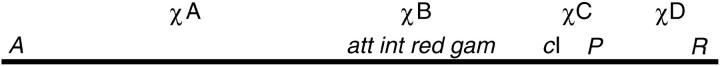WHEN we stumbled over Chi in coliphage λ (in 1972?), it appeared to be a uniquely accessible example of a “recombination initiator,” whose existence was implied by gene-conversion gradients (polarons) of fungi. Hence, it promised to have wide significance for our understanding of meiotic as well as of prokaryotic recombination. For a time, Chi seemed to fulfill its promise, but things turned out otherwise. Nevertheless, Chi did elucidate basic aspects of genetic recombination and genome maintenance, played a role in the development of λ as a cloning vehicle, and continues to bring enzymological surprises.
In this Perspectives, which reflects my rather personal memory of events, literature citations of work from our lab are omitted to improve readability. They can be found in older reviews (e.g., Myers and Stahl 1994; Smith 1998) or electronically. For those in a hurry, here is the bottom line: Escherichia coli's RecBCD enzyme enters duplex DNA at a double-strand break and travels in a destructive mode until it encounters a properly oriented octamer called Chi. This encounter civilizes the enzyme, which keeps on traveling, in a recombinagenic mode, recruiting E. coli's strand-invasion protein, RecA, to effect recombination when a homolog is available. The primary adaptive significance of Chi is likely to concern E. coli DNA replication, when breaks occur at the fork. Since these breaks are repaired by a RecBCD-promoted recombination-like reaction (usually between the two tines of the fork), Chi plays a role in the maintenance of the E. coli genome.
Discovery:
In 1969–1970, Mary Morgan Stahl (Figure 1) and I enjoyed a sabbatical leave with Noreen and Ken Murray in Edinburgh, where we pursued our studies on relationships between DNA replication and genetic recombination in the coliphage λ. During this period, Noreen's undergraduate honors student, David Henderson, encountered a curious phenomenon while trying to “Benzerize” the region of the λ chromosome that contained the known recombination genes (Figure 2). [Seymour Benzer (1961) had created a set of overlapping deletions within the rII region of phage T4 as a device for rapidly mapping any newly arising rII mutation.] Among λ's recombination genes are gam, whose product inactivates E. coli's recombination-related nuclease RecBCD, and red, which supplies recombination enzymes more compatible with λ's life style. Among the deletions that David sought were those that were missing both red and part of gam. Phage missing red and gam had been identified previously as λpbio-transducing phage. These λpbio phage occasionally arise in lysogenic cells when λ prophage, excising itself carelessly from the E. coli chromosome, picks up the E. coli bio gene in place of its own recombination region (see Anderson 1987). Such phage can be selected for by their novel ability to make (pretty-good-sized) plaques on a P2 lysogen of E. coli (the “Spi−” phenotype), a property shown to depend on the loss of both the red and gam gene functions. To select Spi− phage that were pure deletions, instead of bio substitutions, David exploited the observation that λ particles with chromosomes shortened by deletions are relatively resistant to the destabilizing effects of the Mg++-chelating agent, EDTA. Using a phage stock that had been grown in the lytic cycle, rather than having been induced from the prophage state, also helped to avoid bio substitutions. David did obtain a set of red gam deletions but found that they all made tiny (“pin-prick”) plaques. Initial attempts to grow them to a useful titer failed. Undaunted, David kept trying to grow them and succeeded when his Darwinian exercise resulted in derivatives with pretty good plaque size. He showed that each of these variants was the result of one or another single mutation, usually distant from the red gam region, that suppressed specifically the poor growth of the red gam deletion mutants. (These “Henderson suppressors” were the as-yet-unnamed Chi mutations.) David's large-plaque variants still retained the originally selected deletion and were red gam mutants, as indicated by their Spi− phenotype. In his subsequent graduate work with Jon Weil at Vanderbilt, David studied the growth-promoting phenotype of the suppressors and noted their stimulating effect on the ability of the red gam deletion phage to synthesize DNA (Henderson and Weil 1974a,b).
Figure 1.—

Mary Morgan Stahl (1934–1996). Mary was the driving force in the Stahl lab throughout the Chi era.
Figure 2.—
Map of λ showing only the features referred to in this Perspectives. att is the site on the λ chromosome that recombines with a related site on the E. coli chromosome when λ is reduced to the prophage state. The product of the int gene catalyzes that reaction. The recombination genes red and gam are described in the text. Sites of Chi arising spontaneously in λ are marked as χ.
How we got involved:
While Mary and I were in Edinburgh, Ken McMilin, working in Germany with Enzo Russo, demonstrated that the imposition of a total block to λDNA replication did not eliminate the ability of λ to recombine and to make infectious particles. Later, Ken realized that such a strong replication block could be used to survey the distribution of exchanges along the length of the λ chromosome, using density-labeled phage. The density of each recombinant phage particle would be determined by its inheritance of DNA from one conserved parent labeled with heavy isotopes and one carrying ordinary isotopes (Figure 3). He limited his experiment to the host-cell recombination system as it operates on λ chromosomes. To eliminate λ's own recombination genes in one mutational step, Ken used a bio substitution that extended from att through gam (Figure 2), knocking out all the known site-specific and generalized recombination genes. The clustering of genes by function, which characterizes the λ genome, made it plausible that any unidentified recombination genes would be deleted, as well.
Figure 3.—
Replication-blocked λ lytic cycle crosses conducted in the absence of DNA replication produce ample phage particles for genetic analysis. When one of the two parents is heavy labeled and the other carries ordinary isotopes, the density of each of the resulting progeny particles reveals the fraction of its DNA that has been inherited from each of the two infecting parents. Selection against terminally located ts markers (Ats and Rts) allows only crossover particles to plate. If most of the crossover particles have enjoyed but one exchange, the location of that exchange is revealed by the position of the particle in a cesium formate equilibrium density gradient. The cosegregation of the cI marker with the density label implies, for most purposes, the validity of the assumption of single exchanges. Lysates centrifuged to equilibrium in a density gradient are collected as drops emerging successively through a needle hole into 1 ml of broth. Each such sample is assayed by plating at permissive temperature for total phage and at high temperature for A+R+ recombinants. Among the recombinants, the cI marker is scored from the appearance of the plaque. Phage-carrying chromosomes that have recombined by splicing the DNA duplex across the cI gene make sectored colonies interpreted as heteroduplexes (see Stahl 1994).
When Mary and I returned to Eugene, Oregon, we embarked on a parallel project, to determine the distribution of exchanges along λ's chromosome in the presence of each of the combinations of λ's known recombination genes, int, red, and gam (Figure 2). To avoid the small-plaque problem of red gam double mutants, anticipated on the basis of David's work, we used a conditional (amber or sus) mutation of gam. Since the project required that we construct numerous different pairs of parental phage, we reckoned that we should first combine our markers that monitor exchange (Ats, cI, and Rts mutations) and the mutation that blocks replication (Psus) (Figure 3) with one of David's deletions, which extended from att through int, red, and gam. Separately we would construct the seven sets of recombination mutants (int, red, gam, int red, int gam, red gam, and int red gam), screening for which is best done in the absence of the markers to be used for monitoring exchanges. Then we would UV irradiate these recombination-deficient phage “heavily” to reduce their contribution of genes to the progeny of a cross, cross the UV'd phage with the Ats cI Psus80 and Psus80 Rts parents that contained the att-gam deletion, and plate the progeny phage under conditions (low temperature on a Su+ recA mutant host) that select for the loss of the deletion. This would ensure the incorporation of the desired combinations of recombination genes while leaving the Ats, Rts, Psus80, and cI markers undisturbed. The strategy worked. Mary constructed the genotypes and grew the stocks needed for the experiments. Only then did I realize that the procedure used practically guaranteed that every one of the 14 genotypes so constructed would carry a “Henderson suppressor.” Since we had no understanding of whether or not the suppressor might alter genetic recombination, we had no choice but to rebuild all 14 phage using a different strategy (from which I will spare you).
Mary quit the lab in protest of my thoughtlessness.
At about this time, Ken noted that crossing over between his λpbio phage was concentrated in and near the bio substitution! This provoked the hypothesis that bio DNA carried a “Henderson suppressor” and that the suppressor was a hotspot for crossing over.
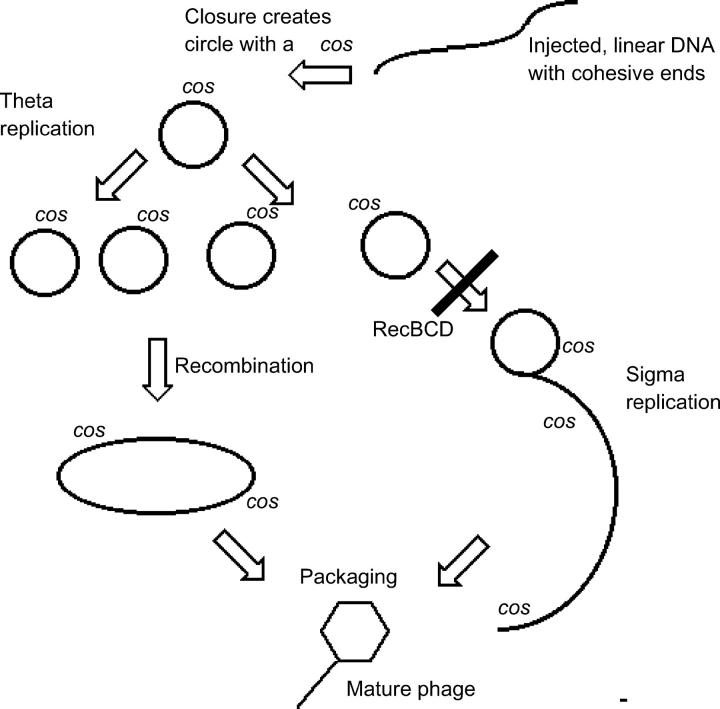
However, an insight was needed to connect plaque size with recombination. The connection was made by the observations of Enquist and Skalka (1973), who noted that recombination-deficient (red gam) phage λ infecting wild-type E. coli appeared to produce predominantly circular DNA monomers rather than the more complex intracellular forms made by recombination-proficient λ. Since λ monomers (unlike dimers and higher multimers) are poor substrates for being packaged into phage particles (Szpirer and Brachet 1970), inefficient packaging could account for the small plaque size of the red gam mutant phage. The inability of these recombination-deficient mutants to produce suitable packaging precursors could, in turn, reflect the monomers' inability to recombine with each other to generate dimers as shown in Figure 4. As described below, we gained support for this possibility. [However, David's earlier observation that the suppressor stimulated DNA replication implied that multimer formation by recombination was not the whole explanation for the phenotype of the suppressor, and subsequent work confirmed his views: the Henderson suppressor does appear to enhance rolling circle replication (Figure 4, and see Dabert et al. 1992).]
Figure 4.—
Replication of λ in its lytic cycle. The linear DNA, injected by a phage particle into E. coli, circularizes, generating the sequence called cos. In wild-type infections, replication in the theta mode, which generates monomer circles, switches to sigma (rolling circle) replication, probably by breakage of replication forks (Enquist and Skalka 1973). Packaging of the DNA into phage heads proceeds efficiently only from dimers or multimers, which contain two (or more) cos sites. Such multimers can arise by recombination or by sigma replication. This simplified figure fails to show the interrelationship between those two processes and unrealistically diagrams the recombination as reciprocal.
Mary's strains prove useful:
On the basis of the considerations above, we guessed that Henderson's suppressor was a cis-acting recombination initiator whose activity was manifest in the absence of λ recombination functions—and we realized that Mary's phage stocks containing the suppressor provided exactly the right material for testing that idea. We suggested to Steve Lam, undergraduate honors student, that he test the idea using the red gam mutant phage set. As expected, Steve found that the stocks did contain a Henderson suppressor (judged by plaque size). The suppressor mapped near the right end of λ's conventional linkage map, and, bingo, the phage carrying it did recombine like gangbusters in that region (Figure 5). Mary forgave me—my career was saved.
Figure 5.—
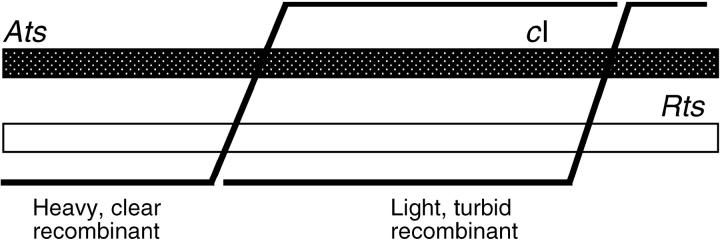
Density-labeled, replication-blocked crosses of red gam mutant λ in the absence of Chi (left) and in the presence of Chi (χD; right). Density-labeled crosses blocked for DNA synthesis (Figure 3) reveal a low, roughly uniform rate of exchanges along the length of the chromosome in the absence of Chi. In the presence of χD, near λ's right end, the rate of exchange is elevated near the Chi, mostly in the interval to the right of the cI marker (open circles), but evidently extending to the left of that marker for about half the length of λ. (The magnitude of the increase implies that Chi civilizes RecBCD thousands of times better than does the average octamer in λ.) In the absence of Chi, sectored plaques (half-solid) were not reliably scored because of the reduced plaque size resulting from the nonconditional red mutation. Due to the poor phage yield of the cross lacking Chi, unadsorbed parental Rts phage make a conspicuous contribution in the light peak of total phage (triangles). The left panel is from Stahl et al. (1974); the right panel is from Lam et al. (1974).
It was about this time, at Joe Bertani's 1973 Summer Solstice Temperate Phages meeting in Sweden, that I first reported our results. Jon Weil was in attendance and was unconvinced that the phenotype of the suppressors was related to recombination hotspot activity. However, he took our tale back to Nashville, and David soon confirmed our results. Over the next year or so, our two labs made discoveries regarding the suppressors that were rarely at variance. They included results showing a coincidence between recombination hotspots and the locations of new suppressor mutations. In our lab, Jean Crasemann spearheaded this work. With these data it was inescapable that the suppressors were, in fact, cis-acting stimulators of recombination, and we named them first chi and then Chi (crossover hotspot instigators), with the former designation being used by Henderson and Weil (1974a)(b) and often by others.
Natural history:
As suggested by the crosses with λpbio phage, described above, Chi is abundantly present in E. coli DNA. This surmise was supported by Bob Malone's finding that λ-transducing phage induced from some other sites on the bacterial chromosome had a Chi+ phenotype like that of λpbio. Daryl Faulds examined red gam mutant derivatives of λ phage into which ∼5-kb EcoRI fragments of bacterial DNA had been cloned (by Ron Davis at Stanford). An increased plaque size and a high recombination rate near the substitution, diagnostic of Chi, characterized about half of these derivatives, allowing the conclusion that there was about one Chi/5 kb of E. coli DNA. This estimate was later confirmed by the genome sequencing project. The same kinds of crosses, involving λ that carried fragments of yeast, revealed that Chi was present in yeast DNA at a similar frequency and could function in red gam mutant λ crosses. However, unpublished experiments by both Lisa Young and me failed to reveal any biological activity for Chi in yeast vegetative or meiotic cells. Much more importantly, Nancy Dower established that Chi could act in a cross between λ prophage carried out by P1 phage-mediated transduction. Since most prophage genes are repressed, this result made it unlikely that Chi activity was peculiar to λ. Then Kathy Triman (1982) obtained results confirming that presumption. She demonstrated Chi activity in P1 transductions involving the E. coli lac gene in the total absence of λ. (In both studies, Chi demonstrably influenced recombination when it was in either the recipient or the donated DNA, raising as-yet-unresolved questions about the uniparental provision of Chi activity.)
The availability of various recombination-defective (Rec−) mutant strains (generously provided by John Clark) allowed the demonstration that Chi was active only in the principal wild-type recombination pathway of E. coli, at that time referred to as the RecBC pathway. The distinctive component of this pathway is RecBCD, a nuclease active on linear double-stranded DNA (and see Gillen and Clark 1974). Since the Gam protein inactivates RecBCD, and the Red proteins provide λ with an alternative pathway, this demonstration implies that Chi in λ could have been discovered easily only in a red gam double mutant.
Properties:
What followed was a geneticists' dream—a joyous cycle of hypotheses, predictions, and experiments. Genetic crosses, with simple variations, revealed the following:
Chi, in an otherwise ordinary red gam mutant λ, stimulated recombination only to its left (on the conventional λ linkage map).
The stimulation extended perceptibly ∼20 kb (i.e., half the length of λ) (Figure 5), making “recombination hotspot” a somewhat misleading description of Chi.
Chi functioned when present in both or when present in only one of the two parents in the phage cross (“cis dominant”).
Chi stimulated recombination when it was in a large heterologous substitution, carried by only one of the two parents in the cross. Chi-stimulated recombination was confined to the region of homology to the left of the substitution and was conspicuous even when that homology was several kilobases distant from the Chi site.
Daryl Faulds, acting against my advice, showed that, when such a Chi-containing substitution was inverted, the Chi was reversibly subdued. (This was the first bit of genetic engineering conducted in our laboratory.) Ezra Yagil and Dhruba Chattoraj helped show that the same orientation dependence of Chi activity applied regardless of where in the λ chromosome the Chi was situated. That demonstration ruled out transcription across Chi as the polarized activator of Chi and pointed toward some event associated with either DNA injection or DNA packaging. Due to the asymmetry of the cohesive end site (cos), the site whose cleavage creates the ends of the λ virion chromosome (Figure 4), each of these processes is unidirectional with respect to the λ map. Ichizo Kobayashi, who had obtained his Ph.D. by studying recombination and packaging of λDNA in Japan, then joined us and showed that the subdued Chi was reactivated by inversion of cos. Chi then acted to its right. On this basis, Chi was predicted to be a nonpalindromic sequence, and, in a pioneering application of DNA sequencing, was shown to be the octomer 5′-GCTGGTGG-3′ (for review, see Smith et al. 1981).
Mary overcame multiple obstacles to show that a subdued Chi can also be activated by a double-strand break introduced in vivo by a restriction enzyme acting far from the Chi. This demonstration provided strong support for the possibility that cos can activate Chi because cos is a double-strand break site, albeit an asymmetric one.
Genetics meets enzymology:
The in vitro properties of the RecBCD enzyme (at that time known as RecBC), combined with a growing understanding of the chromosome-packaging apparatus of λ, provided ways of thinking about the Chi-cos interaction. In vitro, the enzyme could be seen under the electron microscope to invade linear duplex DNA at an end. The enzyme progressed through the duplex, either digesting or not digesting the DNA, depending on the ionic environment (reviewed in Smith 1998). Thus a severed cos or any other double-strand break could be an entry point for the enzyme, with cos being the major one in λ.
Studies on λ packaging revealed that terminase, the enzyme that cuts cos in preparation for λDNA packaging, remains bound at the left end of the λ chromosome (Feiss et al. 1983). This feature could account for the need for proper orientation of Chi, with respect to cos, for Chi to function: one had only to suppose that terminase blocked the entry of RecBCD into the left end of a chromosome about to undergo packaging. The requirement for correct Chi orientation (with respect to cos orientation) implied that an enzyme traveling from cos to Chi must approach the 5′-GCTGGTGG-3′ sequence from the right, as written here, to respond to the Chi.
Does the E. coli chromosome also have characteristic entry sites for RecBCD? A clue came from Ichizo and Mary's discovery that replication of λ increases the activity of a subdued Chi carried by the λ. This finding led Ichizo to a model in which replication forks (being especially vulnerable to breakage?) were entry sites for RecBCD enzyme. This work, combined with Henderson's observation of Chi-stimulated DNA replication, implied mutual stimulation of DNA replication and genetic recombination (as foreseen by Skalka 1974) and contributed to the current view that fork repair is, indeed, the important role of E. coli's RecBCD recombination pathway. Support for this view, and for an important role for Chi in the process, came from the E. coli genome sequencing project, which revealed that the Chi sequences on opposite sides of the replication origin tended to be oppositely oriented, each in the manner that would allow RecBCD to respond to them in its course of fork repair.
What happens when RecBCD meets Chi?
Under in vitro conditions that minimized digestion, a traveling, purified RecBCD introduced a nick in the DNA at Chi (Ponticelli et al. 1985). In the model for recombination that was paradigmatic at the time (Meselson and Radding 1975), such a nick could serve as the recombination-initiating event, and the nick-at-Chi model got a lot of press on that account (e.g., Smith and Stahl 1985). Susan Rosenberg chided me for meekly accepting a model without testing its predictions, and she took the lead in challenging the complacency that resulted from the beguiling congruence of observation and theory. The issue was reopened.
The discovery of mutations that knocked out the previously shadowy RecD subunit of the RecBCD enzyme opened new ways of thinking about Chi (Amundsen et al. 1986; Biek and Cohen 1986). David Thaler and Beth Sampson noted that, in a recD mutant host, a Chi-less red gam λ cross behaved as if there were a Chi sequence at the right end of the λ chromosome (the entry site for RecBCD). Among David's abundant ideas was the suggestion that the role of Chi is to remove RecD activity from the RecBCD enzyme. This change would convert the enzyme from a casual traveler into one dedicated to effecting recombination (and see Koppen et al. 1995).
Contemporaneously with these studies, Ichizo addressed the question of whether or not Chi-induced recombination was reciprocal: Were both complementary products made in a single act? A number of λ crosses had indicated that reciprocality was unlikely, as judged by the relative frequencies of complementary recombinants among mature phage particles. However, Ichizo speculated that the apparent lack of reciprocality reflected the rules of packaging from a dimer formed by Chi-stimulated recombination. Indeed, he designed crosses that separated packaging from the cos cutting that allowed entry of RecBCD; he found that the degree of nonreciprocality was diminished. For simplicity, we took that to mean that the recombination event stimulated by Chi was, in fact, a reciprocal one. Soon thereafter we received a letter, in four colors on a sheet of wrapping paper, from Siberia.
The Siberian connection:
The correspondence initiated by that letter, which described the writer's views of recombination, flowered, and before long we were exchanging personal as well as scientific viewpoints with Andrei Kuzminov of Novosibirsk. Andrei was blunt— he told us that some of our notions about recombination were wrong. In particular, it was unreasonable of us to think that RecBCD-mediated recombination could ever be reciprocal in a simple sense. He pointed out that this enzyme demolishes linear DNA in vivo (a property of the enzyme that we often swept under the carpet) and that the role of Chi must be to stop the demolition. There is no way, he wrote, that the two recombining duplexes could generate both crossover products when the DNA to the right of Chi on one of the participating λ duplexes had been destroyed. In so far as we saw reciprocality, he argued, it must mean that a third duplex got into the act. We put Andrei's view to the test in crosses that varied the relative multiplicity of infection of the two parental, infecting phage. When the Chi-carrying phage was in excess, we got approximate reciprocality; when the Chi-carrying parent was in the minority, the recombinant that would have inherited DNA to the right of Chi from that parent was relatively rare, all as predicted by the proposal of a triparental reaction.
The invitations:
Andrei bemoaned the collapse of science in the USSR, and Mary urged me to invite him to our lab “before Someone Else grabs him.” Andrei responded to our invitation by inviting me to Siberia (expenses paid within the USSR). He said that I should meet him before committing to hiring him. (Sure, right, of course. What other reason could he have had?) Travel within that rapidly disintegrating system was an adventure, but that is another story. When I finally did get to Novosibirsk, I was surprised to find a graduate student, where I had expected to find an established scientist. However, Andrei proved to be as sharp as his letters. By the end of the visit he, too, was convinced that he should come to Eugene. He soon did and in due course took the reins of the Chi research in Eugene.
During his decade in Eugene, Andrei oriented our Chi research toward the role of Chi and RecBCD in maintaining the E. coli chromosome during vegetative growth. His mastery of the literature combined with his meticulous experimentation led to a number of nice articles, including treasured review articles (e.g., Kuzminov 1999, 2001).
Chi, the cis-acting recombinator, can act in trans:
Rik Myers and Andrei teamed up for an elegant set of experiments supporting the view that Chi civilizes RecBCD by removing or inactivating the RecD subunit. They performed red gam mutant λ crosses in cells flooded with accessible Chi sequences (carried on a multicopy plasmid that could be linearized at a cloned cos site). The λ phage recombined as if they were doing so in a recD mutant host. When such crosses were conducted in cells that overproduced RecD protein, the crosses looked like ordinary red gam mutant crosses.
Civilized RecBCD carries on until it has done its duty:
Once the traveling RecBCD exonuclease has been civilized by a cis-acting Chi, what determines where the exchange will occur? Rik answered that question with a set of λ crosses. When Chi, present in only one of two λ parents, is opposite a large heterology, the ability of Chi to act beyond the heterology is influenced by the relative numbers of the two parental phage. When homology at Chi is abundantly available, due to a high multiplicity of the Chi+ parent, the (undetectable) exchanges occur between Chi+ chromosomes. When homology at Chi is scarce due to the low multiplicity of that parent, recombination between the Chi+ and Chi− parents occurs with full force beyond the limit of the heterologous substitution. Thus, a traveling Chi-civilized RecBCD enzyme keeps on traveling until it finds homology and effects recombination.
The “how” of this amusing phenomenology is currently yielding to in vitro analyses in other venues (e.g., Arnold and Kowalczykowski 2000; Taylor and Smith 2003), which are certain to get a big boost from the recently elucidated 3-D structure of the gigantic RecBCD protein (Singleton et al. 2004).
Why a “destructase” plus a “civilizer”?
It has been proposed that the destructive activity of RecBCD is E. coli's weapon against invading DNA and that Chi is E. coli's protection against destruction of self. In a sense, it may be an exonuclease version of the restriction-modification self/nonself systems.
Musing:
If wild-type λ (48.5 kb) contained even one GCTGGTGG (which occurs once every 5 kb in E. coli), Chi might still be undiscovered.
Acknowledgments
Several of my former collaborators corrected my memories and, along with present colleagues, provided valuable editorial advice. The E. coli/λ genetics community was generous throughout the course of this research with their wisdom and their strains.
References
- Amundsen, S. K., A. F. Taylor, A. M. Chaudhury and G. R. Smith, 1986. recD: the gene for an essential third subunit of exonuclease V. Proc. Natl. Acad. Sci. USA 83: 5558–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P., 1987. Twenty years of illegitimate recombination. Genetics 115: 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, D. A., and S. C. Kowalczykowski, 2000. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J. Biol. Chem. 275: 12261–12265. [DOI] [PubMed] [Google Scholar]
- Benzer, S., 1961. On the topography of the genetic fine structure. Proc. Natl. Acad. Sci. USA 47: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek, D. P., and S. N. Cohen, 1986. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J. Bacteriol. 167: 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabert, P., S. D. Ehrlich and A. Gruss, 1992. Chi sequence protects against RecBCD degradation of DNA in vivo. Proc. Natl. Acad. Sci. USA 89: 12073–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist, L. W., and A. Skalka, 1973. Replication of bacteriophage λ DNA dependent on the function of host and viral genes. I. Interaction of red, gam and rec. J. Mol. Biol. 75: 185–212. [DOI] [PubMed] [Google Scholar]
- Feiss, M., I. Kobayashi and W. Widner, 1983. Separate sites for binding and nicking of bacteriophage λ DNA by terminase. Proc. Natl. Acad. Sci. USA 80: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen, J. R., and A. J. Clark, 1974 The RecE pathway of bacterial recombination, pp. 123–136 in Mechanisms in Recombination, edited by R. F. Grell. Plenum, New York.
- Henderson, D., and J. Weil, 1974a chi mutations in phage lambda, pp. 89–94 in Mechanisms in Recombination, edited by R. F. Grell. Plenum, New York.
- Henderson, D., and J. Weil, 1974. b Recombination-deficient deletions in bacteriophage λ and their interaction with chi mutations. Genetics 79: 143–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen, A., S. Krobitsch, B. Thoms and W. Wackernagel, 1995. Interaction with the recombination hot spot chi in vivo converts the RecBCD enzyme of Escherichia coli into a chi-independent recombinase by inactivation of the RecD subunit. Proc. Natl. Acad. Sci. USA 92: 6249–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov, A., 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63: 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov, A., 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98: 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, S. T., M. M. Stahl, K. D. McMilin and F. W. Stahl, 1974. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics 77: 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson, M. S., and C. M. Radding, 1975. A general model for genetic recombination. Proc. Natl. Acad. Sci. USA 72: 358–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, R. S., and F. W. Stahl, 1994. Chi and the RecBCD enzyme of Escherichia coli. Annu. Rev. Genet. 28: 49–70. [DOI] [PubMed] [Google Scholar]
- Ponticelli, A. S., D. W. Schultz, A. F. Taylor and G. R. Smith, 1985. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell 41: 45–51. [DOI] [PubMed] [Google Scholar]
- Singleton, M. R., M. S. Dillingham, M. Gaudier, S. C. Kowalczykowski and D. B. Wigley, 2004. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432: 187–193. [DOI] [PubMed] [Google Scholar]
- Skalka, A., 1974 A replicator's view of recombination and repair, pp. 421–432 in Mechanisms in Recombination, edited by R. F. Grell. Plenum, New York.
- Smith, G. R., 1998 Chi sites and their consequences, pp. 49–66 in Bacterial Genomes: Physical Structure and Analysis, edited by F. J. do Bruijn, J. R. Lupski and G. M. Weinstock. International Thomson, Albany, NY.
- Smith, G. R., and F. W. Stahl, 1985. Homologous recombination promoted by Chi sites and RecBC enzyme of Escherichia coli. BioEssays 2: 244–249. [Google Scholar]
- Smith, G. R., S. M. Kunes, D. W. Schultz, A. Taylor and K. L. Triman, 1981. Structure of Chi hotspots of generalized recombination. Cell 24: 429–436. [DOI] [PubMed] [Google Scholar]
- Stahl, F. W., 1994. The Holliday junction on its thirtieth anniversary. Genetics 138: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, F. W., K. D. McMilin, M. M. Stahl, J. M. Crasemann and S. Lam, 1974. The distribution of crossovers along unreplicated λ bacteriophage chromosomes. Genetics 77: 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpirer, J., and P. Brachet, 1970. Relations physiologiques entre les phages tempérés λ et Φ80. Mol. Gen. Genet. 108: 78–92. [DOI] [PubMed] [Google Scholar]
- Taylor, A. F., and G. R. Smith, 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423: 889–893. [DOI] [PubMed] [Google Scholar]
- Triman, K. L., 1982 Chi stimulation of genetic recombination in Escherichia coli in the absence of bacteriophage lambda. Ph.D. Thesis, University of Oregon, Eugene, OR.