminal half of Mad1p, we focused on more fully investigating the role that this part of the protein plays in the functions of Mad1p. To examine CTF, the stability of a nonessential chromosome fragment (CF) was monitored as previously described (Warren et al. 2002). Colonies that retain the CF are white, while loss of the CF results in red-sectored colonies (ctf phenotype). The CF loss rate in the first mitotic division was determined by half-sector analyses of colonies grown on solid medium. As shown in Figure 5A, a Mad1 1-2-3C-GFP strain showed negligible CF loss (0.5 half-sectored colony per 1000), while the mad1Δ strain exhibited elevated loss of the CF (8.1/1000). Mad1 1-2C-GFP, Mad1 1C-GFP, and Mad1 2C-GFP strains also exhibited elevated CF loss (5.2/1000, 6.2/1000, and 7.4/1000, respectively), similar to mad1Δ cells. In contrast, a Mad1 2-3C-GFP strain exhibits minimal chromosome loss (0.8/1000) as did the Mad1 3C (1.3/1000) strain. This result indicates that the C-terminal half of Mad1p is sufficient for high-fidelity chromosome transmission comparable to that of wild type. As with the checkpoint assays, the strains expressing the natR-marked MAD1 alleles produced results similar to the strains expressing the MAD1-GFP-fusions, further supporting the requirement and sufficiency of the C-terminal half of Mad1p for CTF (Figure 5B).
Figure 5.—
The C-terminal half of Mad1p is necessary and sufficient for chromosome transmission fidelity (CTF). The WT and mad1 mutant strains contain a nonessential chromosome fragment (CF), which bears a suppressor of ade2-101 present in the strains. Strains bearing the CF produce white colonies while the loss of the CF results in colonies with red sectors. (A) The rate of loss of the CF in the first mitotic division, indicated by a half-red and half-white colony, was determined for the MAD1-GFP strains and the resulting half-sector CF loss rate per 1000 divisions was plotted for strains. The strains used were: 1-2-3C-GFP (YMB2634), mad1Δ (YMB1113), 1-2C-GFP (YMB2670), 1C-GFP (YMB2784), 2C-GFP (YMB2817), 2-3C-GFP (YMB2856), and 3C-GFP (YMB2687). (B) The CF loss rate was determined for the MAD1 natR-marked strains. Strains used were: YMB2623 (MAD1 1-2-3C), YMB1113 (mad1Δ), YMB2589 (MAD1 1-2C), YMB2588 (MAD1 1C), and YMB2586 (MAD1 3C).
mad1 1-2C and mad1 3C exhibit distinct genetic interactions:
A genetic interaction is indicated when a double mutant exhibits a phenotype, such as slow growth, that is not observed with either single mutant. Among the genes that interact with MAD1 (MSPs), synthetic lethal screens have identified genes that encode kinetochore components, genes with roles in maintaining cohesion, and genes involved in microtubule function (Lee and Spencer 2004; Tong et al. 2004). Using the SGA technique (Tong et al. 2001, 2004) we generated mad1 1-2C mspΔ and mad1 3C mspΔ double-mutant haploids. As controls, we generated mad1Δ mspΔ and MAD1mspΔ haploids. The size of the double-mutant colonies was analyzed quantitatively (see materials and methods). A representative example is shown in Figure 6. As indicated in Table 2 (see supplementary Table S1 at http://www.genetics.org/supplemental/ for complete SGA results), the mad1Δ mspΔ strains formed small colonies, confirming the genetic interaction of MAD1 with these MSPs, while the MAD1 mspΔ strains formed large colonies, as expected. Combining mad1 1-2C with the mspΔ deletions indicated in Table 2 resulted in small colonies. In contrast, combination of mad1 3C with these mspΔ strains resulted in large colonies. These results are consistent with the ability of the C-terminal half of Mad1p to mediate checkpoint arrest. Further, it is likely that the slow-growth phenotypes of many of the reported mad1Δ mspΔ double mutants (Lee and Spencer 2004; Tong et al. 2004) result from checkpoint deficiency combined with kinetochore, spindle, or cohesion defects.
Figure 6.—
Growth of strains with MAD1, mad1Δ, mad1 1-2C, or mad1 3C combined with chl4Δ. MAD1 and mad1 strains were mated with strains bearing mutations in genes reported to interact synthetically with MAD1 (materials and methods). The growth of double-mutant haploids was compared. The growth of a mad1Δ chl4Δ strain is reduced relative to that of the MAD1 chl4Δ strain, a result consistent with the previously reported synthetic genetic interaction between MAD1 and CHL4 (Lee and Spencer 2004; Tong et al. 2004). The growth of mad1 1-2C chl4Δ is similar to the growth of the mad1Δ chl4Δ strain. In contrast, the growth of mad1 3C chl4Δ is similar to that of the MAD1 chl4Δ strains.
TABLE 2.
Representative results of SGA analysis withMAD1 alleles
| Size of double mutants
|
|||||
|---|---|---|---|---|---|
| mspΔa | Annotationb | MAD1 | mad1Δ | 1-2C | 3C |
| chl4Δ | Central KT | Large | Small | Small | Large |
| bik1Δ | Outer KT | Large | Small | Small | Large |
| cin2Δ | MT function | Large | Small | Small | Large |
| ctf4Δ | Cohesion | Large | Small | Small | Large |
mspΔ, MAD1 synthetic partner gene deletion.
Annotation of MSP function. KT, kinetochore; MT, microtubule.
The C-terminal half of Mad1p mediates enrichment at CEN DNA:
Mad1p was previously shown to associate with centromeres when tested in a one-hybrid assay (Warren et al. 2002). Recently, centromere association of Mad1p was shown to be enhanced upon nocodazole treatment using the ChIP technique (Gillett et al. 2004). On the basis of the requirement for the C-terminal half of Mad1p for spindle checkpoint activation and chromosome transmission fidelity, it seemed likely that this part of Mad1p interacts with CEN DNA. ChIP analyses were used to investigate this possibility.
Chromatin from logarithmic phase and nocodazole-treated wild-type and MAD1-GFP strains was crosslinked in vivo and isolated. A portion of each chromatin preparation was retained as a control (total), while the remainder was immunoprecipitated with anti-GFP antibodies (IP) or not treated with antibody (mock). The chromatin was then subjected to PCR using primers specific for CEN IV or specific to ACT1 as a negative control. For each set of PCRs, three dilutions of total confirmed that the PCR conditions were in the linear range of amplification (Figure 7A, lanes 1–3 and 6–8). In logarithmic cells, Mad1 1-2-3C-GFP showed little association with CEN IV (Figure 7A, lane 4) while in nocodazole-treated cells, Mad1 1-2-3C-GFP exhibited CEN IV enrichment to a level similar to that previously reported (Figure 7A, lane 9). Mad1 1-2-3C-GFP was not enriched at a noncentromere locus, the ACT1 gene, and minimal PCR product was produced when chromatin from a wild-type (untagged) strain was used (Figure 7, A and B). Mad1 1-2C-GFP, Mad1 1C-GFP, and Mad1 2C-GFP did not exhibit significant enrichment at CEN IV in logarithmic or in nocodazole-treated cells, indicating the requirement for the C-terminal half of Mad1p to mediate CEN DNA association. In contrast, Mad1 2-3C-GFP and Mad1 3C-GFP showed association with CEN IV in nocodazole-treated cells. We also observed that Mad1 2-3C-GFP was enriched at CEN IV even in logarithmic cells (Figure 7, A and B). The degree of nocodazole-induced enrichment at CEN IV exhibited by Mad1 3C-GFP was lower than that exhibited by either Mad1 1-2-3C-GFP or Mad1 2-3C-GFP. However, as the abundance of Mad1 3C-GFP is lower as determined by Western blotting (Figure 3), this may indicate a comparable degree of CEN IV association. These data indicate that the C-terminal half of Mad1p is both necessary and sufficient for CEN association and that the N-terminal half has no apparent role in this Mad1p property.
Figure 7.—
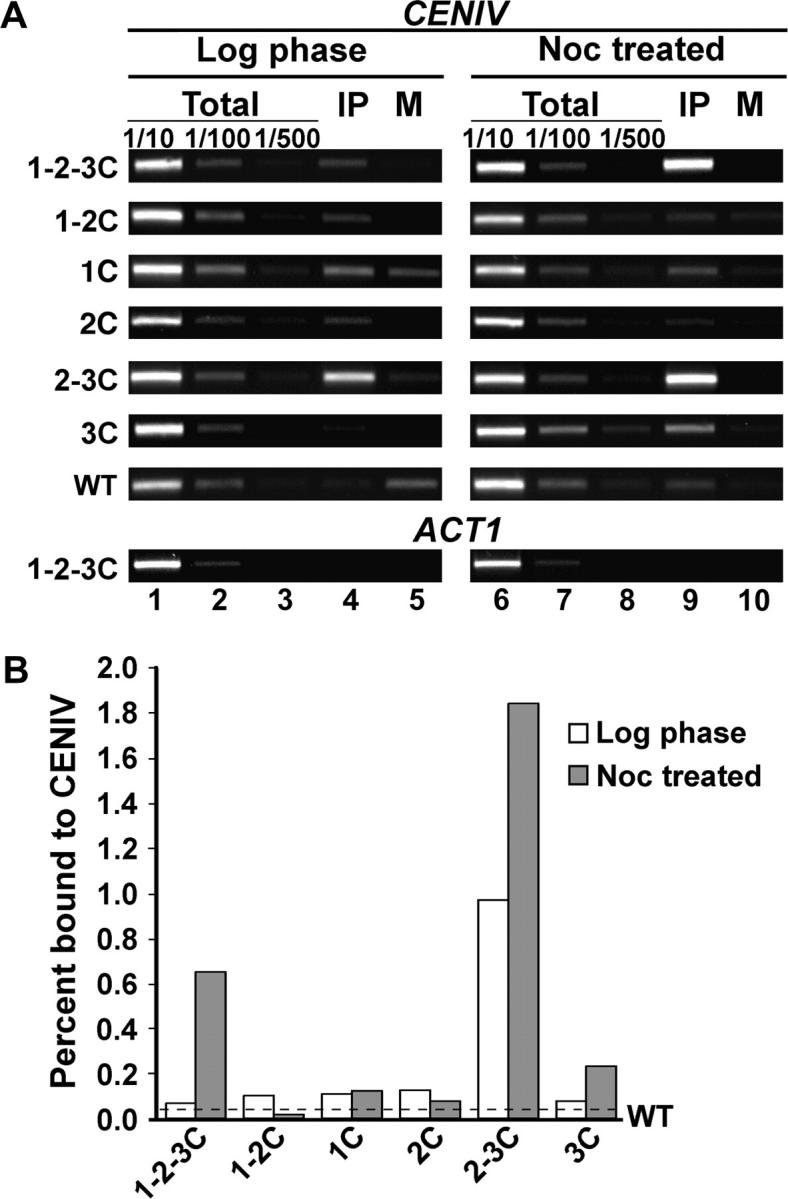
The C-terminal half of Mad1p mediates association with CEN DNA. (A) ChIP experiments were done using MAD1-GFP/HIS5 and wild-type (untagged) strains grown to logarithmic phase (log phase) or log phase cultures treated with nocodazole (noc treated) at 25°. Dilutions of chromatin samples from total (1/10, 1/100, 1/500), immunoprecipitated with anti-GFP (IP), and mock (M) were analyzed using PCR and primers specific for CEN IV or ACT1 as a control. Band intensities were determined with ImageQuant TL software. The PCR yield was proportional to the amount of template. (B) The graph represents the percentage of CEN IV DNA from IP vs. total in log phase and noc-treated samples for each row in A. The dashed line indicates the average percentage of CEN IV DNA detected using a wild-type strain (untagged) in log phase and noc-treated samples and establishes the level of nonspecific signal. Each immunoprecipitation was performed twice and gave similar results.
DISCUSSION
Our studies revealed that the C-terminal half of Mad1p is both necessary and sufficient for checkpoint activation, high-fidelity chromosome transmission, and association with CEN DNA but not robust NPC association. This conclusion is based on our observations that (1) the growth of strains expressing Mad1 3C-GFP was similar to that of wild type on benomyl-containing medium, and these strains arrested when treated with nocodazole; (2) the Mad1 3C-GFP strains also transmitted a CF with high fidelity; and (3) Mad1 3C-GFP associates with CEN DNA upon nocodazole treatment as assayed by ChIP, while live-cell imaging studies indicated that Mad1 3C-GFP does not exhibit robust association with the NPC. A genetic study further confirmed that the C-terminal half of Mad1p contains an activity absent from the N-terminal half. When the C-terminal half of Mad1p is present, deletion of many MSPs does not result in a slow-growth phenotype. In contrast, when this part of Mad1p is absent, deletion of MSPs results in slow growth. Our data point to the importance of the C-terminal half of Mad1p for checkpoint and chromosome transmission functions and indicate that these functions likely involve interaction with kinetochores, but not robust association with NPCs.
Loss of checkpoint activity could cause chromosome loss for cells that fail to arrest upon loss of attachment or tension in an unperturbed cell cycle. Deletion of MAD1, MAD2, BUB1, or BUB3 results in a chromosome loss phenotype; however, mad3Δ strains exhibit little chromosome loss (Warren et al. 2002). In our analyses, strains lacking the C-terminal half of Mad1p grew poorly on medium containing benomyl, failed to arrest efficiently in nocodazole, and exhibited a CF loss phenotype. Conversely, strains that were checkpoint competent transmitted the CF with high fidelity. Consistent with these results, a mad1-1 strain, in which Mad1p has a truncated C terminus, is sensitive to benomyl and exhibits increased loss of chromosome III (Li and Murray 1991; Chen et al. 1999). Our results also agree with a previous study that mapped a minimal region of Mad1p that conferred benomyl resistance to a C-terminal segment (amino acids 392–749) joined to an additional Mad1p segment (amino acids 156–215) expressed from an episome (Chen et al. 1999). We find that a slightly larger Mad1p C-terminal segment (amino acids 334–749) is itself sufficient for benomyl resistance and arrest in nocodazole when expressed as a single-copy gene from the endogenous locus. Taken together, our results indicate that absence of checkpoint function likely contributes to the chromosome loss phenotype of cells lacking the C-terminal half of Mad1p.
Many MSPs encode components of the kinetochore, proteins involved in sister chromatid cohesion, and proteins involved in the function of the mitotic spindle. Deletion of MSPs likely causes deficiencies in kinetochore-spindle attachment and/or tension, and the activity of the spindle checkpoint might be required for the viability of these strains, even during unperturbed growth. When combined with mad1Δ or with mad1 1-2C, mutations that inactivate the spindle checkpoint function of Mad1p, these MSP deletions resulted in growth inhibition. In contrast, combining the checkpoint-competent mad1 3C mutation with many of the MSP deletions did not inhibit growth. These results confirm that synthetic lethality with mad1Δ can be caused by perturbations in kinetochore or spindle function that are lethal in combination with an inability to arrest the cell cycle.
Several spindle checkpoint proteins, including Mad1p, have been localized to CEN DNA using ChIP (Kerscher et al. 2003; Kitagawa et al. 2003; Gillett et al. 2004) and recruitment to kinetochores is believed to be an important event in spindle checkpoint function (Musacchio and Hardwick 2002). Using ChIP, we determined that Mad1-GFP proteins bearing the C-terminal half of Mad1p were enriched at CEN DNA upon nocodazole treatment, while Mad1p-GFP proteins lacking this part of Mad1p were not CEN enriched. The correlation between CEN association and both checkpoint competence and chromosome transmission fidelity indicates that Mad1p function in these processes likely involves interactions with CEN DNA. Interestingly, Mad1 2-3C-GFP exhibits association with CEN DNA even in the absence of nocodazole treatment, and the degree of CEN association under this condition is similar to that of Mad1 1-2-3C-GFP upon nocodazole treatment. However, the CEN association does not appear to indicate checkpoint activation. Strains expressing Mad1 2-3C do not exhibit a marked slow-growth phenotype or increased resistance to benomyl (data not shown), phenotypes that would be expected if the presence of Mad1 2-3C at CEN DNA was sufficient to activate the spindle checkpoint. In addition, Mad1 2-3C-GFP is further enriched at CEN DNA upon nocodazole treatment, potentially indicating a requirement for additional Mad1 2-3C-GFP at the CEN for checkpoint activation.
Xenopus Mad1p (Xmad1) has been reported to localize to kinetochores, and the first 442 amino acids are required for this localization (Chung and Chen 2002). Conversely, a Xmad1 protein containing amino acids 326–718 did not localize to the kinetochore. The requirement for the N terminus for kinetochore localization, rather than the C terminus, contrasts with budding yeast Mad1p and the human homolog of Mad1p, both of which require the C terminus (Figure 5 and Iwanaga et al. 2002). This observation could indicate a difference in the respective Mad1p proteins or may reflect differences in the two systems.
We determined that optimum NPC association of Mad1p required the N-terminal half of Mad1p, but this association is not a requirement for the checkpoint function, CTF, or CEN association of Mad1p. There is limited amino acid sequence conservation between the N-terminus of budding yeast Mad1p and its orthologs (Chen et al. 1998). However coiled coil domains are present in the N-termini of Mad1p's orthologs and it may be these structures, rather than the sequence per se, that mediate NPC association. In vertebrates, NPC association is unlikely a requirement for the checkpoint function, as the nuclear envelope breaks down during mitosis. However, as multiple nucleoporins are recruited to kinetochores in vertebrates (reviewed in Stukenberg and Macara 2003) and S. cerevisiae nup170Δ strains exhibit chromosome loss phenotype (Kerscher et al. 2001), NPC components may make an important contribution to checkpoint control in yeast and animals. Alternatively, it is possible that the NPC association of Mad1p relates to its function in nuclear transport (Iouk et al. 2002) rather than its function in the spindle checkpoint.
The manner in which Mad1p functions in the spindle checkpoint and chromosome transmission is unknown at the molecular level. Our studies will provide a basis for investigation of the mechanisms by which the C-terminal half of Mad1p mediates cell cycle arrest and chromosome transmission fidelity. While our studies indicate that checkpoint competence likely contributes to high-fidelity chromosome transmission it is possible Mad1p may have a role in chromosome transmission that is independent from its checkpoint function. A genetic strategy that targets the C-terminal half of MAD1 might uncover independent checkpoint and chromosome transmission functions.
Acknowledgments
We thank the National Cancer Institute Laboratory of Receptor Biology and Gene Expression Fluorescence Imaging Facility, T. Karpova, and J. McNally for use and assistance with the Delta Vision microscopy system. We thank C. Espelin, Y-J. Kim, L. Crotti, E. Gillett, and P. Sorger for advice on ChIP assays; R. Scott and R. Wozniak for sharing unpublished data; K. Hardwick for strains and reagents; K. Devlin for expert technical assistance during the early course of this work; and N. Harper for tetrad dissections. We acknowledge M. Dasso, M. Lichten, O. Kersher, and M. Nau for insightful comments on the article. We are especially grateful to members of the Basrai and Spencer laboratories, especially W.-C. Au, C. Carter, M. Eckley, Y.-J. Kim, and S. Kokseng, for many fruitful discussions and comments on this article. This work was supported in part by research grants GM50842, GM62368, and HG02432 (F. A. Spencer) and training grant GM007445 (M. S. Lee).
References
- Basu, J., H. Bousbaa, E. Logarinho, Z. Li, B. C. Williams et al., 1999. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146: 13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D. J., D. Dawson and T. Stearns, 2000 Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, pp. 171–181. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Campbell, M. S., G. K. Chan and T. J. Yen, 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114: 953–963. [DOI] [PubMed] [Google Scholar]
- Chen, R. H., A. Shevchenko, M. Mann and A. W. Murray, 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143: 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R. H., D. M. Brady, D. Smith, A. W. Murray and K. G. Hardwick, 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell 10: 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, E., and R. H. Chen, 2002. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell 13: 1501–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix, O., J. M. Peters, M. W. Kirschner and D. Koshland, 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10: 3081–3093. [DOI] [PubMed] [Google Scholar]
- Crotti, L. B., and M. A. Basrai, 2004. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. EMBO J. 23: 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles, M., V. Liberal, M. L. Scott, R. Benezra and P. K. Sorger, 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101: 635–645. [DOI] [PubMed] [Google Scholar]
- Draviam, V. M., S. Xie and P. K. Sorger, 2004. Chromosome segregation and genomic stability. Curr. Opin. Genet. Dev. 14: 120–125. [DOI] [PubMed] [Google Scholar]
- Fabre, E., and E. Hurt, 1997. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet. 31: 277–313. [DOI] [PubMed] [Google Scholar]
- Gillett, E. S., C. W. Espelin and P. K. Sorger, 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter, P., C. Mann, M. Snyder and R. W. Davis, 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40: 381–392. [DOI] [PubMed] [Google Scholar]
- Hoyt, M. A., L. Totis and B. T. Roberts, 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517. [DOI] [PubMed] [Google Scholar]
- Ikui, A. E., K. Furuya, M. Yanagida and T. Matsumoto, 2002. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 115: 1603–1610. [DOI] [PubMed] [Google Scholar]
- Iouk, T., O. Kerscher, R. J. Scott, M. A. Basrai and R. W. Wozniak, 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159: 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga, Y., T. Kasai, K. Kibler and K. T. Jeang, 2002. Characterization of regions in hsMAD1 needed for binding hsMAD2. A polymorphic change in an hsMAD1 leucine zipper affects MAD1–MAD2 interaction and spindle checkpoint function. J. Biol. Chem. 277: 31005–31013. [DOI] [PubMed] [Google Scholar]
- Kalitsis, P., E. Earle, K. J. Fowler and K. H. Choo, 2000. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 14: 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., P. Hieter, M. Winey and M. A. Basrai, 2001. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics 157: 1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., L. B. Crotti and M. A. Basrai, 2003. Recognizing chromosomes in trouble: association of the spindle checkpoint protein Bub3p with altered kinetochores and a unique defective centromere. Mol. Cell. Biol. 23: 6406–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, K., R. Abdulle, P. K. Bansal, G. Cagney, S. Fields et al., 2003. Requirement of Skp1-Bub1 interaction for kinetochore-mediated activation of the spindle checkpoint. Mol. Cell 11: 1201–1213. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., and A. M. Rose, 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1: 514–521. [DOI] [PubMed] [Google Scholar]
- Lamb, J. R., W. A. Michaud, R. S. Sikorski and P. A. Hieter, 1994. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 13: 4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. S., and F. A. Spencer, 2004. Bipolar orientation of chromosomes in Saccharomyces cerevisiae is monitored by Mad1 and Mad2, but not by Mad3. Proc. Natl. Acad. Sci. USA 101: 10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., and A. W. Murray, 1991. Feedback control of mitosis in budding yeast. Cell 66: 519–531. [DOI] [PubMed] [Google Scholar]
- Liu, S. T., G. K. Chan, J. C. Hittle, G. Fujii, E. Lees et al., 2003. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol. Biol. Cell 14: 1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma, S., V. M. Stucke and E. A. Nigg, 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270. [DOI] [PubMed] [Google Scholar]
- Musacchio, A., and K. G. Hardwick, 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3: 731–741. [DOI] [PubMed] [Google Scholar]
- Pangilinan, F., and F. Spencer, 1996. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol. Biol. Cell 7: 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R. V., and P. Hieter, 1998. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet. 32: 307–337. [DOI] [PubMed] [Google Scholar]
- Spencer, F., S. L. Gerring, C. Connelly and P. Hieter, 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg, T. P., and G. Macara, 2003. The kintochore NUPtials. Nat. Cell Biol. 5: 945–947. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Warren, C. D., D. M. Brady, R. C. Johnston, J. S. Hanna, K. G. Hardwick et al., 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13: 3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E., and M. Winey, 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14: 706–714. [DOI] [PubMed] [Google Scholar]