Abstract
We have used affinity chromatography to identify two proteins that bind to the SH3 domain of the actin cytoskeleton protein Rvs167p: Gyp5p and Gyl1p. Gyp5p has been shown to be a GTPase activating protein (GAP) for Ypt1p, a Rab GTPase involved in ER to Golgi trafficking; Gyl1p is a protein that resembles Gyp5p and has recently been shown to colocalize with and belong to the same protein complex as Gyp5p. We show that Gyl1p and Gyp5p interact directly with each other, likely through their carboxy-terminal coiled-coil regions. In assays of GAP activity, Gyp5p had GAP activity toward Ypt1p and we found that this activity was stimulated by the addition of Gyl1p. Gyl1p had no GAP activity toward Ypt1p. Genetic experiments suggest a role for Gyp5p and Gyl1p in ER to Golgi trafficking, consistent with their biochemical role. Since Rvs167p has a previously characterized role in endocytosis and we have shown here that it interacts with proteins involved in Golgi vesicle trafficking, we suggest that Rvs167p may have a general role in vesicle trafficking.
THE actin cytoskeleton provides the structural basis for cell polarity in Saccharomyces cerevisiae and other eukaryotes. Three types of actin structures are found in vegetative yeast cells: actin cables, cortical actin patches, and the cytokinetic ring. Actin cables are long bundles of actin filaments that are believed to function as tracks for polarized transport of organelles and vesicles (Novick and Botstein 1985). Cortical actin patches are punctate cytoskeletal bodies found in polarized clusters at regions of cell growth. They exhibit great biochemical complexity and are dynamic in composition (for review see Pruyne and Bretscher 2000; Munn 2001). Actin patches are thought to be sites of endocytosis (for review see Engqvist-Goldstein and Drubin 2003); however, the mechanism of this is only beginning to be understood (Kaksonen et al. 2003). The cytokinetic ring consists of an actomyosin-based contractile ring assembled on a septin scaffold at the cell division site (for review see Tolliday et al. 2001).
Protein transport along secretory and endocytic pathways in eukaryotic cells is primarily mediated by transport vesicles that bud from a donor compartment and then fuse with a specific target compartment. Transport between organelles is directional and vesicle budding, targeting, and fusion must be tightly regulated to ensure specificity of fusion. Specificity of fusion is controlled by proteins on the surface of vesicles and target membranes, v-SNAREs and t-SNAREs (vesicle and target membrane soluble N-ethylmaleimide-sensitive factor attachment protein receptors), as well as small Rab-type GTPases, also known as Ypt GTPases, that are specific for each type of traffic (for review see Bonifacino and Glick 2004). Ypt GTPases confer specificity by tethering vesicles to their target membranes. Like other small GTPases, Ypt proteins cycle between an active GTP-bound and an inactive GDP-bound state. This cycling is regulated by GTPase activating proteins (GAPs) and GTP exchange factors. The role of GTP hydrolysis in vesicle fusion is somewhat unclear; it has been associated with membrane fusion itself, recycling of the GTPase, or timing of vesicle fusion (for review see Segev 2001).
A number of pieces of evidence point to a role for the actin cytoskeleton in vesicle trafficking. Mutants with a temperature-sensitive allele of the gene encoding actin, act1-1, accumulate post-Golgi secretory vesicles and are partially defective in secretion of invertase (Novick and Botstein 1985). Actin cables function as tracks along which type V myosins travel carrying post-Golgi vesicles to sites of polarized secretion (for review see Pruyne and Bretscher 2000). There is some evidence that intact actin patches are required for proper vesicle trafficking. Cells lacking the actin patch proteins Myo3p and Myo5p accumulate vesicles and have a partial block in secretion of invertase (Goodson et al. 1996). In addition, mutants with the act1-1 allele and mutants lacking the actin patch protein Sla2p accumulate vesicles that contain the Golgi GTPase Ypt1p, suggesting that actin patch proteins may be required for some step in vesicle trafficking between Golgi and plasma membrane (Mulholland et al. 1997).
Two proteins that have been localized to cortical actin patches are Rvs167p and Rvs161p (Balguerie et al. 1999). RVS167 and RVS161, which encode closely related proteins, were first identified in a screen for mutants that exhibited reduced viability upon starvation (Bauer et al. 1993). Mutation of RVS167 or RVS161 causes a phenotype consistent with a role for the Rvs proteins in cortical actin cytoskeleton organization and endocytosis: loss of viability and unusual cell morphology in poor growth medium or salt-containing medium, delocalized actin distribution under suboptimal growth conditions, abnormal (random) budding in diploids, and defects in endocytosis and sporulation (Bauer et al. 1993). Consistent with a requirement for Rvs167p and Rvs161p in vesicle trafficking, ultrastructural studies have revealed that rvs mutants accumulate late secretory vesicles at sites of membrane and cell wall construction (Breton et al. 2001).
Rvs167p and Rvs161p are members of a family of proteins that include amphiphysins, which are proteins involved in endocytosis of synaptic vesicles in nerve terminals (for review see Zhang and Zelhof 2002). Proteins in this family are characterized by the presence of an N-terminal BAR domain and it is through their respective BAR domains that Rvs167p interacts with Rvs161p (Navarro et al. 1997; Sivadon et al. 1997; Colwill et al. 1999). The crystal structure of the BAR domain of Drosophila amphiphysin has recently been solved (Peter et al. 2004). It is a crescent-shaped dimer, in which each monomer forms a coiled coil. The BAR domain binds preferentially to highly curved negatively charged membranes, and this property is thought to promote membrane deformation leading to vesicle biogenesis (Lee and Schekman 2004; Peter et al. 2004). The central portion of Rvs167p consists of a region rich in glycine, proline, and alanine (the GPA region) and is thought to play a role in Rvs regulation since it is phosphorylated in vivo (Friesen et al. 2003). At its carboxy terminus, Rvs167p, like the amphiphysins, has an Src homology 3 (SH3) domain, a protein module well defined for binding proline-rich sequences (Pawson and Scott 1997). Large-scale two-hybrid and phage display screens have identified a number of proteins that bind to the SH3 domain of Rvs167p (Bon et al. 2000; Uetz et al. 2000; Drees et al. 2001; Ito et al. 2001; Tong et al. 2002; Talarek et al. 2005); however, few of these interactions have been confirmed. Domain mapping of Rvs167p has revealed that the GPA region and the SH3 domain are largely dispensable for all Rvs167p functions tested (Colwill et al. 1999). Thus, although the SH3 domain is conserved among amphiphysins, and several biologically important ligands bind to the SH3 domain in genetic or biochemical screens, its biological function remains unknown.
In this work we have used affinity chromatography to identify two proteins that bind to the SH3 domain of the actin cytoskeleton protein Rvs167p: Gyp5p and Gyl1p. Gyp5p has been shown to be a GAP for Ypt1p (De Antoni et al. 2002), a Rab GTPase involved in ER to Golgi trafficking (Bacon et al. 1989; Segev 1991; for review see Lazar et al. 1997). Gyl1p is a protein with sequence similarity to Gyp5 and has been shown to colocalize with Gyp5p (Chesneau et al. 2004). We show that Gyp5p and Gyl1p interact directly with each other as well as with Rvs167p. In vitro, recombinant Gyl1p stimulates the GAP activity of Gy5p toward Ypt1p. In vivo, co-overexpression of GYL1 and GYP5 is toxic in the absence of SEC22 and in the absence of RUD3, two genes involved in ER to Golgi trafficking that have a synergistic growth defect in combination with RVS167. We suggest that Rvs167p may play a role in vesicle trafficking in several systems, including ER to Golgi trafficking.
MATERIALS AND METHODS
Yeast strains and procedures:
Yeast strains are described in Table 1. Strains were derived using standard yeast genetic techniques, from either BY263, an S288C-derived strain (Measday et al. 1994), or BY4741, the S288C-derived strain from which the set of yeast gene-deletion mutants was made (Brachmann et al. 1998; Winzeler et al. 1999). Strains with deletions and strains with genes tagged at their endogenous locus were constructed as described (Longtine et al. 1998). For spot assays, cultures grown to midlog phase were serially diluted and spotted onto the appropriate plates and incubated for 3 days at 30°.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY263a | MATatrp1Δ63 ura3-52 lys2-801 ade2-107 his3Δ200 leu2-Δ1 | Measday et al. (1997) |
| BY264a | BY263 a/α diploid | This study |
| BY508a | BY263 rvs167Δ::TRP1 | Lee et al. (1998) |
| BY1179a | BY263 GYP5-myc::kan | This study |
| BY1177a | BY263 GYL1-myc::kan | This study |
| BY1185a | BY263 GYP5-HA::kan | This study |
| BY1313a | BY263 GYP5-HA::kan GYL1-myc::kan | This study |
| BY1191a | BY263 gyp5Δ::His5 | This study |
| BY1189a | BY263 gyl1Δ::TRP1 | This study |
| BY1254a | BY263 gyp5Δ::His5 gyl1Δ::TRP1 | This study |
| BY4741a | MATahis3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | Brachmann et al. (1998) |
| BY4741a gyp5Δkanb | Deletion consortium strain | |
| BY4741a sec22Δ::kanb | Deletion consortium strain | |
| BY4741a rud3Δ::kanb | Deletion consortium strain | |
| BY4741a rvs167Δ::kanb | Deletion consortium strain |
Except as noted, strains are isogenic to the parent strain, BY263, an S288C derivative.
Strains from the deletion consortium are isogenic to the parent strain, BY4741, which is also derived from S288C (Brachmann et al. 1998).
DNA manipulations:
Plasmids are listed in Table 2. Standard procedures were used for recombinant DNA manipulations (Ausubel et al. 1994). Details of construction of plasmids are available upon request. PCR reactions were done with Pfx polymerase (Invitrogen, Burlington, Ontario) as recommended by the manufacturer. The integrity of all PCR products was confirmed by sequencing. Oligonucleotides were synthesized by Invitrogen and sequences are available upon request.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pAR100-RVS-SH3 (BA1267) |
RVS167 SH3 (codons 423–482) with an N-terminal 6-His tag under the control of the T7 promoter |
This work |
| pAR100-RVS-P473L (BA1268) |
Same as pAR100-RVS-SH3 but encoding a protein with leucine substituted for proline at position 473 |
This work |
| pDN108 (BA1269) | fyn SH3 (codons 84–142) with a C-terminal Flag epitope followed by a 6-His tag under the control of the T7 promoter |
Maxwell and Davidson (1998) |
| pGPA-SH3-His (BA1344) |
RVS167 GPA-SH3 (codons 282–482) with a C-terminal 6-His tag under the control of the T7 promoter |
Friesen et al. (2003) |
| pRSET-B-GYL1 (BA1612) | GYL1 with a 6-His tag under the control of the T7 promoter | This work |
| pET28 + GYP5 (BA1677) | Full-length GYP5 (residues 1–894) with an N-terminal 6-His tag under the control of the T7 promoter |
This work |
| pET28 + GYP5ΔN (BA1679) |
GYP5 residues 403–894 with an N-terminal 6-His tag under the control of the T7 promoter |
This work |
| pET28 + GYP5ΔC (BA1680) |
GYP5 residues 1–690 with an N-terminal 6-His tag under the control of the T7 promoter |
This work |
| pET19 + GYP5 (BA1343) |
GYP5 missing the first 15 amino acids with an N-terminal 6-His tag in pET19b |
This work |
| pGEX + GYL1 (BA1334) | GYL1 with an N-terminal GST tag in pGEX-3X | This work |
| pET15-SEC4 | SEC4 with an N-terminal 6-His tag in pET15b | Du et al. (1998) |
| pET15-YPT1 | YPT1 with an N-terminal 6-His tag in pET15b | Du et al. (1998) |
| pET15-GYP1 | GYP1 with an N-terminal 6-His tag in pET15b | Du et al. (1998) |
| p426-GAL | High-copy URA3 plasmid with GAL promoter | Ronicke et al. (1997) |
| pAcGHLT-RVS167 | RVS167 with N-terminal GST + His tag used for insect cell infection | Friesen et al. (2003) |
| pAcHLT-RVS161 | RVS161 with N-terminal His tag used for insect cell infection | This work |
| p426-GAL-GYL1-Flag (BA1691) |
GYL1 with a C-terminal Flag tag under control of the GAL promoter on high-copy URA3 plasmid |
This work |
| p415-GAL-GYP5-Flag (BA1699) |
GYP5 with a C-terminal Flag tag under control of the GAL promoter on CEN LEU2 plasmid |
This work |
Affinity chromatography:
SH3 domain proteins containing 6-His tags at their carboxy termini were purified from strain GJ1158 (Bhandari and Gowrishankar 1997) using Ni-NTA agarose (QIAGEN, Valencia, CA) following a denaturing purification procedure recommended by the manufacturer. Proteins were covalently coupled to Affi-Gel 10 Resin (Bio-Rad, Mississauga, Ontario) in coupling buffer (20 mm HEPES, pH 8.0, 300 mm NaCl, 10% glycerol) according to the manufacturer's recommendations. The concentration of coupled protein on the resin was ∼10 μm. Twenty-microliter columns were made in Marsh pipette tips (ABgene, Rochester, NY) with a 10-μl frit made of 150- to 212-μm glass beads (Sigma, Oakville, Ontario). Yeast extracts were made and chromatography was done as described by Ho et al. (1997) using frozen pellets from 1.5 liter of log-phase cells (strain BY264). We typically loaded 20 mg of yeast extract per column. Columns were washed with SB (20 mm HEPES, pH 7.2, 10% glycerol, 0.1 mm DTT, 0.1 mm PMSF) + 100 mm NaCl and bound proteins were eluted with SB + 1 m NaCl and then with SB + 1% SDS. Eluates were separated on Novex 4–12% gradient gels (Invitrogen) using MOPS SDS running buffer recommended by the manufacturer. Gels were stained with Coomassie blue (Bio-Rad), destained, and then stained with silver using a low concentration of formaldehyde (Shevchenko et al. 1996). Bands representing proteins that bound to wild-type Rvs167p SH3, but not to Rvs167p SH3-P473L or to fyn SH3, were cut out of the gel. Tryptic peptides were isolated using an in-gel digestion procedure as described by Figeys et al. (2001). Masses of peptides were identified by matrix-assisted laser desorption ionization time of flight spectrometry using a PerSeptive DESTR at Borealis (Toronto), and proteins were identified using ProFound (http://prowl.rockefeller.edu/cgi-bin/ProFound).
Antibodies, immunoprecipitations, Western blots, and Far Western assays:
Standard procedures were used for yeast cell extract preparation and immunoblotting (Lee et al. 1998). Antibodies used were polyclonal α-Rvs167 (Lee et al. 1998), monoclonal α-myc (9E10, produced by University of Toronto monoclonal antibody facility), monoclonal α-HA (12CA5, Sigma), and monoclonal α-Flag M2 (Sigma). For co-immunoprecipitation, cells from 100 ml of log-phase culture were vortexed 9 × 1 min with glass beads in lysis buffer [250 mm NaCl, 50 mm Tris-Cl, pH 7.5, 5 mm EDTA, 0.1% NP40, 0.5 mm DTT, 20 mm NaF, 20 mm β-glycerophosphate, 1 mm PMSF, and protease inhibitor cocktail lacking EDTA (Boehringer Mannheim, Indianapolis)] and clarified by centrifugation at 16,000 × g for 10 min. Protein (3 mg) was incubated with 2 μl 9E10 monoclonal anti-myc or 2 μl affinity purified anti-Rvs167 antibodies for 1 hr on ice and then 20 μl protein A Sepharose (Pharmacia, Piscataway, NJ) was added and the mixture was incubated for another hour on a Nutator. The beads were washed five times in 0.8 ml cold lysis buffer and suspended in 20 μl 2× sample buffer, and 7 μl was analyzed on 7.5% polyacrylamide gels. Far Western hybridization was done as described by Guichet et al. (1997) using 35S-labeled Gyl1p, Gyp5p, Gyp5pΔN, or Gyp5pΔC that had been synthesized using a coupled T7 polymerase-reticulocyte lysate system (Promega, Madison, WI) primed with plasmids pRSET-B-YMR192W (BA1612), pET28 + GYP5 (BA1677), pET28 + GYP5ΔN (BA1679), and pET28 + GYP5ΔC (BA1680).
Protein purification:
His-tagged Gyp5p was partially purified from Escherichia coli containing pET19 + GYP5 using Novagen His Bind resin (Novagen, Madison, WI) following the manufacturer's instructions. Similarly, GST-tagged Gyl1p was partially purified from E. coli containing pGEX + GYL1 using glutathione Sepharose (Amersham, Piscataway, NJ). Protein was dialyzed against 20 mm Tris-Cl, pH 8.0, 0.3 m NaCl, 1 mm MgCl2, 10% glycerol and stored at −80°. Protein was quantified by comparison of the band representing full-length protein to bands on a gel with dilutions of bovine serum albumin. For purification of Rvs167p-Rvs161p, Hi5 insect cells were co-infected with recombinant baculoviruses expressing GST-His-RVS167 and His-RVS161. After ∼48 hr, cells were harvested, washed, and lysed in insect cell lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.1% NP40, 5 mm NaF, 0.5 mm MgCl2, and 1 mm DTT). Following centrifugation at 10,000 × g for 20 min, cell extracts were incubated with glutathione Sepharose beads for 1 hr at 4°. Beads were washed four times in 10 volumes of lysis buffer and GST-His-Rvs167p and associated His-Rvs161p were eluted with lysis buffer containing 50 mm glutathione.
GAP assays:
Assays for GAP activity were done as described by Du et al. (1998). Briefly, 1 μm His-Ypt1p or His-Sec4p was preloaded with 5 μm GTP spiked with [γ-32P]GTP for 30–60 min at room temperature and then Ypt1p-GTP or Sec4p-GTP was diluted 10-fold and added, in the presence of a 200-fold excess of unlabeled GTP, to His-Gyp5p, GST-Gyl1p, GST-His-Rvs167p-His-Rvs161p, or combinations of these proteins that had been premixed and left on ice for 20 min. Samples were taken in duplicate at 0, 1, 2, 5, 10, 30, and 60 min and GTP hydrolysis (release of 32P) was measured by the charcoal-binding method (Du et al. 1998). Protein binding to GTP was measured using a filter-binding assay at 0 and 60 min after addition of GAPs.
RESULTS
Identification of two novel Rvs167p-interacting proteins using affinity chromatography:
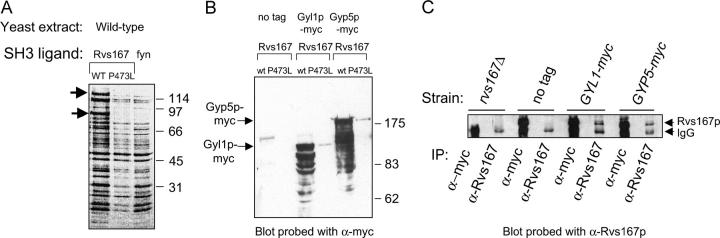
Rvs167p plays an important role in regulating the actin cytoskeleton; however, the SH3 domain is not required to complement any defects seen in an rvs167Δ strain (Colwill et al. 1999). Large-scale two-hybrid screens (Bon et al. 2000; Uetz et al. 2000; Drees et al. 2001; Ito et al. 2001; Tong et al. 2002) and phage display studies (Tong et al. 2002) have identified a number of proteins that bind to Rvs167p and specifically to the SH3 domain of Rvs167p. Many of these proposed SH3 interactions have been seen only in a two-hybrid assay, however, and it seems likely that a significant fraction will prove to be artifacts of overexpression or due to indirect interactions in these relatively artificial systems. To identify proteins in yeast extract that bind to the SH3 domain of Rvs167p we performed affinity chromatography using Rvs167p SH3 with a carboxy-terminal 6-histidine tag as ligand. As a control for the Rvs167p SH3 we used Rvs167p SH3-P473L, which contains an amino acid substitution that has been shown to cause defects in binding the putative Rvs167p ligands Abp1p and Las17p in a two-hybrid assay (Colwill et al. 1999). As a control for specificity of binding we used an unrelated SH3 domain from the chicken fyn tyrosine kinase (Maxwell and Davidson 1998). We passed extract made from our standard wild-type diploid strain (BY264) over the columns, eluted the bound proteins with 1% SDS, separated the proteins on gels, and looked for bands that were present in eluates from wild-type Rvs167p SH3 columns but not mutant Rvs167p SH3 or fyn SH3 (Figure 1A). Using mass spectrometry, we identified two proteins that bound specifically to the wild-type Rvs167p SH3 domain: Ypl249p, known as Gyp5p, and Ymr192p, known as Gyl1p.
Figure 1.—
Rvs167p SH3 domain binds to Gyp5p and Gyl1p. (A) Affinity chromatography using wild-type Rvs167p SH3 domain, a mutant Rvs167p SH3 domain with a P473L substitution, and the unrelated fyn SH3 domain as ligands. Extract from wild-type log-phase yeast cells was passed over the columns; the columns were washed; and bound proteins were eluted with 1% SDS, run on a 4–12% acrylamide gradient gel, and stained with silver. Arrows on the left point to bands representing two proteins that bound specifically to wild-type Rvs167p SH3 but not to mutant SH3 or to fyn SH3. These bands were cut out of the gel and the proteins were digested with trypsin and analyzed by mass spectroscopy, which identified them as Ypl249p (Gyp5p) and Ymr192p (Gyl1p). Position of protein molecular weight markers (in kDa) is shown on the right. (B) Gyl1p-myc and Gyp5p-myc bind to wild type but not mutant Rvs167p SH3 in a pulldown assay. Extracts from yeast cells with no tag, cells expressing GYL1-myc, and cells expressing GYP5-myc were passed over microcolumns of wild-type Rvs167p SH3 or mutant Rvs167p SH3. The bound proteins were eluted with 1% SDS and run on an SDS gel, the gel was transferred to nitrocellulose, and the blot was probed with α-myc antibodies. Arrows on the left point to the predicted positions of full-length Gyp5p-myc and Gyl1p-myc. (C) Co-immunoprecipitation of Rvs167p with Gyl1p-myc or Gyp5p-myc. Western blot analysis of immunoprecipitations from yeast extract using α-Rvs167p antibodies. Lysates from BY508 (rvs167Δ), BY263 (wt), BY1177 (GYL1-myc), and BY1179 (GYP5-myc) were incubated with α-Rvs167p or α-myc antibodies as indicated and the resulting immunoprecipitates were analyzed by Western blot. The positions of migration of Rvs167p and IgG are shown on the right.
We next used a pulldown assay to confirm that the proteins we had identified by mass spectrometry were Gyp5p and Gyl1p. We constructed yeast strains with carboxy-terminal 13-myc tags at the endogenous gene loci, made extract from log-phase cells, and monitored binding to the Rvs167p SH3 columns by Western blot (Figure 1B). In this assay, Gyp5p-myc and Gyl1p-myc from yeast extract bound to wild-type Rvs167p SH3 but not to Rvs167p SH3-P473L. A number of faster-migrating bands that hybridized to the α-myc antibody were seen in extract from strains producing both Gyp5p-myc and Gyl1p-myc (Figure 1B). These may be degradation products of Gyp5p and Gyl1p.
Co-immunoprecipitation of Rvs167p with Gyp5p and Gyl1p:
To test whether we could detect an interaction between Rvs167p and Gyp5p and between Rvs167p and Gyl1p under physiological conditions, we used a co-immunoprecipitation assay. We immunoprecipitated either with α-Rvs167 or with α-myc antibodies, separated the immunoprecipitated proteins on an SDS gel, immunoblotted, and probed the blot with α-Rvs167 antibodies (Figure 1C). In this experiment, Rvs167p was co-immunoprecipitated with α-myc in cells expressing GYL1-myc (Figure 1C, lane 6) or GYP5-myc (Figure 1C, lane 8) but not in an untagged strain (Figure 1C, lane 4) nor in an rvs167Δ strain (Figure 1C, lane 2). Interactions between Rvs167p and both Gyp5p and Gyl1p have been detected previously in large-scale screens with overexpressed proteins (see discussion); however, we present here evidence that Rvs167p and Gyp5p as well as Rvs167p and Gyl1p interact when the genes are expressed from their endogenous promoters.
Sequence comparison of Gyp5p and Gyl1p:
Gyp5p has been identified as a GTPase-activating protein for the Rab GTPase Ypt1p, which is required for vesicle trafficking from ER to Golgi (De Antoni et al. 2002). Gyl1p has been found to colocalize with Gyp5p (Chesneau et al. 2004) and GYL1 has synthetic interactions with FUS2 (Talarek et al. 2005). When we examined these two proteins in a BLAST search (Karlin and Altschul 1990, 1993) we found that Gyp5p and Gyl1p are similar in primary sequence to each other (also noted by Chesneau et al. 2004; Talarek et al. 2005). Indeed the genes encoding Gyl1p and Gyp5p are part of a proposed whole-genome duplication in an ancestor of Saccharomyces (Wolfe and Shields 1997; Kellis et al. 2004). Over almost the entire length of Gyl1p (amino acids 21–720), the two proteins are 29% identical and 51% similar (Figure 2A). Gyp5p has a 185-residue extended region at its amino terminus with no homology. Gyl1p was originally given the name App2p in the Saccharomyces Genome Database (http://www.yeastgenome.org); however, for reasons we explain below, it has been renamed Gyl1p for Gyp5-like protein (http://www.yeastgenome.org).
Figure 2.—

Sequence comparison of Gyl1p and Gyp5p. (A) Diagram of Gyl1p and Gyp5p showing predicted domain structure and sequence similarity. Four PXXP motifs (P) are found in the N-terminal portions of each protein. The GYP domain is located in the central portion of Gyp5p and a region with 35% sequence identity to this is located within the central portion of Gyl1p. The conserved finger arginine in Gyp5p is marked “R”. The C-terminal region of both proteins is predicted to contain coiled coils (C-C). Gyl1p and Gyp5p are 29% identical over almost the entire length of Ymr192p. (B) Alignment and predicted secondary structure of the GYP domain of Gyp5p (amino acids 412–690) and the corresponding region of Gyl1p (amino acids 262–535) and secondary structure of Gyp1p observed in the crystal structure. Protein sequence alignments of the GYP domain of Gyp1p, Gyp5p, and Gyl1p were done with ClustalW and edited manually. Amino acid residues are colored as follows: red, hydrophobic (A, V, F, P, M, I, L, W, including aromatic Y); blue, acidic (D, E); magenta, basic (R, H, K); green, other (hydroxyl + amine + basic; S, T, Y, H, C, N, G, Q). Symbols under the alignment indicate level of conservation: an asterisk means that the residues or nucleotides in that column are identical in all sequences in the alignment; a colon means that conserved substitutions have been observed; a period indicates semiconserved substitutions. The GYP fingerprint sequences are highlighted by a yellow background and the absolutely conserved residues are shown in boldface type. The strings of H labeled “Gyl1 jpred” and “Gyp5 jpred” represent α-helices predicted by the secondary structure prediction program Jpred for the GYP region of Gyl1p and Gyp5p. Dashed lines indicate regions with no predicted secondary structure. The strings of H that are labeled “Gyp1 observed” represent α-helices seen in the crystal structure of Gyp1p (Rak et al. 2000).
Since we had identified Gyp5p and Gyl1p as proteins that bound to the SH3 domain of Rvs167p, we looked in the proteins for PXXP sequences, which are SH3-binding motifs (for review see Mayer 2001). Both Gyp5p and Gyl1p have several PXXP motifs in their amino-terminal portion, including one PXXP that fits the consensus derived by Tong et al. (2002) for binding to the Rvs167p SH3 domain in a phage display assay. The PXXP motif that fits the consensus for binding to the Rvs167p SH3 in Gyp5p begins at residue 282 (PPLPPR, PXXP underlined) and in Gyl1p at residue 125 (PPLPPR). In addition, comparison of the sequence of Gyl1p and Gyp5p to a database of known parallel two-stranded coiled coils (http://www.ch.embnet.org/software/COILS; Lupas et al. 1991) revealed that each protein has a predicted coiled-coil region in its carboxy terminus (residues 750–870 in Gyp5p, also noted by De Antoni et al. (2002), and residues 590–700 in Gyl1p).
A number of GAPs for Ypt/Rab-specific GTPases have been identified biochemically, by their ability to stimulate the GTPase activity of one or more Ypt-type GTPases (Albert and Gallwitz 1999, 2000; Albert et al. 1999). The catalytic domain, known as the GYP domain, for GAP for Ypt protein, has been delimited in vitro for some of these Ypt GAPs (Albert and Gallwitz 1999). Within this GYP domain there are three absolutely conserved “GYP fingerprint” sequences, RXXXW, IXXDXXR, and YXQ (Figure 2B; Neuwald 1997), which were originally used to identify Gyp5p as a putative member of the class (De Antoni et al. 2002). The conserved arginine in the RXXXW motif is thought to be important for maintaining the structure of the GYP domain since substitutions of this arginine to alanine in Gyp1p and Gyp7p rendered the protein unstable during purification (Albert and Gallwitz 1999). The conserved arginine within the IXXDXXR sequence motif has been shown to be critical for GAP catalytic function in all Rab GTPases tested, including Gyp5p (Albert et al. 1999; De Antoni et al. 2002). This arginine is thought to act in catalysis like the “finger arginine” of Ras-GAP (Albert et al. 1999; De Antoni et al. 2002). The crystal structure of the GYP domain of Gyp1p, a GAP with high levels of in vitro GAP activity toward the GTPases Ypt51p and Sec4p, has been solved (Rak et al. 2000). The GYP domain of Gyp1p contains 16 α-helices and the IXXDXXR sequence, containing the finger arginine, extends from within helix 5 into the following loop (Rak et al. 2000). Surprisingly, although Gyl1p is 35% identical and 54% similar to Gyp5p throughout the GYP domain of Gyp5p, Gyl1p is missing two of the three GYP fingerprint sequences, IXXDXXR and YXQ (Figure 2B).
Because of the high degree of conservation between Gyp5p and Gyl1p (Figure 2A) and because Gyp5p has GAP activity in vitro (De Antoni et al. 2002), we were interested in whether they would have the same predicted secondary structure as Gyp1p. We used the secondary structure prediction server Jpred (Cuff and Barton 2000) to predict the secondary structure elements of the GYP domain of Gyp5p and the analogous region of Gyl1p. All of the α-helices observed in the crystal structure of Gyp1p had corresponding α-helices predicted for Gyp5p (Figure 2B), consistent with the in vitro activity of Gyp5p as a GAP for Ypt1p. In contrast, the predicted secondary structure of Gyl1p had 15/16 α-helices but was missing helix 5, which overlaps the finger arginine (Figure 2B). No secondary structure element was predicted in this region for Gyl1p. Thus, although Gyl1p contains a region that is similar in primary sequence to the GYP domain of other Rab GAPs, it lacks a catalytic arginine and probably an important helix required for activity and/or GTPase binding. It is not clear from this analysis whether Gyl1p would have GAP activity.
Gyp5p and Gyl1p interact directly with each other:
Recently Chesneau et al. (2004) have shown that Gyp5p and Gyl1p can be co-immunoprecipitated from a subcellular fraction containing plasma membrane and cytoskeleton and organelle membranes (P13) as well as the fraction containing late Golgi and vesicle membranes (P100). We confirmed this interaction using a co-immunoprecipitation assay from whole-cell extract. We found that in a strain in which Gyp5p was tagged with HA and Gyl1p was tagged with myc we could efficiently co-immunoprecipitate the two proteins by immunoprecipitating with either α-HA or α-myc (Figure 3A).
Figure 3.—
Gyp5p and Gyl1p interact directly. (A) Co-immunoprecipitation of Gyl1p-myc with Gyp5p-HA. Lysates from BY1177 (GYL1-myc), BY1185 (GYP5-HA), and BY1313 (GYL1-myc GYP5-HA) were incubated with α-HA or α-myc antibodies as indicated and the resulting immunoprecipitates were analyzed by Western blot, hybridizing with α-myc or α-HA as shown at the top. The positions of Gyl1p-myc and Gyp5p-HA are shown on the sides. (B) Far Western analysis of Gyl1p, Gyp5p, and truncated versions of Gyp5p binding to Gyl1p, Gyp5p, and Rvs167p. Partially purified His-Gyl1p, His-Gyp5p, and Rvs167 GPA-SH3-His were run on SDS gels and transferred to nitrocellulose. The membranes were hybridized with in vitro translated Gyl1p, Gyp5p, Gyp5pΔC, and Gyp5pΔN; washed; and analyzed by autoradiography.
Interactions between proteins that are detected by co-immunoprecipitation could be bridged by a third protein. To test whether the interaction that we saw between Gyp5p and Gyl1p was direct, we did a Far Western assay. We partially purified His-Gyl1p and His-Gyp5p from E. coli, separated the proteins on an SDS gel, and transferred the proteins to nitrocellulose. As a positive control for binding, we also included Rvs167p GPA-SH3. [We used the GPA-SH3 region of Rvs167p even though the SH3 domain alone could bind to Gyp5p and Gyl1p (Figure 1B) because the 6-kDa SH3 peptide does not bind efficiently to nitrocellulose in a Western blot.] We hybridized the Western blots with 35S-labeled in vitro-translated Gyl1p and Gyp5p (Figure 3B, 35S-labeled Gyl1p and Gyp5p). In this experiment, 35S-labeled Gyl1p bound to Gyp5p and to Rvs167p GPA-SH3 but not to itself (Figure 3B, 35S-labeled Gyl1p). Gyl1p also bound to lower molecular weight bands, which we believe are Gyp5p degradation products (Figure 3B, 35S-labeled Gyl1p). Likewise, 35S-labeled Gyp5p bound to Gyl1p and to Rvs167p GPA-SH3 but not to itself (Figure 3B, 35S-labeled Gyp5). We conclude that the Rvs167p SH3 domain interacts directly with both Gyp5p and Gyl1p and that Gyp5p and Gyl1p interact directly with each other in this assay and do not homodimerize.
To identify the portion of Gyp5p that interacts with Gyl1p we constructed genes that encoded truncations of GYP5, in vitro translated these in the presence of [35S]methionine, and used the labeled proteins to probe Western blots (Figure 3B, 35S-labeled Gyp5pΔC and Gyp5pΔN). A version of Gyp5p lacking the C-terminal 204 amino acids (and so lacking the predicted coiled-coil domain) was still able to bind to Rvs167p but could not bind to Gyl1p (Figure 3B, 35S-labeled Gyp5pΔC). Conversely, a version of Gyp5p lacking the N-terminal 402 amino acids (and so lacking the several PXXP motifs) could bind to Gyl1p but could not bind to Rvs167 GPA-SH3 (Figure 3B, 35S-labeled Gyp5pΔN). This finding, that Gyp5p requires its C-terminal 204 residues to bind to Gyl1p, suggests that the two proteins may interact through their coiled-coil regions.
Biochemical activity of Gyp5p and Gyl1p:
Gyp5p has been identified biochemically as a Ypt GAP. Purified Gyp5p accelerated the intrinsic GTP hydrolysis rate of Ypt1p significantly and had a smaller effect on Sec4p (De Antoni et al. 2002). For ease of purification De Antoni et al. (2002) used an N-terminally truncated version of Gyp5p, extending from residue 400 to 892, in their assays. Because we wanted to test for interaction with Rvs167p, which we have shown requires the N-terminal portion of Gyp5p (Figure 3B), we have assayed partially purified full-length Gyp5p and Gyl1p, which had been expressed in E. coli. Consistent with previous reports (Albert and Gallwitz 1999), we found that both protein preparations contained a substantial fraction of truncated peptides (data not shown). We assayed for GAP activity on Sec4p and Ypt1p that had been prebound to [γ-32P]GTP, using single-round turnover conditions (Du et al. 1998). In this assay the putative GAP is added to the [32P]GTP-bound GTPase in the presence of a large excess of unlabeled GTP such that only a single round of GTP hydrolysis is measured. Release of 32P was determined by the charcoal-binding method (Du et al. 1998). As a positive control we tested Gyp1p, a previously characterized GAP that has been shown to have GAP activity on both Sec4p and Ypt1p (Du et al. 1998). In these experiments we observed only a low level of GAP activity by Gyp5p on Sec4p (Figure 4A). This was consistent with published results; even with the truncated version of Gyp5p, De Antoni et al. (2002) detected only marginal GAP activity on Sec4p. Full-length Gyp5p had concentration-dependent GAP activity on Ypt1 (Figure 4B). Under single-turnover conditions, concentrations of Gyp5p as low as 5 nm were sufficient to enhance the GTPase activity of 100 nm GTP-Ypt1p (Figure 4B), and the initial slopes of the curves are directly proportional to the Gyp5p concentration, clearly indicating the catalytic nature of the interaction (Figure 4, B and C).
Figure 4.—
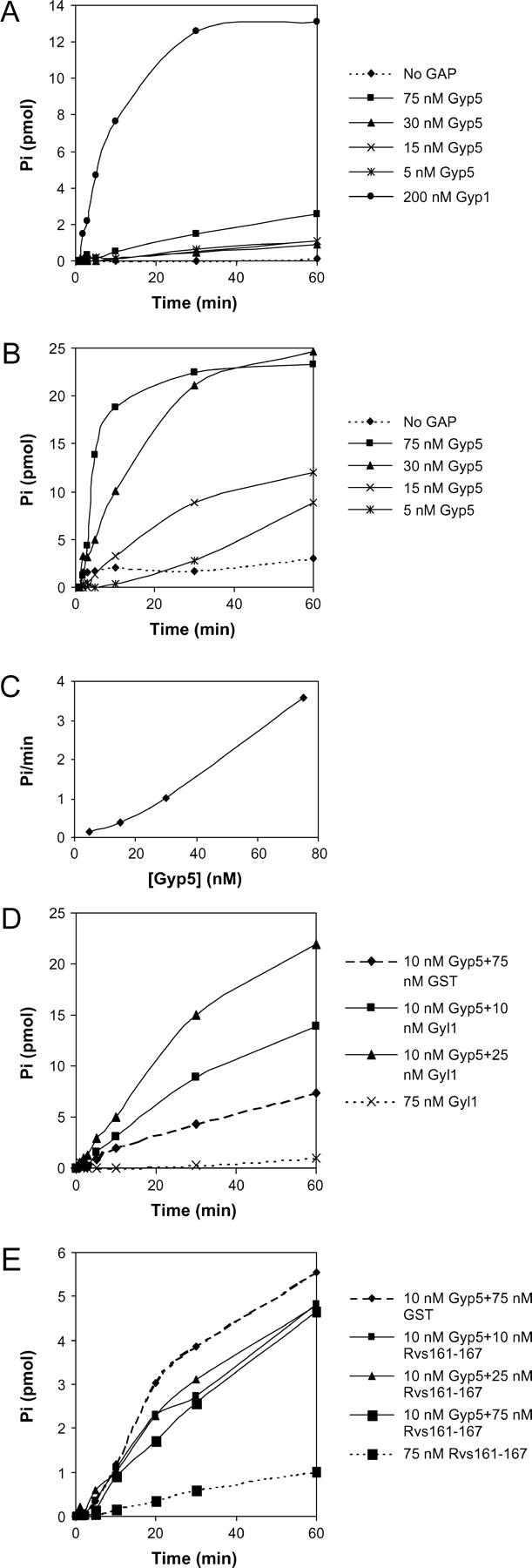
Single-round turnover assays of GAP activity on Sec4p and Ypt1p. One micromolar Ypt GTPases His-Sec4p and His-Ypt1p were prebound with [γ-32P]GTP for 30–60 min and then diluted 10-fold into reactions containing a 200-fold excess of cold GTP and His-Gyp1p, His-Gyp5p, GST, GST-Gyl1p, GST-His-Rvs167p-His-Rvs161p, or combinations of these that had been allowed to incubate together on ice for 20 min. Samples were taken in duplicate at 0, 1, 2, 5, 10, 30, and 60 min and GTP hydrolysis was assayed using the charcoal-binding method. Values shown represent average Pi released in a 100-μl reaction after zero time values have been subtracted. (A) Assay of GAP activity of various concentrations of Gyp5p on the GTPase Sec4. (B) Assay of GAP activity of various concentrations of Gyp5p on the GTPase Ypt1. (C) The initial rates of Pi release are plotted against concentration of Gyp5p for B. (D) Effect of adding Gyl1p and Gyp5p to Ypt1p. (E) Effect of adding Rvs167p-Rvs161p dimer on Gyp5p GAP activity toward Ypt1p.
Next we tested for GAP activity by Gyl1p on Ypt1p. Full-length Gyl1p had no GAP activity on Ypt1p (Figure 4D). Since Gyp5p and Gyl1p interact with each other, we tested whether Gyp5p-dependent activation of Ypt1p GTPase activity was affected by the addition of Gyl1p. We chose to assay a low level of Gyp5p so that the GAP reaction would not be saturated. Addition of 10 nm or 25 nm Gyl1p to 10 nm Gyp5p in an assay on Ypt1p-GTP led to stimulation of the GAP activity of Gyp5p (Figure 4D). Although in repeated assays we have seen that Gyl1p can stimulate the GAP activity of Gyp5p on Ypt1p, the effects were variably dose dependent (data not shown). We attribute this to variable degradation of the GST-Gyl1p protein.
Since Gyp5p binds to the SH3 domain of Rvs167p we were interested in whether Rvs167p would affect GAP activity of Gyp5p. Because Rvs167p forms a heterodimer with Rvs161p in log-phase cells (Navarro et al. 1997; Colwill et al. 1999), we tested the effect of adding Rvs167p-Rvs161p dimer, which had been expressed in insect cells. Addition of Rvs167p-Rvs161p had little or no effect on Gyp5p GAP activity (Figure 4E).
Phenotype of gyp5Δ gyl1Δ cells:
Rvs167p is implicated in control of the actin cytoskeleton; cells lacking RVS167 display defects associated with actin loss of function, namely delocalized actin patches, random budding in diploids, defects in endocytosis and sporulation, and defective growth on medium containing salt. Because we had seen that Gyp5p and Gyl1p interact with Rvs167p, we asked whether gyp5Δ and gyl1Δ strains had any defects associated with the actin cytoskeleton. The fact that Gyp5p and Gyl1p were 29% identical to each other in primary structure suggested that they might have a redundant function so we also examined a strain deleted for both GYP5 and GYL1. We tested for defects in actin polarization, salt sensitivity, bud site selection, fluid-phase endocytosis, and sporulation. In every test, including assays in which rvs167Δ cells were defective, gyp5Δ cells, gyl1Δ cells, and the gyp5Δ gyl1Δ double mutant appeared similar to wild type (data not shown). This suggested either that Rvs167p was not playing its role in the actin cytoskeleton through its interaction with Gyp5p or Gyl1p or that the genes encoding these two proteins were redundant with other genes.
A number of synthetic lethal interactions with RVS167 have been identified, using a candidate approach (Lila and Drubin 1997) and using the synthetic genetic array approach (Tong et al. 2001, 2004). We tested whether GYP5, GYL1, or both genes had genetic interactions with four actin cytoskeleton genes that had a previously identified synthetic interaction with RVS167: SLA1, SLA2, SAC6, and SRV2 (Lila and Drubin 1997). The gyp5Δ and gyl1Δ single mutants and the double mutant showed no growth defect in combination with deletion of any of these actin cytoskeleton genes (data not shown). We conclude that GYP5 and GYL1 likely do not have a primary role in the actin cytoskeleton.
Does Gyl1p have a redundant function with App1p?
Gyl1p/Ymr192p had been tentatively named App2p in the Saccharomyces Genome Database (http://www.yeastgenome.org) on the basis of a computational analysis of protein-protein interactions in large-scale studies, which suggests a possible role in actin filament organization (Samanta and Liang 2003). Samanta and Liang (2003) predicted that if two proteins have a significantly larger than random number of common interaction partners, they are likely to have a close functional association. Close functional association could mean that the two proteins are part of a complex (e.g., Rvs167p and Rvs161p) or that they have a parallel function (e.g., Cln1p and Cln2p; Samanta and Liang 2003). When a network-based statistical algorithm was used to compare protein-protein interaction data from large-scale studies, Gyl1p/Ymr192p was found to be clustered with Ynl094p [named App1p for actin patch protein 1 because it was localized to cortical actin patches (Drees et al. 2001)] and with Las17p, another actin patch protein, and therefore was tentatively named App2p (Samanta and Liang 2003). Gyl1 and App1 have no apparent sequence similarity, except that both proteins have PXXP motifs that could bind to SH3 domains; they were clustered together solely because of their common interacting partners in large-scale screens. To test whether GYL1/YMR192w and APP1 had a redundant function, we constructed a double-mutant strain and assayed it for growth defects using spot dilution assays. The double-mutant strain was able to grow as efficiently as either of the single mutants (or as wild type) on rich medium containing either glucose or galactose as carbon source (data not shown). A very subtle growth defect in the double mutant was detectable when the strains were assayed on synthetic growth medium (data not shown). This was no worse in the triple-mutant app1Δ gyl1Δ gyp5Δ strain and was not exacerbated by high temperature (data not shown). In addition, the app1Δ gyl1Δ double mutant had no defects in polarizing its actin cytoskeleton (data not shown). We were not able to test for a synthetic interaction between GYL1 and LAS17, the gene encoding the other protein clustered with Gyl1p, because las17Δ mutant cells are extremely slow growing on their own.
Genetic interactions with GYP5 and GYL1:
Because we had identified Gyp5p and Gyl1p as proteins that interact with Rvs167p, we thought they were likely to have some cellular role in common with Rvs167p. Since we had seen that Gyp5p and Gyl1p had GAP activity on the small GTPase Ypt1p (Figure 4), which is involved in ER to Golgi trafficking (Bacon et al. 1989; Segev 1991; for review see Lazar et al. 1997), it seemed likely that they might have genetic interactions with genes involved in vesicle trafficking from ER to Golgi. We have recently screened for genes that have synergistic growth defects in combination with RVS167 and identified two genes involved in ER to Golgi vesicle trafficking: SEC22 and RUD3 (Tong et al. 2004). SEC22 encodes a v-SNARE found on the surface of vesicles and is required for fusion of ER-derived vesicles with the Golgi (Lian and Ferro-Novick 1993; Parlati et al. 2000). RUD3 encodes a matrix protein that is involved in the structural organization of the cis-Golgi (Kim et al. 1999). SEC22 and RUD3 have genetic interactions with each other: a rud3 sec22 double mutant is slow growing and overproduction of RUD3 from a multicopy plasmid suppresses the temperature sensitivity of a sec22-3 strain (Kim et al. 1999). These genetic interactions suggest that SEC22 and RUD3 function in the same or parallel pathways, presumably to regulate vesicle fusion at the Golgi. Since SEC22 and RUD3 are important in the absence of RVS167, Rvs167p function may be needed when ER to Golgi trafficking is compromised. Because Gyp5p and Gyl1p have a physical interaction with Rvs167p, we looked for genetic interactions with SEC22 and RUD3. Neither SEC22 nor RUD3 had a synthetic lethal interaction with GYP5 or GYL1 (data not shown), indicating that Sec22p and Rud3p likely do not function in a redundant pathway with these Rvs167p-interacting proteins.
A second possibility was that Rvs167p was acting in opposition to Gyp5p and Gyl1p. In this case we might expect to see synthetic dosage lethality between GYP5 or GYL1 and SEC22 or RUD3. To test this we transformed cells with plasmids containing GYP5 and GYL1 with carboxy-terminal FLAG epitopes, under the control of the GAL promoter, and assayed for a phenotype upon overexpression of these genes in the various genetic backgrounds. In wild-type cells, overexpression of GYP5 had no effect on growth and overexpression of GYL1 was slightly toxic (Figure 5A, top). Because Gyp5p and Gyl1p interacted with each other, it was possible that any effect of overexpression of one would be limited by the concentration of the other. To address this possibility, we also tried co-overexpressing GYL1 and GYP5, which had a small inhibitory effect on growth of wild-type cells (Figure 5A). We next looked at the effects of co-overexpression of GYL1 and GYP5 in the absence of SEC22 or RUD3. In both a sec22Δ strain and a rud3Δ strain, co-overexpression of GYL1 and GYP5 was highly toxic (Figure 5A, middle). Overexpression of GYL1 alone inhibited growth somewhat, as seen in wild-type cells, but in a rud3Δ and a sec22Δ strain, maximal toxicity of GYL1 overexpression required GYP5 to be overexpressed as well (Figure 5A). These findings suggest that Gyl1p, together with its binding partner Gyp5p, acts in a manner antagonistic to that of Sec22p and Rud3p. Since RUD3 and SEC22 both have synthetic lethal interactions with RVS167, they are likely involved in a pathway parallel to RVS167 in vesicle trafficking from ER to Golgi. The finding that GYL1 and GYP5 co-overexpression is toxic in the absence of SEC22 and RUD3 suggests that RVS167 is also working antagonistically to GYL1 and GYP5. Rvs167p interacts directly with Gyl1p and Gyp5p, indicating that these proteins are in the same pathway. Our model predicts that even though Gyl1p and Gyp5p are acting in the opposite direction to Rvs167p in ER to Golgi trafficking, overexpression of GYL1 and GYP5 would not be toxic in an rvs167Δ strain because these genes are all in the same pathway. As predicted, overexpression of GYL1 and GYP5 was no more toxic in rvs167Δ cells than in wild type (Figure 5A, bottom), suggesting that the overexpression phenotype may require RVS167. We confirmed by Western blot that Gyl1p-Flag and Gyp5p-Flag were being produced in all the strains tested (Figure 5B). Overexpression of GYL1 and GYP5 had no effect on polarization of the actin cytoskeleton or on endocytosis in any of the strains assayed (data not shown).
Figure 5.—
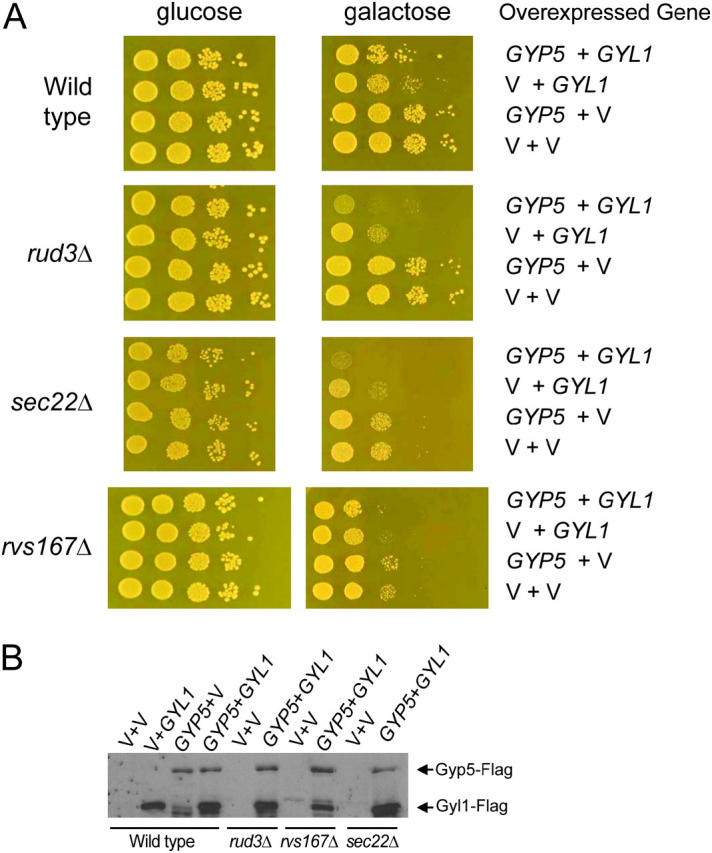
Toxicity of co-overexpression of GYL1 and GYP5 in strains compromised for ER to Golgi trafficking. (A) Tenfold serial dilutions of log-phase cultures of BY4741a (wild type), rud3Δ, sec22Δ, and rvs167Δ strains from the deletion consortium containing p426-GAL-GYL1-Flag, p415-GAL-GYP5-Flag, or vectors were spotted on synthetic glucose medium lacking uracil and leucine and on synthetic galactose medium lacking uracil and leucine. Plates were incubated at 30° for 3 days. Because rvs167Δ cells have a growth defect on galactose, 10-fold more of these cells were spotted in the first dilution than were spotted for the other strains. (B) Western blot showing Gyl1p-Flag and Gyp5p-Flag in strains shown in A. Strains (indicated below blot) containing plasmids (indicated above blot) were grown to log phase in synthetic medium lacking uracil and leucine and containing raffinose. Galactose was added to 2% and cells were grown for 3 hr. Extracts were made and analyzed by Western blot with α-Flag antibodies.
DISCUSSION
In this study we have identified two proteins that bind to the SH3 domain of Rvs167p: Gyp5p, a protein previously identified as a GAP for the Golgi GTPase Ypt1p, and Gyl1p, a protein that resembles Gyp5p. We show that Rvs167p can be co-immunoprecipitated with either Gyl1p or Gyp5p (Figure 1). Furthermore, Gyl1p and Gyp5p interact directly with each other, likely through their carboxy-terminal coiled-coil regions (Figure 3). In assays of GAP activity, Gyp5p had GAP activity toward Ypt1p, the Rab-type GTPase involved in ER to Golgi trafficking (as reported by De Antoni et al. 2002), and this activity was stimulated by the addition of Gyl1p. Gyl1p had no GAP activity toward Ypt1p (Figure 4). Genetic experiments suggest a role for Gyl1p and Gyp5p in ER to Golgi trafficking, consistent with their biochemical role.
Two-hybrid screens (Bon et al. 2000; Uetz et al. 2000; Tong et al. 2002; Talarek et al. 2005) and a large-scale co-immunoprecipitation study (Ho et al. 2002) have identified Gyp5p and Gyl1p/Ymr192p as proteins that interact with overproduced Rvs167p. The data from these high-throughput screens have not been confirmed, however, and likely contain a significant fraction of false positive interactions. We present here the first evidence that Rvs167p and Gyp5p as well as Rvs167p and Gyl1p interact under conditions where neither protein is overproduced.
The reported localization of these proteins is consistent with the interactions we describe here. Using immunofluorescence, Chesneau et al. (2004) found that Gyp5p and Gyl1p partially colocalize at the site of bud emergence, the bud tip, and the bud neck. Both Gyp5p-GFP and Gyl1p/Ymr192p-GFP localize to buds and bud necks as well as having a general cytoplasmic localization (Huh et al. 2003; Chesneau et al. 2004; Talarek et al. 2005; cytoplasmic localization also reported by De Antoni et al. 2002). The localization of Gyp5p and Gyl1p is consistent with sites enriched for Golgi compartments, near the bud site in G1 cells and at the site of septum formation around the time of cytokinesis (Preuss et al. 1992; Rossanese et al. 2001). Balguerie et al. (1999) found that Rvs167p-GFP localizes to sites of bud emergence and to the bud neck as well as to cortical actin patches. Thus, the reported localization of the three proteins overlaps. It is clear, however, that Rvs167p localizes to cortical actin patches whereas Gyp5p and Gyl1p do not (Chesneau et al. 2004); thus a significant fraction of the Rvs167p in the cell would not be able to bind to Gyp5p and Gyl1p.
Gyl1p is not an actin patch protein:
Deletion of GYL1 is of little phenotypic consequence. A similar pattern of protein-protein interactions from large-scale screens suggested that GYL1 might have a role similar to that of the actin patch protein APP1 (Samanta and Liang 2003). We found that deletion of GYL1 had little or no synergistic growth defect in combination with deletion of APP1. Furthermore Gyl1p-GFP was localized to bud and bud neck, as well as having a more general cytoplasmic localization, but was not found in cortical actin patches (Huh et al. 2003; Chesneau et al. 2004). In addition we have detected no defects in actin metabolism, endocytosis, or growth on salt in cells deleted for GYL1, GYP5, or both genes, suggesting that these genes are not required for a functional actin cytoskeleton.
We have shown that Gyp5p and Gyl1p interact directly with each other and that this interaction requires the C-terminal portion of Gyp5p, which contains a predicted coiled-coil region. Given that Gyp5p and Gyl1p interact with each other, the question arises as to why both proteins interact with Rvs167p. One possibility is that Gyp5p and Gyl1p form a dimer that can be recruited by Rvs167p binding to either Gyp5p or Gyl1p. In this case Rvs167p binding sites on both Gyp5p and Gyl1p would serve to increase the affinity of the Gyp5p-Gyl1p complex for Rvs167p and ensure that the relatively weak interaction with the Rvs167p SH3 domain is maintained. SH3 domains typically bind their ligands with Kd's in the micromolar range (for review see Mayer 2001). Another possibility is that Gyp5p and Gyl1p have distinct biological roles and only one ligand is bound by Rvs167p in a given cellular context. In a previous study we found that phosphorylation of Rvs167p by the cyclin-dependent kinase Pcl2p-Pho85p inhibits the interaction between Rvs167p and Gyl1p/Ymr192p in a Far Western assay (Friesen et al. 2003). Thus the interaction of the SH3 domain of Rvs167p with its ligands may be regulated, perhaps in a cell-cycle-dependent manner.
Primary structure of Gyl1p:
Gyl1p and Gyp5p have 29% identity and 51% similarity over the entire length of Gyl1p. Although the GAP domain of Gyp5p is 35% identical (and 54% similar) to the analogous region in Gyl1p, two of the three absolutely conserved “GYP fingerprint” motifs found in Gyp5p and other Ypt GAPs are not found in Gyl1p. From a sequence comparison of Ymr192p/Gyl1p with several known yeast Ypt GAPs, Talarek et al. (2005) suggested that R354 could be the catalytic arginine of Gyl1p. After a careful examination of its predicted primary and secondary structure (Figure 2B), we conclude that this conserved motif is not present in Gyl1p. Although Gyl1p is likely to have the same overall structure as Gyp5p and other Gyp's, differences in active site residues suggest that Gyl1p is likely to have different activity. Under standard assay conditions we saw no GAP activity by Gyl1p toward Ypt1p, although Gyl1p was able to stimulate the GAP activity of Gyp5p (Figure 4D).
Genetics of GYL1 and GYP5:
Deletion of both GYP5 and GYL1 results in no obvious phenotype. Genes encoding eight Ypt GAPs have been identified in yeast and in most cases deletion of one or more of them has no effect on growth (Bi et al. 2000; Rak et al. 2000; De Antoni et al. 2002). Genetic evidence supports the biochemical finding that Gyp5p has a role in ER to Golgi vesicle trafficking: deletion of GYP5 in a protease-deficient strain expressing a GTPase-deficient version of YPT1 (Ypt1Q67L) leads to a cold-sensitive phenotype (De Antoni et al. 2002). However, a fraction of Gyp5p is able to co-immunoprecipitate with Sec4p and a fraction of Gyp5p is present in post-Golgi vesicles at the plasma membrane (Chesneau et al. 2004). These results suggest that Gyp5p may have roles both in ER to Golgi trafficking, acting on Ypt1p, and in exocytosis, acting on Sec4p. De Antoni et al. (2002) found that a truncated version of Gyp5p stimulated GTP hydrolysis of Ypt1p 150-fold but stimulated Sec4p only 24-fold. We have found that full-length Gyp5p has only very weak GAP activity on Sec4p compared to Ypt1p in vitro (Figure 4, A and B). Furthermore, we have found that co-overexpression of GYL1 and GYP5 is severely toxic in two genetic backgrounds compromised for ER to Golgi trafficking: sec22Δ and rud3Δ (Figure 5A). This genetic result provides evidence that the Gyp5p-Gyl1p interaction with Ypt1p that we detect biochemically is biologically relevant. We see no significant effect of Rvs167p-161p on GAP activity of Gyp5p (Figure 4). Gyp5p-GFP has been localized to the bud tip and bud neck and this localization is dependent on the presence of the SH3 domain of Rvs167p (Talarek et al. 2005). This suggests that the interaction with Rvs167p may be responsible for localization or stability of Gyp5p. Since we could detect Gyp5p-Flag in our rvs167Δ strain (Figure 5B), it seems likely that Gyp5p stability is not affected. However, if Gyp5p requires Rvs167p for proper localization, this could explain why overproduction of Gyl1p and Gyp5p was not toxic in an rvs167Δ strain.
A connection between vesicle trafficking and a cortical actin patch protein:
Because Gyp5p and Gyl1p interact with the SH3 domain of Rvs167p, there must be a biological connection between these proteins. One possibility is that the interaction may reveal a link between cortical actin patches and the Golgi. Mulholland et al. (1997) proposed that the actin cytoskeleton and, indeed, actin patches are required for some specific step in the latter part of the secretory pathway, in trafficking from late Golgi to the plasma membrane. How this requirement for the actin cytoskeleton might work is not clear.
An alternative explanation for why the cortical actin patch protein Rvs167p physically interacts with proteins involved in ER to Golgi vesicle trafficking is that the Rvs proteins have a general role in vesicle biogenesis or fusion. The structure of Drosophila amphiphysin, a crescent-shaped dimer that binds preferentially to highly curved negatively charged membranes, suggests a role for BAR domain proteins in generating the membrane-bending events during vesicle formation or fusion. Rvs167p and Rvs161p, which form a heterodimer, are the only BAR domain-containing proteins found in S. cerevisiae according to the Simple Modular Architecture Research Tool SMART (Letunic et al. 2004). Several pieces of evidence point to a role for the Rvs proteins in different types of vesicle trafficking. First, Rvs167p and Rvs161p have a well-defined role in endocytosis and have been shown to localize to cortical actin patches, which are thought to be sites of endocytosis. Second, in this article we provide physical and genetic evidence that Rvs167p has a role in vesicle trafficking from ER to Golgi. In addition to this work, two large-scale screens have suggested physical interaction between Rvs167p-Rvs161p and the COPI vesicle coat involved in retrograde trafficking from Golgi to ER: a two-hybrid interaction with SEC21 (Bon et al. 2000) and an affinity precipitation interaction with Sec27p (Ho et al. 2002). Third, RVS167 and RVS161 have synthetic lethal interactions with VPS21 (Singer-Kruger and Ferro-Novick 1997), a gene encoding the Rab-type GTPase involved in vesicle trafficking between the early and late endosome and to the vacuole (Prescianotto-Baschong and Riezman 2002). Fourth, Bon et al. (2000) observed two-hybrid interactions between RVS167 and both SEC8 and EXO70, two genes encoding components of the exocyst complex, which is required for exocytosis. In addition, rvs167 and rvs161 mutants accumulate secretory vesicles (Breton et al. 2001). Thus, besides the well-known role for Rvs167p-Rvs161p in endocytosis, we have provided evidence for a role in trafficking from ER to Golgi; other large-scale screens have found a role in trafficking from early to late endosome and in secretion, suggesting a role for Rvs167p-Rvs161p in vesicle trafficking in many different cellular compartments. Since Rvs167p has been reported to bind to actin in a two-hybrid assay (Amberg et al. 1995; Lombardi and Riezman 2001), this proposed general role in vesicle trafficking might require actin. We suggest that proteins binding to the SH3 domain of Rvs167p, such as Gyp5p and Gyl1p, may help to direct the vesicle trafficking machinery to sites of vesicle fusion.
Acknowledgments
We thank Peter Novick for plasmids and protocols used in GAP assays. We are grateful to Arianna Rath and Alan Davidson for constructing the pAR100-RVS-SH3 plasmids and to the lab of Mike Tyers for pGAL-GYP5-Flag and for constructing earlier versions of pGAL-GYL1-Flag. We thank Jon Millman for helpful discussions and advice and Christine Humphries and Dongqing Huang for critical reading of the manuscript. This work was supported by an operating grant from the National Cancer Institute of Canada with funds from the Canadian Cancer Society. Our Synthetic Genetic Array laboratories are supported by a Collaborative Genomics Special Project grant from the Canadian Institutes of Health Research and by funds from Genome Canada through the Ontario Genomics Institute.
References
- Albert, S., and D. Gallwitz, 1999. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J. Biol. Chem. 274: 33186–33189. [DOI] [PubMed] [Google Scholar]
- Albert, S., and D. Gallwitz, 2000. Msb4p, a protein involved in Cdc42-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol. Chem. 381: 453–456. [DOI] [PubMed] [Google Scholar]
- Albert, S., E. Will and D. Gallwitz, 1999. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 18: 5216–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D. C., E. Basart and D. Botstein, 1995. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2: 28–35. [DOI] [PubMed] [Google Scholar]
- Ausubel, R., R. Brent, R. Kingston, D. Moore, J. Seidman et al., 1994 Current Protocols in Molecular Biology, pp. 3.01–3.19.7. John Wiley & Sons, New York.
- Bacon, R. A., A. Salminen, H. Ruohola, P. Novick and S. Ferro-Novick, 1989. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J. Cell Biol. 109: 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balguerie, A., P. Sivadon, M. Bonneu and M. Aigle, 1999. Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J. Cell Sci. 112: 2529–2537. [DOI] [PubMed] [Google Scholar]
- Bauer, F., M. Urdaci, M. Aigle and M. Crouzet, 1993. Alteration of a yeast SH3 protein leads to conditional viability and defects in cytoskeletal and budding patterns. Mol. Cell. Biol. 13: 5070–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari, P., and J. Gowrishankar, 1997. An Escherichia coli host strain useful for efficient overproduction of cloned gene products with NaCl as the inducer. J. Bacteriol. 179: 4403–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E., J. B. Chiavetta, H. Chen, G. C. Chen, C. S. Chan et al., 2000. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell 11: 773–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon, E., P. Recordon-Navarro, P. Durrens, M. Iwase, A. Toh-E et al., 2000. A network of proteins around Rvs167p and Rvs161p, two proteins related to the yeast actin cytoskeleton. Yeast 13: 1229–1241. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J. S., and B. S. Glick, 2004. The mechanisms of vesicle budding and fusion. Cell 116: 153–166. [DOI] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Breton, A. M., J. Schaeffer and M. Aigle, 2001. The yeast Rvs161 and Rvs167 proteins are involved in secretory vesicles targeting the plasma membrane and in cell integrity. Yeast 18: 1053–1068. [DOI] [PubMed] [Google Scholar]
- Chesneau, L., S. Dupre, A. Burdina, J. Roger, S. LePanse et al., 2004. Gyp5p and Gyl1p are involved in the control of polarized exocytosis in budding yeast. J. Cell Sci. 117: 4757–4767. [DOI] [PubMed] [Google Scholar]
- Colwill, K., D. Field, L. Moore, J. Friesen and B. Andrews, 1999. In vivo analysis of the domains of yeast Rvs167p suggests Rvs167p function is mediated through multiple protein interactions. Genetics 152: 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff, J. A., and G. J. Barton, 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40: 502–511. [DOI] [PubMed] [Google Scholar]
- De Antoni, A., J. Schmitzova, H. H. Trepte, D. Gallwitz and S. Albert, 2002. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J. Biol. Chem. 277: 41023–41031. [DOI] [PubMed] [Google Scholar]
- Drees, B. L., B. Sundin, E. Brazeau, J. P. Caviston, G. C. Chen et al., 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154: 549–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L.-L., R. N. Collins and P. J. Novick, 1998. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J. Biol. Chem. 273: 3253–3256. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E. Y., and D. G. Drubin, 2003. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19: 287–332. [DOI] [PubMed] [Google Scholar]
- Figeys, D., L. D. McBroom and M. F. Moran, 2001. Mass spectrometry for the study of protein-protein interactions. Methods 24: 230–239. [DOI] [PubMed] [Google Scholar]
- Friesen, H., K. Murphy, A. Breitkreutz, M. Tyers and B. Andrews, 2003. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Mol. Biol. Cell 7: 3027–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson, H. V., B. L. Anderson, H. M. Warrick, L. A. Pon and J. A. Spudich, 1996. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 133: 1277–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichet, A., J. W. R. Copeland, M. Erdelyi, D. Hlousek, P. Zavorsky et al., 1997. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature 385: 548–552. [DOI] [PubMed] [Google Scholar]
- Ho, Y., S. Mason, R. Kobayashi, M. Hoekstra and B. Andrews, 1997. Role of the casein kinase I isoform, HRR25, and the cell cycle regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94: 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore et al., 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori et al., 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen, M., Y. Sun and D. G. Drubin, 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115: 475–487. [DOI] [PubMed] [Google Scholar]
- Karlin, S., and S. F. Altschul, 1990. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. USA 87: 2264–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin, S., and S. F. Altschul, 1993. Applications and statistics for multiple high-scoring segments in molecular sequences. Proc. Natl. Acad. Sci. USA 90: 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., B. W. Birren and E. S. Lander, 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- Kim, D. W., M. Sacher, A. Scarpa, A. M. Quinn and S. Ferro-Novick, 1999. High-copy suppressor analysis reveals a physical interaction between Sec34p and Sec35p, a protein implicated in vesicle docking. Mol. Biol. Cell 10: 3317–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, T., M. Gotte and D. Gallwitz, 1997. Vesicular transport: How many Ypt/Rab GTPases make a eukaryotic cell? Trends Biochem. Sci. 22: 468–472. [DOI] [PubMed] [Google Scholar]
- Lee, J., K. Colwill, V. Aneliunas, C. Tennyson, L. Moore et al., 1998. Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr. Biol. 8: 1310–1321. [DOI] [PubMed] [Google Scholar]
- Lee, M. C., and R. Schekman, 2004. Cell biology. BAR domains go on a bender. Science 303: 479–480. [DOI] [PubMed] [Google Scholar]
- Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks et al., 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32: D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, J. P., and S. Ferro-Novick, 1993. Bos1p, an integral membrane protein of the endoplasmic reticulum to Golgi transport vesicles, is required for their fusion competence. Cell 73: 735–745. [DOI] [PubMed] [Google Scholar]
- Lila, T., and D. Drubin, 1997. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell 8: 367–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi, R., and H. Riezman, 2001. Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J. Biol. Chem. 276: 6016–6022. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lupas, A., M. Van Dyke and J. Stock, 1991. Predicting coiled coils from protein sequences. Science 252: 1162–1164. [DOI] [PubMed] [Google Scholar]
- Maxwell, K. L., and A. R. Davidson, 1998. Mutagenesis of a buried polar interaction in an SH3 domain: sequence conservation provides the best prediction of stability effects. Biochemistry 37: 16172–16182. [DOI] [PubMed] [Google Scholar]
- Mayer, B. J., 2001. SH3 domains: complexity in moderation. J. Cell Sci. 114: 1253–1263. [DOI] [PubMed] [Google Scholar]
- Measday, V., L. Moore, J. Ogas, M. Tyers and B. Andrews, 1994. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science 266: 1391–1395. [DOI] [PubMed] [Google Scholar]
- Measday, V., L. Moore, R. Retnakaran, J. Lee, M. Donoviel et al., 1997. A family of cyclin-like proteins that interact with the cyclin-dependent kinase, Pho85. Mol. Cell. Biol. 17: 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland, J., A. Wesp, H. Riezman and D. Botstein, 1997. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol. Biol. Cell 8: 1481–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn, A., 2001. Molecular requirements for the internalisation step of endocytosis: insights from yeast. Biochim. Biophys. Acta 1535: 236–257. [DOI] [PubMed] [Google Scholar]
- Navarro, P., P. Durrens and M. Aigle, 1997. Protein-protein interaction between the RVS161 and RVS167 gene products of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1343: 187–192. [DOI] [PubMed] [Google Scholar]
- Neuwald, A. F., 1997. A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase activators. Trends Biochem. Sci. 22: 22–23. [DOI] [PubMed] [Google Scholar]
- Novick, P., and D. Botstein, 1985. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell 40: 405–416. [DOI] [PubMed] [Google Scholar]
- Parlati, F., J. A. McNew, R. Fukuda, R. Miller, T. H. Sollner et al., 2000. Topological restriction of SNARE-dependent membrane fusion. Nature 407: 194–198. [DOI] [PubMed] [Google Scholar]
- Pawson, T., and J. D. Scott, 1997. Signaling through scaffold, anchoring, and adaptor proteins. Science 278: 2075–2080. [DOI] [PubMed] [Google Scholar]
- Peter, B. J., H. Kent, I. G. Mills, Y. Vallis, P. J. Butler et al., 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499. [DOI] [PubMed] [Google Scholar]
- Prescianotto-Baschong, C., and H. Riezman, 2002. Ordering of compartments in the yeast endocytic pathway. Traffic 3: 37–49. [DOI] [PubMed] [Google Scholar]
- Preuss, D., J. Mulholland, A. Franzusoff, N. Segev and D. Botstein, 1992. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol. Biol. Cell 3: 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., and A. Bretscher, 2000. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 113: 571–585. [DOI] [PubMed] [Google Scholar]
- Rak, A., R. Fedorov, K. Alexandrov, S. Albert, R. S. Goody et al., 2000. Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins. EMBO J. 19: 5105–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronicke, V., W. Graulich, D. Mumberg, R. Muller and M. Funk, 1997. Use of conditional promoters for expression of heterologous proteins in Saccharomyces cerevisiae. Methods Enzymol. 283: 313–322. [DOI] [PubMed] [Google Scholar]
- Rossanese, O. W., C. A. Reinke, B. J. Bevis, A. T. Hammond, I. B. Sears et al., 2001. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell. Biol. 153: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta, M. P., and S. Liang, 2003. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. USA 100: 12579–12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev, N., 1991. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science 252: 1553–1556. [DOI] [PubMed] [Google Scholar]
- Segev, N., 2001. Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13: 500–511. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., M. Wilm, O. Vorm and M. Mann, 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger, B., and S. Ferro-Novick, 1997. Use of a synthetic lethal screen to identify yeast mutants impaired in endocytosis, vacuolar protein sorting and the organization of the cytoskeleton. Eur. J. Cell Biol. 74: 365–375. [PubMed] [Google Scholar]
- Sivadon, P., M. Crouzet and M. Aigle, 1997. Functional assessment of the yeast Rvs161 and Rvs167 protein domains. FEBS Lett. 417: 21–27. [DOI] [PubMed] [Google Scholar]
- Talarek, N., A. Balguerie, M. Aigle and P. Durrens, 2005. A novel link between a Rab GTPase and Rvs proteins: the yeast amphiphysin homologues. Cell Biochem. Funct. 23 (in press). [DOI] [PubMed]
- Tolliday, N., N. Bouquin and R. Li, 2001. Assembly and regulation of the cytokinetic apparatus in budding yeast. Curr. Opin. Microbiol. 4: 690–695. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Tong, A. H. Y., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tong, A. H. Y., B. Drees, G. Nardelli, G. D. Bader, B. Brannetti et al., 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295: 321–324. [DOI] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T. Mansfield, R. S. Judson et al., 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Winzeler, E., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Zhang, B., and A. C. Zelhof, 2002. Amphiphysins: raising the BAR for synaptic vesicle recycling and membrane dynamics. Bin-Amphiphysin-Rvsp. Traffic 3: 452–460. [DOI] [PubMed] [Google Scholar]