Abstract
Most organisms use crossovers (chiasmata) to maintain physical connections between homologous chromosomes that ensure their proper segregation at the first meiotic division. The fission yeast Schizosaccharomyces pombe has a residual ability to segregate homologous chromosomes in the absence of meiotic recombination (achiasmate segregation). Using cytologically tagged chromosomes, we established a role for the microtubule motor dynein in meiotic chromosome segregation. Dhc1, the motor subunit of dynein, is required for chromosome segregation in both the presence and the absence of recombination. Dlc1, a member of the Tctex-1 dynein light-chain family, preferentially affects the segregation of achiasmate chromosomes. Dlc1 is the first identified protein, outside of Drosophila, that preferentially affects achiasmate chromosome segregation. We discuss possible roles of the dynein motor in this process.
SEXUALLY reproducing organisms must produce gametes that contain precisely half the somatic number of chromosomes. This is accomplished by a specialized form of cell division, meiosis, that consists of one round of DNA replication followed by two successive rounds of chromosome segregation. After meiotic DNA replication, homologous chromosomes (homologs), each consisting of two sister chromatids, find each other, align, and recombine during an extended prophase. At the first meiotic division (MI) homologs segregate to opposite poles, halving the number of chromosomes. For this reason MI is called a reductional division. At the second meiotic division (MII) sister chromatids segregate to opposite poles, producing four haploid nuclei that differentiate into gametes.
One of the hallmarks of meiosis is an elevated level of genetic recombination. The immediate role of recombination in meiosis is to provide the physical connection between homologs generally required to ensure proper homolog segregation at MI. Meiotic recombination is initiated by a developmentally regulated program that involves the formation of DNA double-strand breaks (DSB) by Rec12, the Schizosaccharomyces pombe ortholog of Spo11 (Keeney et al. 1997; Cervantes et al. 2000). The DNA DSBs are then repaired via an interaction with a homolog, frequently resulting in the formation of crossovers (reviewed in Roeder 1997; Keeney 2001). Although the initiating DNA DSB is crucial, to recombine homologs must also be in close proximity. Clustering of telomeres is observed in meiotic prophase of many organisms and is thought to facilitate the initial interaction between homologs (reviewed in Scherthan 2001; Yamamoto and Hiraoka 2001). In the fission yeast S. pombe, telomeres cluster tightly at the spindle pole body (SPB). Telomere clustering is followed, in S. pombe, by a telomere-led oscillatory nuclear movement (horsetail movement) that continues throughout prophase (Chikashige et al. 1994). Horsetail movement depends completely on the microtubule motor, dynein (Yamamoto et al. 1999). Mutations that reduce or eliminate either telomere clustering or horsetail movement also reduce pairing and meiotic recombination (Shimanuki et al. 1997; Cooper et al. 1998; Nimmo et al. 1998; Yamamoto et al. 1999; Ding et al. 2004).
While crossovers are generally required for proper MI segregation, some organisms exhibit a robust ability to segregate recombinationless, or achiasmate, chromosomes. Male fruit flies, Drosophila melanogaster, complete a faithful meiosis in the absence of meiotic recombination (reviewed in Hawley 1989; McKee 1998), but female Drosophila and the budding yeast Saccharomyces cerevisiae require recombination to properly complete meiosis (Hall 1972; Klein et al. 1999). In the latter cases, however, if only one or two pairs of achiasmate chromosomes per meiosis are present, they are segregated faithfully (reviewed in Dawson et al. 1986; Guacci and Kaback 1991; Hawley and Theurkauf 1993). In S. pombe, mutants that lack meiotic recombination display a significant residual ability to segregate homologs (achiasmate segregation; Molnar et al. 2001a,b; Sharif et al. 2002; Davis and Smith 2003).
Segregation of achiasmate chromosomes in Drosophila has been extensively characterized. Drosophila females possess two distinct achiasmate segregation mechanisms (reviewed in Hawley and Theurkauf 1993). One mechanism of achiasmate segregation requires pairing of homologous centromere-proximal heterochromatin (Dernburg et al. 1996; Karpen et al. 1996). In the second mechanism two achiasmate chromosomes, heterologous in this case, segregate from each other with high fidelity but pairing is not observed (Dernburg et al. 1996). Additionally, several proteins in Drosophila, including α-tubulin 67C (Matthies et al. 1999), the kinesin-like protein Nod (Zhang and Hawley 1990; Zhang et al. 1990), Axs (Whyte et al. 1993; Kramer and Hawley 2003), and Mtrm (Harris et al. 2003), are required preferentially for achiasmate segregation. Some of these proteins are conserved among diverse eukaryotes, allowing the possibility that their role in achiasmate chromosome segregation is also conserved.
In humans, decreased meiotic recombination is associated with MI missegregation events (reviewed in Hassold and Hunt 2001). The resulting aneuploid gametes are associated with ∼35% of lost pregnancies (reviewed in Hassold and Hunt 2001). Additionally, aneuploidy, including trisomy 21, which results in Down syndrome, is the leading known cause of mental retardation. Down syndrome is also associated with an increased incidence of leukemia (Iselius et al. 1990; Lorber et al. 1992; Minelli et al. 2001). While it is not known whether humans have a system to segregate achiasmate chromosomes (reviewed in Koehler and Hassold 1998), greater knowledge of the processes that promote proper meiotic segregation may help us to understand the origins of human meiotic aneuploidy.
We thought it likely that the mechanism of achiasmate segregation would require that homologs be brought into close proximity during meiotic prophase. In S. pombe rec12 mutants, in which meiotic recombination is essentially eliminated (DeVeaux et al. 1992; Davis and Smith 2003), pairing is reduced but not abolished (Nabeshima et al. 2001; Ding et al. 2004). The residual pairing in the rec12 mutant is most pronounced at the telomeres and centromeres (Ding et al. 2004) and, at least at the centromeres, is dependent on Dhc1, the dynein heavy chain (Ding et al. 2004). We therefore assayed segregation genetically in several mutants that might, like dhc1 mutants, have reduced recombination-independent pairing, in both rec+ and rec12Δ genetic backgrounds. Most mutations had little or no effect on achiasmate segregation (see supplementary materials at http://www.genetics.org/supplemental/). However, as reported here, dlc1Δ and dhc1Δ had a strong achiasmate segregation defect when assayed cytologically. Our results with these two mutants have demonstrated that the dynein light chain, Dlc1, was required preferentially for achiasmate segregation, while the dynein heavy chain was required for meiotic chromosome segregation in both the presence and the absence of recombination.
MATERIALS AND METHODS
Yeast strains, media, and culture conditions:
Strains were grown at 32° on yeast extract agar (YEA) plus histidine (50 μg/ml), leucine (100 μg/ml), lysine (100 μg/ml), and uracil (100 μg/ml) (YEA + 4S; Gutz et al. 1974); YEA + 4S plus adenine (100 μg/ml) (YEA + 5S); or supplemented Edinburgh minimal medium 2 (EMM2 as modified in Nurse 1975) solid media. Liquid cultures were grown at 30° in yeast extract liquid + 5S [YEL (Gutz et al. 1974) plus five supplements as in YEA + 5S]. Sporulation was at 25° on supplemented sporulation agar (SPA; Gutz et al. 1974) for 2–4 days. The yeast strains and mutant alleles used are described below or in references in Table 1.
TABLE 1.
S. pombe strains
| Strain | Genotype |
|---|---|
| GP4 | h+ ade6-M210 |
| GP2106 | h− ade6-M216 his2 leu1-32 |
| GP3714 | h− ade6-M210 lys1-37 |
| GP3716 | h+ ade6-M216 |
| GP3721 | h+ ade6-M210 lys1-37 rec12-169::kanMX6 |
| GP3723 | h− ade6-M216 rec12-169::kanMX6 |
| GP3845 | h− ade6-M216 ura4-D18 dhc1-D2::ura4+ |
| GP3894 | h+ ade6-M210 lys1-37 ura4-D18 dlc1::ura4+ |
| GP3898 | h− ade6-M216 ura4-D18 dlc1::ura4+ |
| GP4000 | h− ade6-M210 lys1-37 ura4-D18 rec12-171::ura4+ |
| GP4002 | h+ ade6-M216 ura4-D18 rec12-171::ura4+ |
| GP4080 | h+ ade6-M210 lys1-37 ura4-D18 dhc1-D126::kanMX6 |
| GP4081 | h+ ade6-M210 lys1-37 ura4-D18 dhc1-D126::kanMX6 rec12-171::ura4+ |
| GP4082 | h− ade6-M216 ura4-D18 dhc1-D126::kanMX6 |
| GP4083 | h− ade6-M216 ura4-D18 dhc1-D126::kanMX6 rec12-171::ura4+ |
| GP4249 | h+ ade6-M216 ura4-D18 dlc1::ura4+ rec12-171::ura4+ |
| GP4250 | h− ade6-M210 lys1-37 ura4-D18 dlc1::ura4+ rec12-171::ura4+ |
| GP4957 | h+ leu1-32 lacO@lys1+ GFP-lacI@his7+ |
| GP4958 | h− leu1-32 lacO@lys1+ GFP-lacI@his7+ |
| GP4959 | h+ leu1-32 ura4-D18 lacO@lys1+ GFP-lacI@his7+ rec12-171::ura4+ |
| GP4960 | h− leu1-32 ura4-D18 lacO@lys1+ GFP-lacI@his7+ rec12-171::ura4+ |
| GP4961 | h+ leu1-32 ura4-D18 lacO@lys1+ GFP-lacI@his7+ dlc1::ura4+ |
| GP4962 | h− leu1-32 ura4-D18 lacO@lys1+ GFP-lacI@his7+ dlc1::ura4+ |
| GP4963 | h+ leu1-32 ura4-D18 lacO@lys1+ GFP-lacI@his7+ dlc1::ura4+ rec12-171::ura4+ |
| GP4964 | h− leu1-32 ura4-D18 lacO@lys1+ GFP-lacI@his7+ dlc1::ura4+ rec12-171::ura4+ |
| GP5099 | h+ leu1-32 ura4-D18 lys1-37 GFP-lacI@his7+ rec12-171::ura4+ |
| GP5101 | h+ leu1-32 ura4-D18 lys1-37 GFP-lacI@his7+ dlc1::ura4+ |
| GP5103 | h+ leu1-32 ura4-D18 lys1-37 GFP-lacI@his7+ dlc1::ura4+ rec12-171::ura4+ |
| GP5117 | h− leu1-32 lacO@lys1+ GFP-lacI@his7+ dhc1-D126::kanMX6 |
| GP5118 | h+ leu1-32 lacO@lys1+ GFP-lacI@his7+ dhc1-D126::kanMX6 |
| GP5120 | h+ leu1-32 lys1-37 GFP-lacI@his7+ dhc1-D126::kanMX6 |
| GP5121 | h− leu1-32 ura4-D18 lacO@lys1+ GFP-lacI@his7+ dhc1-D126::kanMX6 rec12-171::ura4+ |
| GP5122 | h+ leu1-32 ura4-D18 lacO@lys1+ GFP-lacI@his7+ dhc1-D126::kanMX6 rec12-171::ura4+ |
| GP5124 | h+ leu1-32 ura4-D18 lys1-37 GFP-lacI@his7+ dhc1-D126::kanMX6 rec12-171::ura4+ |
The strains were derived from lab stocks by meiotic crosses. Complete genealogies are available upon request. Alleles are commonly used auxotrophic markers, the mating-type locus and dhc1-D2::ura4+ (Yamamoto et al. 1999), dhc1-D126::kanMX6 (see materials and methods), dlc1::ura4+ (Miki et al. 2002), lys1-37::lacO-lys1+ (lacO@lys1+; Nabeshima et al. 1998), rec12-169::3HA6His-kanMX6, rec12-171::ura4+, and his7-366::GFP13-lacI12-NLS-his7+ (GFP-lacI@his7+; Davis and Smith 2003).
A complete replacement of the dhc1 coding sequence with 3HA-6His-kanMX was performed as follows. Plasmid pFA6a-3HA-6His-kanMX6 (Davis and Smith 2003) was used as template in a polymerase chain reaction (PCR) to generate the dhc1-D126::kanMX6 allele using the method of Bahler et al. (1998). The forward and reverse primers in this reaction contained homology to the 5′ and 3′ ends of dhc1+ corresponding, respectively, to nucleotides 7022–7101 of cosmid c30C2 (GenBank accession no. AL355652) and 12826–12905 of cosmid c1093 (GenBank accession no. AL132839). The resulting PCR product was used to transform S. pombe strain GP3845 (h− ade6-M216 ura4-D18 dhc1-D2::ura4+), which contained a ura4+ insertion in the dhc1 gene. The resulting G418-resistant transformants were screened for the loss of the ura4+ marker. Deletion of dhc1 was confirmed by a PCR.
Detecting aneuploid meiotic products:
The frequency of aneuploid meiotic products was determined by random spore analysis. Spores were liberated from asci, and vegetative cells killed, by treatment with glusulase and ethanol (Ponticelli and Smith 1989). Spore suspensions were plated on YEA + 5S and replicated to supplemented SPA after 3–4 days to detect I2-staining spore colonies (mat1-P/mat1-M heterozygous diploids; Bresch et al. 1968). Spore suspensions were also plated on YEA + 4S + guanine (100 μg/ml) (YEAG), which inhibits uptake of adenine (Cummins and Mitchison 1967), to detect adenine-prototrophic spores and on YEA + 4S to determine the total number of viable spores. Both ade6-M210/ade6-M216 heterozygous diploids and heterozygous chromosome III (ChrIII) disomes grow on YEAG. Heterozygous diploids form large colonies after 3 days, while heterozygous ChrIII disomes form small colonies only after 4 or more days. Only the small colonies (ChrIII disomes) were counted on YEAG.
Microscopy:
For detection of lacO-tagged chromosomes in live cells we used a previously described variant of the green fluorescent protein–LacI–nuclear localization signal fusion (GFP13–LacI12–NLS; Straight et al. 1998), adapted for S. pombe (Davis and Smith 2003). Cells were mated on supplemented SPA and, after 18–24 hr, zygotes were examined. By this time, many zygotes had undergone both MI and MII. Only those zygotes with four nuclei, as detected by background GFP13–LacI12–NLS fluorescence, were counted. In crosses homozygous for the lacO array, zygotes with segregation patterns consistent with premature segregation of sister chromatids at MI were eliminated from the analysis. Stacks of images from 24 focal planes separated by 0.2 μm were captured using SoftWoRx software and a DeltaVision microscope system (Applied Precision), which included an Olympus IX70 microscope and an Olympus UPlan Apo 100× 1.35 NA objective.
RESULTS
Visualization of ChrI segregation indicates a role for both Dhc1 and Dlc1 in achiasmate segregation of homologs at MI:
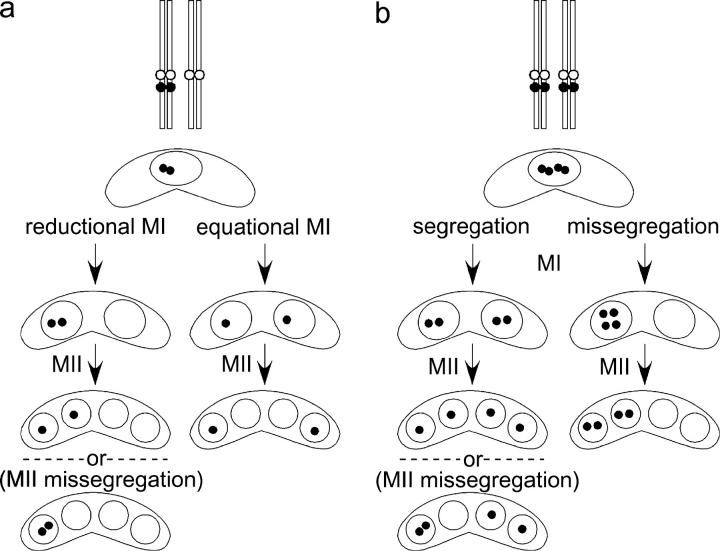
To determine the frequency of homolog segregation at MI, we used strains in which chromosome I (ChrI) was marked near the centromere with a tandem array of lacO DNA (Nabeshima et al. 1998). This array can be visualized by fluorescence microscopic observation of the GFP–LacI–NLS fusion protein that binds it (Straight et al. 1996). We performed crosses homozygous for the lacO array in both a rec+ and rec12Δ background and examined zygotes that had undergone MII. Faithful MI homolog segregation would result in one lacO-containing chromatid in each of the four nuclei. As the nuclei in S. pombe asci are ordered (Kitajima et al. 2003), missegregation of a homolog at MI would result in four lacO-containing chromatids in one pair of sister nuclei (the two nuclei at one end of a zygote) and none in the other pair (see Figure 1).
Figure 1.—
Characteristic patterns of GFP–LacI–NLS staining in strains heterozygous (a) and homozygous (b) for a lacO-tagged ChrI. Solid circles represent the lacO array. Open circles represent the centromere of ChrI. See results for description. Not all missegregation patterns are illustrated.
In a rec+ background, the dlc1Δ mutant showed high levels of proper homolog segregation (99%, Table 2) that were not significantly different from wild type (χ2 = 0.20; P > 0.5). In contrast, the frequency of proper homolog segregation in the dhc1Δ mutant (91%, Table 2) was reduced relative to that in wild type (χ2 = 12.9; P < 0.0005). The frequency of proper segregation in a rec12Δ background, 66%, was reduced significantly by both the dlc1Δ and dhc1Δ mutations (52 and 53%, Table 2; χ2 = 8.32, P < 0.005; χ2 = 6.07, P < 0.02, respectively). Segregation was not significantly different from random, 50%, in either double mutant (χ2 = 0.10, P > 0.7 and χ2 = 0.22, P > 0.5, respectively). These data indicate that, while Dlc1 played a role preferentially in achiasmate segregation, Dhc1 was required for chromosome segregation in both the presence and the absence of recombination.
TABLE 2.
Dlc1 and Dhc1 are required for achiasmate segregation
| Homologous chromosomes at MI
|
|||
|---|---|---|---|
| Parental genotype | na | Proper segregation (%)b |
Missegregation (%)b |
| rec+ | 225 | 99 | 1 |
| dhc1Δ | 157 | 91 | 9 |
| dlc1Δ | 136 | 99 | 1 |
| rec12Δ | 324 | 66 | 34 |
| dhc1Δ rec12Δ | 148 | 53 | 47 |
| dlc1Δ rec12Δ | 180 | 52 | 48 |
Crosses homozygous for the lacO array near centromere I were performed, and zygotes were examined by fluorescence microscopy after 18–24 hr on sporulation medium. Zygotes with four nuclei, as detected by background GFP13–LacI12–NLS fluorescence, were scored for the number and location of lacO signals. The segregation pattern of homologs at MI was inferred as described in Figure 1 and results.
Number of zygotes counted.
See Figure 1 for a description of the segregation patterns.
We occasionally observed segregation patterns consistent with premature segregation of sister chromatids (PSSC) in MI and also missegregation of sister chromatids at MII (data not shown). In these crosses, which were homozygous for the lacO array, it was not always possible to distinguish these two segregation patterns. To more precisely determine the frequency of other types of segregation errors, we examined the behavior of sister chromatids in crosses heterozygous for the lacO array. The results of the heterozygous crosses (below) were consistent with the observed frequency of PSSC and MII missegregation in homozygous crosses.
Visualization of ChrI segregation indicates that the absence of Rec12, Dlc1, or Dhc1 modestly affects the cosegregation of sister chromatids at MI:
MI of wild-type meiosis is reductional, that is, sister chromatids move together to the same pole and homologs move to opposite poles. To determine whether sister chromatids move together to the same pole at MI (cosegregate) in dlc1Δ, dhc1Δ, and rec12Δ mutant meioses, we performed crosses heterozygous for the lacO array. We examined zygotes that had undergone MII and determined whether or not lacO-containing sister chromatids were in sister nuclei. If the first division is reductional-like, both sister chromatids containing the array will be in one pair of sister nuclei and none in the other pair. In contrast, one lacO-containing sister chromatid in each pair of sister nuclei indicates an equational first division (see Figure 1). In wild-type crosses, lacO cosegregated in 87% of MI divisions (Table 3). The lacO array is integrated at the lys1 gene, ∼4 cM from the centromere in wild-type meiosis (Watanabe and Nurse 1999). The frequency of apparent sister chromatid segregation at MI in wild type can be explained by recombination between the centromere and the lacO array and is consistent with previously published results (Watanabe and Nurse 1999). In rec12Δ, dlc1Δ, dhc1Δ, rec12Δ dlc1Δ, and rec12Δ dhc1Δ mutants, sister chromatids cosegregated at MI at least 92% of the time (Table 3). Because meiotic recombination is reduced or abolished in these mutants (DeVeaux et al. 1992; Yamamoto et al. 1999; Miki et al. 2002; Davis and Smith 2003), recombination between the centromere and the lacO array is unlikely to significantly influence these results. These data indicate that Rec12, Dlc1, and Dhc1 play a small but significant role in ensuring that sister chromatids cosegregate at MI.
TABLE 3.
Sister chromatids predominantly cosegregate at MI indlc1Δ,dhc1Δ, andrec12Δ mutants
| Sister chromatids
|
||||
|---|---|---|---|---|
| Parental genotypea | nb | MI cosegregation (%)c | MI missegregation (%)c | MII missegregation (%)c |
| rec+ | 183 | 87 | 13 | 2 |
| dhc1Δ | 157 | 92 | 8 | 2 |
| dlc1Δ | 63 | 94 | 6 | 2 |
| rec12Δ | 236 | 94 | 6 | 8 |
| dhc1Δ rec12Δ | 121 | 93 | 7 | 9 |
| dlc1Δ rec12Δ | 44 | 98 | 2 | <7 |
Crosses heterozygous for the lacO array near centromere I were performed, and zygotes were examined by fluorescence microscopy after 18–24 hr on sporulation medium. Zygotes with four nuclei, as detected by background GFP13–LacI12–NLS fluorescence, were scored for the number and location of lacO signals. The segregation pattern of sister chromatids at MI was inferred as described in Figure 1 and results.
Crosses were homozygous for the indicated mutations, except the rec+ cross was heterozygous for the rec12Δ allele.
Number of zygotes counted.
See Figure 1 for a description of the segregation patterns.
We were also able to detect MII errors. If sister chromatids cosegregate at MI but missegregate at MII, both lacO-containing sister chromatids would be contained in a single nucleus (see Figure 1). While sister chromatid segregation at MII in dlc1Δ or dhc1Δ single mutants was not different from that in wild type, the frequency of MII missegregation in rec12Δ and rec12Δ dhc1Δ mutants was 8 and 9%, respectively (Table 3). These data indicate that, although none of the mutants tested were severely impaired, sister chromatid missegregation at MII was elevated in rec12Δ relative to wild type (χ2 = 5.85, P < 0.02). A similar conclusion, based on genetic assays of segregation, was reached by Sharif et al. (2002).
Genetic assays of segregation suggest a role for Dhc1 in the presence of recombination:
In the first meiotic division, homologous chromosomes segregate to opposite poles. In the absence of recombination between a heterozygous marker and the centromere, this reductional segregation eliminates heterozygosity in the products of meiosis. However, if homologous chromosomes missegregate at MI (i.e., if both homologs remain together), heterozygosity is retained. Because S. pombe has only three chromosomes, even mutants that segregate homologs at random are expected to produce a significant number of viable progeny. Nonetheless, if homologs missegregate at MI, an elevated frequency of heterozygous meiotic products is expected (Figure 2 and Davis and Smith 2003). These meiotic products include heterozygous diploids and heterozygous ChrIII disomes, the only aneuploids that have been propagated in S. pombe (Niwa and Yanagida 1985; Molnar and Sipiczki 1993). These features of S. pombe biology allowed us to characterize meiotic segregation by genetic analysis of the products of meiosis.
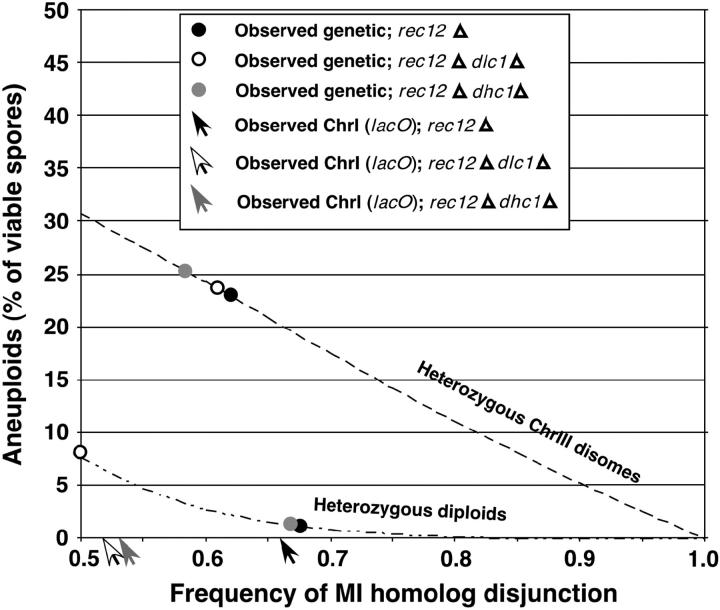
Figure 2.—
Expected frequency of heterozygous meiotic products at homolog segregation frequencies of 50–100%. The formulas used to construct the graph have been described (Davis and Smith 2003). The observed frequencies of heterozygous spore colonies characteristic of MI homolog missegregation (Table 4) are plotted on the appropriate expectation curve and are indicated by solid circles (rec12Δ), open circles (rec12Δ dlc1Δ), and shaded circles (rec12Δ dhc1Δ). The solid (rec12Δ), open (rec12Δ dlc1Δ), and shaded (rec12Δ dhc1Δ) arrows indicate the frequency of proper segregation by ChrI as observed cytologically (Table 2).
Mutations that specifically abolish achiasmate segregation by definition would not alter segregation nor, therefore, the frequency of heterozygous meiotic products, in the presence of meiotic recombination. To determine if this was true of dhc1Δ and dlc1Δ mutants, we analyzed their segregation phenotype in a rec12+ (chiasmate) background. Crosses heterozygous for the complementing ade6 alleles, M210 and M216, allowed missegregation of ChrIII to be assayed by formation of Ade+ colonies (Moreno et al. 1991). Because formation of a heterozygous ChrIII disome requires only one missegregation event (that of ChrIII), the frequency of heterozygous ChrIII disomes among viable spores increases linearly as the frequency of MI missegregation increases (Figure 2 and Davis and Smith 2003). The frequency of heterozygous ChrIII disomes is predicted to increase from 0.06 ± 0.01% of viable spores, as observed in wild-type meioses (Table 4), to 30.7% of viable spores if homolog segregation at MI is random (Figure 2 and Davis and Smith 2003), an increase of 500-fold. The observed ChrIII disome frequencies of dlc1Δ mutant crosses were 1.4 ± 0.1%, ∼25 times higher than that of wild type (Table 4). The dhc1Δ mutant was more severely affected, with a ChrIII disome frequency (9.1 ± 0.7%; Table 4) that, although >150 times higher than that of wild type, was lower than that of the rec12Δ mutant (23 ± 1.7%; Table 4). These data suggest that Dhc1 plays a significant role in MI segregation even in the presence of recombination.
TABLE 4.
Genetic assays of segregation defects inrec+ andrec12Δ background
| Genotype | Strains crossed | Heterozygous ChrIII disomes (% of viable spores)a |
Heterozygous diploids (% of viable spores)b |
|---|---|---|---|
| Wild type | c | 0.06 ± 0.01 (8) | 0.9 ± 0.3 (8) |
| dhc1Δ | GP4080 × GP4082 | 9.1 ± 0.7 (4) | <0.3 (4) |
| dlc1Δ | GP3894 × GP3898 | 1.4 ± 0.1 (3) | 1.8 ± 0.4 (3) |
| rec12Δ | d | 23.0 ± 1.7 (14) | 1.1 ± 0.2 (14) |
| dhc1Δ rec12Δ | GP4081 × GP4083 | 25.1 ± 1.9 (4) | 1.5 ± 0.3 (4) |
| dlc1Δ rec12Δ | GP4249 × GP4250 | 23.5 ± 2.0 (3) | 8.1 ± 0.7 (3) |
Spores were plated on YEAG to assay ChrIII heterozygosity. Small Ade+ colonies were counted as ChrIII disomes (see materials and methods). Values are mean ± SEM (from n experiments).
Spores were plated on YEA + 5S. Colonies were replicated to supplemented SPA to assay heterozygous diploids by staining with I2 to determine the frequency of Spo+ (mat1-P/mat1-M heterozygous) spores. Values are mean ± SEM (from n experiments). Where no heterozygous diploid spores were observed, the upper 95% confidence limit based on the Poisson distribution is given.
Data are from five GP3714 × GP3716 and three GP4 × GP2106 crosses.
Data are from six GP3721 × GP3723 and eight GP4000 × GP4002 crosses.
Formation of a heterozygous diploid requires three missegregation events (those of ChrI, II, and III). Therefore, the frequency of heterozygous diploids among viable spores does not increase markedly until the frequency of MI missegregation is >∼30% (Figure 2 and Davis and Smith 2003). The frequencies of heterozygous diploids (assayed as Spo+, resulting from mat1-P/mat1-M heterozygosity on ChrII) among viable spores in the dhc1Δ and dlc1Δ mutant crosses was low (Table 4), consistent with ChrIII disome results.
Genetic assays of segregation suggest a role for Dlc1 specific to achiasmate segregation:
Mutations that specifically abolish achiasmate segregation should result in an increased frequency of heterozygous meiotic products only when combined with a mutation, such as rec12Δ, that eliminates meiotic recombination. To determine if dhc1Δ and dlc1Δ had a more dramatic effect on MI segregation in the absence of recombination, we analyzed their segregation phenotype in a rec12Δ (achiasmate) background. As above, crosses were performed heterozygous for the complementing ade6 alleles, M210 and M216. In a rec12Δ background the formation of heterozygous ChrIII disomes among viable spores is not predicted to be very sensitive to increased missegregation because the missegregation frequency is already high (Figure 2 and Davis and Smith 2003). Accordingly, neither the rec12Δ dhc1Δ nor the rec12Δ dlc1Δ crosses had a frequency of heterozygous ChrIII disomes significantly higher than that in the rec12Δ single mutant (Table 4).
In contrast, the formation of heterozygous diploid spores in a rec12Δ background is predicted to be very sensitive to increased missegregation. The frequency of heterozygous diploids is predicted to increase from 1.1 ± 0.2% of viable spores, as observed in rec12Δ meioses (Table 4), to 7.7% of viable spores if homolog segregation at MI is random (Figure 2 and Davis and Smith 2003). In a rec12Δ mutant background dhc1Δ mutant crosses produced no more heterozygous diploid meiotic products than did the rec12Δ single-mutant cross (Table 4). The dlc1Δ rec12Δ double-mutant crosses resulted in 8.1 ± 0.7% heterozygous diploid meiotic products (Table 4), consistent with random segregation of homologs at MI (7.7% heterozygous diploids predicted; Figure 2). Together with the data from rec12+ meioses, these data suggest that Dlc1 is required preferentially for achiasmate segregation. Interestingly, while dlc1Δ rec12Δ double mutants yielded elevated frequencies of heterozygous diploid spores, the dhc1Δ rec12Δ crosses did not result in more heterozygous diploid spores than the rec12Δ crosses (1.5 ± 0.3% and 1.1 ± 0.2%, respectively; Table 4). The discrepancy between the genetic results, which indicate no significant role for Dhc1 in achiasmate segregation, and the cytological results is discussed below.
DISCUSSION
While it appears to be generally true that reciprocal recombination is required for proper MI segregation, in many organisms recombination-independent mechanisms can promote the proper segregation of achiasmate chromosomes. Here, we provide evidence that the microtubule motor dynein promotes meiotic chromosome segregation in both the presence and the absence of recombination. Additionally, we identify dlc1Δ as the first mutation, outside Drosophila, that preferentially affects the segregation of achiasmate chromosomes.
The dynein light chain, Dlc1, is required for achiasmate segregation:
Analysis of dlc1Δ meioses indicated that chromosome missegregation was only modestly elevated by the absence of Dlc1 in rec+ (chiasmate) meioses. The frequency of ChrIII disomes (Table 4) observed in dlc1Δ crosses was consistent with a homolog missegregation frequency of ∼3% (Figure 2). The frequency of missegregation in dlc1Δ measured by direct observation of lacO-tagged ChrI, 1% (Table 2), was not significantly different from that expectation (χ2 = 0.81; P > 0.3). Miki et al. (2002) measured crossovers in dlc1Δ mutants in five nonoverlapping intervals that constitute ∼4% of the genome; recombination frequencies were reduced <10-fold relative to wild type. If recombination were decreased by a factor of five uniformly throughout the genome, the mean number of crossovers per homolog would be 3.8, 3, and 2.2 for chromosomes I, II, and III, respectively (Munz 1994). Since S. pombe lacks crossover interference (Munz 1994), the frequency of achiasmate ChrI, -II, and -III would be expected to be the corresponding Poisson null terms of 0.02, 0.05, and 0.11, respectively. Missegregation of the infrequent achiasmate chromosomes thus appears to be sufficient to explain the segregation defect we observed in dlc1Δ (rec+) meioses. Our data indicate that when crossovers are present, Dlc1 plays at most a modest role in faithful chromosome segregation at MI. In contrast, dlc1Δ had a more severe effect in a rec12Δ background. Direct observation of lacO-tagged ChrI showed that segregation of homologs was reduced from 66% in rec12Δ to 52% in dlc1Δ rec12Δ, i.e., nearly random (Table 2). The frequency of proper segregation in dlc1Δ rec12Δ crosses was significantly different from that of rec12Δ (χ2 = 8.32; P < 0.005) but was not significantly different from random (χ2 = 0.10; P > 0.7). Our genetic results also are consistent with completely random segregation of homologs at MI; the level of heterozygous diploid meiotic products seen in dlc1Δ rec12Δ meioses (8.1%, Table 4) was not significantly different from that predicted for random segregation (7.7%; χ2 = 0.08, P > 0.7). Our data indicate that Dlc1 is required preferentially for achiasmate segregation.
There are two ways to think about the role of Dlc1 in achiasmate segregation. Dlc1 may play a novel role in achiasmate segregation that is distinct from its minor role in wild-type meiosis. For instance, in rec12Δ, but not rec12+, meioses, Dlc1 may be required for homologous association of centromeres (discussed below). Alternatively, Dlc1 may play the same role as in wild-type meiotic chromosome segregation but this role becomes crucial only in the absence of recombination. For instance, the aberrant horsetail movement observed in the absence of Dlc1 (Miki et al. 2002) may not perturb homologous interactions that are stabilized by crossovers (rec12+), but it may severely perturb homologous interactions that are not stabilized by crossovers (rec12Δ).
Cytological data indicate that the dynein heavy chain, Dhc1, is required for achiasmate segregation:
Analysis of dhc1Δ meioses indicates that horsetail movement or some other function of Dhc1 is required for faithful MI homolog segregation in rec+ meiosis. The observed ChrIII disome frequency (Table 4), our most sensitive genetic assay, is consistent with a homolog missegregation frequency of ∼15% (Figure 2). The frequency of missegregation in dhc1Δ measured by direct observation of lacO-tagged ChrI, 9% (Table 2), is not significantly different from that expectation (χ2 = 2.21; P > 0.10). The level of missegregation we observed in dhc1Δ meioses cannot be explained by the modest decrease in recombination observed by others in dhc1 mutants (Yamamoto et al. 1999; see above). In fact, meiotic recombination is reduced less in the dhc1Δ mutant than in the dlc1Δ mutant in all three genetic intervals tested in both mutants (Yamamoto et al. 1999; Miki et al. 2002). The level of missegregation that we observe in the dhc1Δ mutant suggests that, in addition to its essential role in horsetail movement (Yamamoto et al. 1999), Dhc1 may be directly involved in the segregation of homologs on the MI spindle. Alternatively, the recombination events in dhc1Δ meioses may not efficiently hold homologs together. Strikingly, while direct observation of lacO-tagged ChrI indicated that Dhc1 was required for achiasmate segregation, the genetic assays did not suggest a role for Dhc1 in achiasmate segregation (Table 2). In the genetic assay, heterozygous diploid formation in the dhc1Δ rec12Δ mutant was not significantly different from that in the rec12Δ mutant (Table 4). This is in contrast to direct observation using lacO-tagged ChrI in which proper segregation of homologs was reduced from 66% in rec12Δ to 53% in dhc1Δ rec12Δ, a significant difference (Table 2; χ2 = 6.07, P < 0.02). Segregation in dhc1Δ rec12Δ was not significantly different from random (χ2 = 0.22; P > 0.5). Because, unlike the genetic assays, observation of lacO-tagged ChrI is a direct measure of homolog segregation, we consider the cytological data to indicate an important role for Dhc1 in achiasmate segregation.
A likely cause of the disparity between our genetic and cytological results for the dhc1Δ rec12Δ mutant (as well as the failure to measure an increase, relative to rec12Δ, in ChrIII disome formation in dhc1Δ rec12Δ and dlc1Δ rec12Δ mutants) is the indirect nature of the genetic assays. While genetic analysis can easily generate large data sets, it is limited to viable meiotic products. Additionally, the genetic assays (heterozygous diploid and ChrIII disomic spore formation; Figure 2, Table 4) are interpretable only if several assumptions are made (see Davis and Smith 2003). Two of these assumptions are particularly relevant:
The only type of segregation error is MI homolog missegregation. In the case of the mutants tested in this article by direct observation using lacO-tagged ChrI, we know that this is not strictly true. Sister chromatids occasionally segregated from each other at MI in the mutants tested, and rec12Δ mutants occasionally missegregated sister chromatids at MII (Table 3).
Heterozygous diploids and ChrIII disomes are as likely to form a colony as a haploid cell. This is most likely to be a problem for mutants with a mitotic phenotype. Several mutants of S. pombe are viable as haploids but are not stable as diploids (Bernard et al. 1998; Catlett and Forsburg 2003). Both Dhc1 and Dlc1 play a role in attachment of chromosomes to the mitotic spindle (referenced in Yamamoto and Hiraoka 2003). Perhaps dhc1Δ diploids (as well as dhc1Δ and dlc1Δ ChrIII disomes) are less stable than wild type. While it is difficult to predict the degree to which a deviation from these assumptions might influence the results of the genetic assays, these caveats complicate interpretation of the genetic assays and may explain the difference between the dhc1Δ rec12Δ genetic and cytological results.
Because of the ease of assaying many meioses with the genetic assays, we tested other likely candidates for missegregation in the presence and absence of Rec12. These candidates include Taz1 (Cooper et al. 1998; Nimmo et al. 1998; Ding et al. 2004), Kms1 (Shimanuki et al. 1997; Niwa et al. 2000), Meu13 (Nabeshima et al. 2001), Hopl (Lorenz et al. 2004), and Mad2 (He et al. 1997). The results are provided in the supplementary materials (Tables S1–S3; http://www.genetics.org/supplemental/) and suggest that some of these proteins may play a role in achiasmate segregation.
The role of dynein in achiasmate segregation:
Taken together, our results indicate that Dhc1 and Dlc1 promote the proper segregation of achiasmate chromosomes. Because both Dhc1 and Dlc1 are required for proper horsetail movement (Yamamoto et al. 1999; Miki et al. 2002), the recent examination of the role of recombination and horsetail movement in homologous association during prophase by Hiraoka and colleagues (Ding et al. 2004) is directly relevant. In brief, telomere clustering and horsetail movement are required to bring chromosomes into close alignment; recombination then stabilizes homologous associations. Significant levels of homologous association are retained in rec12 mutants, especially at the centromeres and telomeres. Importantly, homologous association of centromeres is eliminated in the dhc1 rec12 mutant (Ding et al. 2004), and proper segregation in the dhc1Δ rec12Δ mutant was reduced to random (Table 2). It is interesting to note that pairing of centric heterochromatin is required in one of the two distinct achiasmate segregation mechanisms possessed by Drosophila females (Dernburg et al. 1996; Karpen et al. 1996). Additionally, centromere pairing of achiasmate chromosomes has recently been observed in S. cerevisiae (Kemp et al. 2004). These data lead us to suggest that pairing of centric heterochromatin is important for achiasmate segregation in S. pombe. A conserved mechanism of centromere pairing may be widely important in the segregation of achiasmate chromosomes.
Other, though not necessarily mutually exclusive, mechanisms may promote achiasmate segregation in S. pombe. One such mechanism is suggested by the second achiasmate system in Drosophila females. As mentioned above, one mechanism of achiasmate segregation in Drosophila females requires pairing of homologous centromere-proximal heterochromatin (Dernburg et al. 1996; Karpen et al. 1996). However, in the second mechanism two achiasmate chromosomes, heterologous in this case, segregate from each other with high fidelity. Interestingly, pairing is not observed (Dernburg et al. 1996), and the heterologous chromosomes are thought to orient toward the less crowded pole, thus assuring that one will go to each pole. It is interesting to speculate that this type of mechanism might be especially effective in an organism, like S. pombe, that makes only a small number of microtubule connections per kinetechore (Ding et al. 1993). A mechanism, such as this, that relies on spatial or numerical constraints might be more effective if fewer chromosomes were achiasmate. This possibility has not been sufficiently tested in S. pombe.
This type of mechanism might be especially sensitive to perturbation of meiotic spindle function. Given the localization of Dlc1 to the spindle pole body at MI (Miki et al. 2002), and the function of dynein in spindle assembly and mitotic chromosome movement in metazoans (reviewed in Banks and Heald 2001), mutations in Dlc1 or Dhc1 may result in perturbation of meiotic spindle function. In fact, regardless of the underlying mechanism promoting achiasmate segregation, altered spindle function in the absence of Dlc1 or Dhc1 could lead to the observed failure of achiasmate segregation.
Acknowledgments
We are grateful to Julia Cooper, Yasushi Hiraoka, Osami Niwa, Hiroshi Nojima, Shelly Sazer, Takahiro Tougan, and Mitsuhiro Yanagida for providing strains; to Takahiro Tougan, Hiroshi Nojima, Ayumu Yamamoto, and Yasushi Hiraoka for sharing unpublished data; and to Sue Amundsen, Sue Biggins, Gareth Cromie, Joseph Farah, and Andrew Taylor for helpful comments on the manuscript. This work was supported by National Institutes of Health research grant GM32194 to G.R.S. and postdoctoral fellowship F32-GM20125 to L.D.
References
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, III et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Banks, J. D., and R. Heald, 2001. Chromosome movement: dynein-out at the kinetochore. Curr. Biol. 11: R128–R131. [DOI] [PubMed] [Google Scholar]
- Bernard, P., K. Hardwick and J. P. Javerzat, 1998. Fission yeast Bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresch, C., G. Muller and R. Egel, 1968. Genes involved in meiosis and sporulation of a yeast. Mol. Gen. Genet. 102: 301–306. [DOI] [PubMed] [Google Scholar]
- Catlett, M. G., and S. L. Forsburg, 2003. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol. Biol. Cell 14: 4707–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes, M. D., J. A. Farah and G. R. Smith, 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5: 883–888. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., D. Q. Ding, H. Funabiki, T. Haraguchi, S. Mashiko et al., 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science 264: 270–273. [DOI] [PubMed] [Google Scholar]
- Cooper, J. P., Y. Watanabe and P. Nurse, 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392: 828–831. [DOI] [PubMed] [Google Scholar]
- Cummins, J. E., and J. M. Mitchison, 1967. Adenine uptake and pool formation in the fission yeast Schizosaccharomyces pombe. Biochim. Biophys. Acta 136: 108–120. [DOI] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2003. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163: 857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, D. S., A. W. Murray and J. W. Szostak, 1986. An alternative pathway for meiotic chromosome segregation in yeast. Science 234: 713–717. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., J. W. Sedat and R. S. Hawley, 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135–146. [DOI] [PubMed] [Google Scholar]
- DeVeaux, L. C., N. A. Hoagland and G. R. Smith, 1992. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics 130: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D. Q., A. Yamamoto, T. Haraguchi and Y. Hiraoka, 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 6: 329–341. [DOI] [PubMed] [Google Scholar]
- Ding, R., K. L. McDonald and J. R. McIntosh, 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120: 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci, V., and D. B. Kaback, 1991. Distributive disjunction of authentic chromosomes in Saccharomyces cerevisiae. Genetics 127: 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Loprieno, 1974 Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics, edited by R. C. King. Plenum Press, New York.
- Hall, J. C., 1972. Chromosome segregation influenced by two alleles of the meiotic mutant c(3)G in Drosophila melanogaster. Genetics 71: 367–400. [DOI] [PubMed] [Google Scholar]
- Harris, D., C. Orme, J. Kramer, L. Namba, M. Champion et al., 2003. A deficiency screen of the major autosomes identifies a gene (matrimony) that is haplo-insufficient for achiasmate segregation in Drosophila oocytes. Genetics 165: 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold, T., and P. Hunt, 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2: 280–291. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., 1989 Exchange and chromosomal segregation in eucaryotes, pp. 497–525 in Genetic Recombination, edited by R. Kucherlapati and G. R. Smith. American Society for Microbiology, Washington, DC.
- Hawley, R. S., and W. E. Theurkauf, 1993. Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet. 9: 310–317. [DOI] [PubMed] [Google Scholar]
- He, X., T. E. Patterson and S. Sazer, 1997. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94: 7965–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iselius, L., P. Jacobs and N. Morton, 1990. Leukaemia and transient leukaemia in Down syndrome. Hum. Genet. 85: 477–485. [DOI] [PubMed] [Google Scholar]
- Karpen, G. H., M. H. Le and H. Le, 1996. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science 273: 118–122. [DOI] [PubMed] [Google Scholar]
- Keeney, S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kemp, B., R. M. Boumil, M. N. Stewart and D. S. Dawson, 2004. A role for centromere pairing in meiotic chromosome segregation. Genes Dev. 18: 1946–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima, T. S., S. Yokobayashi, M. Yamamoto and Y. Watanabe, 2003. Distinct cohesin complexes organize meiotic chromosome domains. Science 300: 1152–1155. [DOI] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S. B. Buonomo, C. Michaelis et al., 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Koehler, K. E., and T. J. Hassold, 1998. Human aneuploidy: lessons from achiasmate segregation in Drosophila melanogaster. Ann. Hum. Genet. 62: 467–479. [DOI] [PubMed] [Google Scholar]
- Kramer, J., and R. S. Hawley, 2003. The spindle-associated transmembrane protein Axs identifies a membranous structure ensheathing the meiotic spindle. Nat. Cell Biol. 5: 261–263. [DOI] [PubMed] [Google Scholar]
- Lorber, B. J., S. B. Freeman, T. Hassold, A. H. Ragab, R. A. Vega et al., 1992. Characterization and molecular analysis of nondisjunction in 18 cases of trisomy 21 and leukemia. Genes Chromosomes Cancer 4: 222–227. [DOI] [PubMed] [Google Scholar]
- Lorenz, A., J. L. Wells, D. W. Pryce, M. Novatchkova, F. Eisenhaber et al., 2004. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 117: 3343–3351. [DOI] [PubMed] [Google Scholar]
- Matthies, H. J., L. G. Messina, R. Namba, K. J. Greer, M. Y. Walker et al., 1999. Mutations in the α-tubulin 67C gene specifically impair achiasmate segregation in Drosophila melanogaster. J. Cell Biol. 147: 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee, B. D., 1998. Pairing sites and the role of chromosome pairing in meiosis and spermatogenesis in male Drosophila. Curr. Top. Dev. Biol. 37: 77–115. [DOI] [PubMed] [Google Scholar]
- Miki, F., K. Okazaki, M. Shimanuki, A. Yamamoto, Y. Hiraoka et al., 2002. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell 13: 930–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli, A., C. Morerio, E. Maserati, C. Olivieri, C. Panarello et al., 2001. Meiotic origin of trisomy in neoplasms: evidence in a case of erythroleukaemia. Leukemia 15: 971–975. [DOI] [PubMed] [Google Scholar]
- Molnar, M., and M. Sipiczki, 1993. Polyploidy in the haplontic yeast Schizosaccharomyces pombe: construction and analysis of strains. Curr. Genet. 24: 45–52. [DOI] [PubMed] [Google Scholar]
- Molnar, M., J. Bahler, J. Kohli and Y. Hiraoka, 2001. a Live observation of fission yeast meiosis in recombination-deficient mutants: a study on achiasmate chromosome segregation. J. Cell Sci. 114: 2843–2853. [DOI] [PubMed] [Google Scholar]
- Molnar, M., S. Parisi, Y. Kakihara, H. Nojima, A. Yamamoto et al., 2001. b Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Munz, P., 1994. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137: 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima, K., T. Nakagawa, A. F. Straight, A. Murray, Y. Chikashige et al., 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9: 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima, K., Y. Kakihara, Y. Hiraoka and H. Nojima, 2001. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20: 3871–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo, E. R., A. L. Pidoux, P. E. Perry and R. C. Allshire, 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828. [DOI] [PubMed] [Google Scholar]
- Niwa, O., and M. Yanagida, 1985. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr. Genet. 9: 463–470. [Google Scholar]
- Niwa, O., M. Shimanuki and F. Miki, 2000. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19: 3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse, P., 1975. Genetic control of cell size at cell division in yeast. Nature 256: 547–551. [DOI] [PubMed] [Google Scholar]
- Ponticelli, A. S., and G. R. Smith, 1989. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics 123: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621. [DOI] [PubMed] [Google Scholar]
- Scherthan, H., 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell. Biol. 2: 621–627. [DOI] [PubMed] [Google Scholar]
- Sharif, W., G. Glick, M. Davidson and W. Wahls, 2002. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki, M., F. Miki, D. Q. Ding, Y. Chikashige, Y. Hiraoka et al., 1997. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet. 254: 238–249. [DOI] [PubMed] [Google Scholar]
- Straight, A. F., A. S. Belmont, C. C. Robinett and A. W. Murray, 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Straight, A. F., J. W. Sedat and A. W. Murray, 1998. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., and P. Nurse, 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400: 461–464. [DOI] [PubMed] [Google Scholar]
- Whyte, W. L., H. Irick, T. Arbel, G. Yasuda, R. L. French et al., 1993. The genetic analysis of achiasmate segregation in Drosophila melanogaster. III. The wild-type product of the Axs gene is required for the meiotic segregation of achiasmate homologs. Genetics 134: 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., and Y. Hiraoka, 2001. How do meiotic chromosomes meet their homologous partners?: lessons from fission yeast. BioEssays 23: 526–533. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., and Y. Hiraoka, 2003. Cytoplasmic dynein in fungi: insights from nuclear migration. J. Cell Sci. 116: 4501–4512. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., R. R. West, J. R. McIntosh and Y. Hiraoka, 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145: 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., and R. S. Hawley, 1990. The genetic analysis of distributive segregation in Drosophila melanogaster. II. Further genetic analysis of the nod locus. Genetics 125: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., B. A. Knowles, L. S. Goldstein and R. S. Hawley, 1990. A kinesin-like protein required for distributive chromosome segregation in Drosophila. Cell 62: 1053–1062. [DOI] [PubMed] [Google Scholar]