Abstract
We used quantitative trait loci (QTL) mapping to evaluate the age specificity of naturally segregating alleles affecting life span. Estimates of age-specific mortality rates were obtained from observing 51,778 mated males and females from a panel of 144 recombinant inbred lines (RILs). Twenty-five QTL were found, having 80 significant effects on life span and weekly mortality rates. Generation of RILs from heterozygous parents enabled us to contrast effects of QTL alleles with the means of RIL populations. Most of the low-frequency alleles increased mortality, especially at younger ages. Two QTL had negatively correlated effects on mortality at different ages, while the remainder were positively correlated. Chromosomal positions of QTL were roughly concordant with estimates from other mapping populations. Our findings are broadly consistent with a mix of transient deleterious mutations and a few polymorphisms maintained by balancing selection, which together contribute to standing genetic variation in life span.
ELUCIDATION of key metabolic pathways in age-dependent physiological deterioration will have great medical benefit. Recent discoveries of genes that extend longevity in model species suggest a remarkable degree of conservation between taxa (Partridge and Gems 2002). In Drosophila, the insulin signaling pathway and intermediary metabolism; response to oxidative, heat, and other stresses; biogenic amine synthesis; steroid hormones; membrane and mitochondrial function; DNA repair and replication; and telomere length have all been implicated (see Geiger-Thornsberry and Mackay 2004 for a review). The next significant challenge will be to identify mutations that increase longevity not only in the lab-specific genotypes and environments, but also in natural populations (Spencer et al. 2003).
Most populations harbor substantial genetic variation for life span, with heritability estimates ranging from 0.1 to 0.3 (Curtsinger et al. 1995; Mackay 2002). Mutations with adverse effects on mortality—either unconditionally deleterious (Medawar 1952) or under balancing selection (Williams 1957)—contribute to the heritable variation. Which genes are responsible for this variation? Two approaches have been taken to answer the question. Geiger-Thornsberry and Mackay (2004) used quantitative complementation tests to establish that 5 of 16 a priori selected genes that regulate longevity in various lab populations also contribute to naturally occurring variation in life span. This study nicely bridges the gap between molecular genetics—focusing on strong phenotypes with little evolutionary relevance—and evolutionary biology, which is sometimes guilty of disregarding molecular mechanisms (Flatt 2004).
Another approach is to map quantitative trait loci (QTL) affecting life span of different Drosophila populations, including standard lines (Nuzhdin et al. 1997) and populations selected for long life via delayed reproduction (Curtsinger et al. 1998; Resler et al. 1998; Curtsinger and Khazaeli 2002; Forbes et al. 2004; Khazaeli et al. 2004; Valenzuela et al. 2004), and then compare QTL positions with those of candidate genes. Life span QTL mapped by Mackay's lab are close to metabolic enzymes Adh and Pgm, the insulin degradation metalloproteinase gene Ide, proteins essential for protein synthesis EF1α, and genes involved in the elimination of reactive oxygen species Sod and Cat (Leips and Mackay 2002). The Sod/Cat region also turns up in studies from the Curtsinger lab, although there is no evidence for differences in Sod activity (Curtsinger and Khazaeli 2002). Low xanthine dehydrogenase (lxd) also maps nearby and potentially contributes to life span differences (Tahoe et al. 2002).
More refined deficiency and linkage disequilibrium mapping have established Dopa decarboxylase (Ddc) as a strong candidate modifier of life span (Pasyukova et al. 2000; DeLuca et al. 2003). Remarkably, DeLuca et al. (2003) were able to estimate contributions to life span of natural alleles in, or immediately adjacent to, this locus. No association was found between the frequency of haplotypes and their effects on life span. This observation and the pattern of DNA polymorphisms in this locus pointed to balancing selection maintaining variation. Repeating the study in different populations could determine whether the longevity modification by Ddc is common.
The average life span characteristic of a particular genotype is, in one sense, merely a sum of its mortality trajectory across ages. However, there is a fundamental difference between mean life spans and age-specific mortality rates: the former give no information about the timing of genetic effects. Two populations could have identical mean life spans but exhibit very different age-specific mortality rates. The distinction is important, because the evolutionary theory of senescence is entirely dependent upon the assumption of genetic variants that have age-specific effects on survival (Medawar 1952; Williams 1957; Charlesworth 1994). Age-specific mortality rates are also used by gerontologists and demographers to quantify senescence in populations, compare species survival patterns, and infer species-specific life span limits (Carey 2002). However, information about age-specific effects of natural genetic variants and spontaneous mutations is very limited (reviewed by Curtsinger et al. 2005).
To date, only one study in Drosophila has documented age-specific effects of segregating alleles that modify life span. Curtsinger and Khazaeli (2002) found evidence for QTL that alter weekly mortality rates in a population of recombinant inbred lines (RILs) and also have positive pleiotropic association with midlife fertility and resistance to oxidative stress. The former effects could be by-products of the selection regime used to establish long-lived flies, as they were selected for the ability to survive to old age and then reproduce (Curtsinger and Khazaeli 2002). The age-specific properties of mortality alleles segregating in natural populations are not known.
It might be possible to use quantitative complementation methods to fill the gap, but a problem looms. One would produce two genotypes for each of two compared alleles—a heterozygote with the null (or deletion) and a heterozygote with the wild type (usually on a balancer chromosome; Mackay and Fry 1996). Significance of genotype-by-allele interaction would indicate a lack of complementation (Pasyukova et al. 2000). While this approach could be conducted across ages to address age-specific mortality, such a test would be cumbersome. There are reasons to believe that the dominance of mortality alleles changes with age (see the review by Promislow and Pletcher 2002), which would directly affect the interaction term. Further, the deletion of a life span regulator could overwhelm the effect being studied. Testing for possibly small age-specific mortality effects in a genetic background of strong-effect deletions is probably not desirable. Here we employ a somewhat less powerful approach, but one that avoids the deletion problem. We use QTL mapping in RILs derived from flies caught in nature (Kopp et al. 2003). We have previously mapped QTL affecting mean life span and stress resistance in these RILs (Wang et al. 2004). Here, we examine survival of 51,778 flies to evaluate the age specificity of QTL effects.
MATERIALS AND METHODS
RILs:
A panel of 144 RILs was generated by crossing a single virgin female from the F1 progeny of a fertilized female caught in the wild (Winters, CA) to a single male from the F1 progeny of a different fertilized female caught at the same location. The genetic crosses employed and methods for genotyping the lines are described in detail by Kopp et al. (2003). Briefly, recombinant F2 genomes were isogenized by 25 generations of full-sib inbreeding. We assayed positions of roo transposable elements by in situ hybridization to polytene salivary gland chromosomes with a biotinylated DNA probe (Shrimpton et al. 1986). Hybridization was detected using the Elite Vectastain ABC kit (Vector Laboratories, Burlingame, CA) and visualized with diaminobenzidine. Locations were determined at the level of cytological bands on the standard Bridges' map in five individuals per line. A marker was recorded as present if detected in all larvae, absent if not detected in any larva, and otherwise segregating. Parental haplotypes for marker alleles were inferred from residual linkage disequilibrium in the population of RILs (Proc CORR, SAS Institute 1988). In some cases, roo elements either were uninformative (present in more than one complementation group) or gave a hybridization signal that was too weak to be scored reliably. One hundred twenty-three markers were retained for the analysis, and recombination distances between them were calculated as in Mezey et al. (2005). Two of the founding third chromosomes appeared to be identical except for chromosome tips (Kopp et al. 2003); thus we assumed three parental haplotypes for the X chromosome (marked by the presence of roo in the following positions: 1C, 1D, 3A, 4C, 5C, 9C, 11F, 12C, 14B, 17E, and 19AB; 1E, 4B, 5A, 6E, 7F, 8D, 9A, and 16F; 5D, 11CD, and 13A); four haplotypes for the second chromosome (21C, 22C, 33F, 48D, 53E, 55A, 55D, and 58D; 22A, 23F, 30D, 31F, 36F, 37C, 49F, 51A, 54A, 54C, 55C, 57B, and 59D; 22B, 24CD, 25D, 28C, 33B, 38A, 41CE, 41F, 42B, 47E, and 48A; 30B, 34EF, 37B, 38CD, 44C, 49B, 50C, and 53A); and three haplotypes of the third chromosome (61C, 63F, 64D, 64E, 66A, 66B, 67D, 67F, 69A, 70C, 76B, 79A, 79C, 79F, 83D, 84DE, 85D, 86D, 87A, 87B, 87E, 87F, 89AB, 94D, 96C, 97B, and 100C; 63E, 65AB, 67C, 73C, 76C, 77B, 77D, 82C, 84F, 85E, 86B, 88E, 89EF, 90EF, and 92E; 62EF, 64B, 64C, 65C, 65E, 73D, 77E, 78E, 83F, 86C, 86E, 88C, 90D, 93B, 94B, 98F, and 100B). Note that some of markers reported by Kopp et al. (2003) were dropped from our current analysis as estimates for recombination distances between them and nearby markers were inconsistent. The fourth chromosome was marked by 102EF. This marker was not significantly associated with any of the traits analyzed here.
Life span measurements:
Life span measurements, all of which were done at the Minnesota laboratory, were obtained in two experimental blocks. In the first block, every RIL was expanded in half-pint bottles under controlled larval density using the method of Fukui et al. (1993). Adult flies of both sexes emerging within 24 hr were collected and numbers were estimated by weight using an electronic balance (Mettler, no. BB2400). For each RI line ∼225 flies of both sexes emerging from a single 24-hr cohort were transferred into a single 3.8-liter population cage specially designed for survival studies (Fukui and Kirshner 1993; Promislow et al. 1996). A total of 31,315 flies were studied. In conjunction with ongoing studies of the relationship between life span and metabolic rates of individual flies (Van Voorhies et al. 2003, 2004a,b; Khazaeli et al. 2005), additional survival data are also available for some of the lines studied in block 1. Because it is not feasible to study metabolism of individual flies at multiple ages from each of 144 lines, with replication, we chose 50 lines at random from the top, middle, and bottom deciles of the life span distribution observed in block 1, thereby reducing the number of lines studied while preserving the full range of variation. Experimental material was expanded and collected as described above and then placed in one (16 lines) or two (34 lines) population cages for survival measurements. A total of 20,463 flies were studied.
Mixed-sex population cages were assigned to random locations in a walk-in incubator maintained at 24°, with constant illumination and 60–70% humidity. The mouth of each cage was covered with fine mosquito netting. The cages were inverted over an 11-cm diameter disk of cooked medium, which was replaced every other day. Flies were transferred, without anesthesia, to clean population cages every 10 days. Dead flies were removed by suction, sexed, and recorded daily until the last death. All RILs were measured contemporaneously in each experimental block.
Survival and variance analyses:
Survival analyses, including estimation of mortality and hazard functions, were implemented with SAS procedures LIFETEST (option “lifetable”) and LIFEREG, with no censoring, and with the sexes treated separately. For the analysis of variance, we first log-transformed the data for normality (tested with procedure UNIVARIATE; SAS Institute 1988, option “normal”). Two-way ANOVAs were then performed with effects due to sex (fixed), line (random), and line × sex interaction (random). Statistical significance of components of variance was assayed with SAS procedure GLM. Genetic correlations between traits and sexes were approximated as correlations between line means and estimated with the SAS procedure CORR.
Single-marker survival analysis:
For each marker locus, the population of RILs was stratified into two groups, one with the marker allele containing the roo insert, and the other having no roo insert at that particular chromosomal location. Differences in survival between groups were tested in each sex using log-rank and Wilcoxon tests (SAS procedure LIFETEST). The log-rank test places more weight on long-term survival, while Wicoxon is more sensitive to early survival. In addition to these nonparametric tests, we analyzed the data assuming a Weibull distribution (SAS procedure LIFEREG).
The structure of the data is not in exact accordance with the assumptions underlying survival analyses. In particular, survival analysis typically assumes independence of every observation, but in our study multiple flies were confined in population cages, and the survival of cage mates may not be independent. Since cages are organized by genotypes, this potentially inflates the significance of associations between survival and marker alleles. To overcome this problem, we estimated an empirical distribution of the test statistics under the null hypothesis (no association between any of the markers and survival patterns). This was done by randomly permuting genotypes among cages 1000 times. For each permutation, we calculated probability for marker-survival associations across all markers and retained the most significant. Statistics from the original data are significant at P = 0.05 and were exceeded <50 times by those most significant associations from the permutations (Doerge et al. 1997).
Composite interval mapping:
Our study makes use of wild, outbred founder flies. There are three X and third chromosome and four second chromosome parental haplotypes segregating among RILs. We adapted standard QTL-mapping software that was designed for homozygous founders to analyze our data (see Wang et al. 2004 for a detailed explanation). Briefly, we test whether an allele from one haplotype encodes a trait value significantly different from the average trait due to the combined effects of other haplotypes. The number of different allelic effects that we can independently estimate is equal to the number of segregating haplotypes minus one. Using haplotype-specific marker alleles, we generated a separate likelihood profile with the composite interval mapping (CIM) procedure in QTL Cartographer (Basten et al. 1999). Options in the CIM module were set to two background parameters, window size of 30 cM, and Kosambi mapping function. Significance thresholds were determined by 1000 permutations for each trait and chromosome.
RESULTS
We measured complete adult life spans of 51,778 Drosophila melanogaster of known relatedness to assess quantitative trait loci affecting age-specific survival. Summary statistics are shown in Table 1. There were in excess of 100 flies per RIL per sex in the first experimental block and ∼200 flies per line per sex in the second block (Table 1). Maximum individual life spans were 104 days for males and 81 days for females.
TABLE 1.
Descriptive statistics for adult life span
| Block 1
|
Block 2
|
|||
|---|---|---|---|---|
| Males | Females | Males | Females | |
| No. of assayed linesa | 140 | 140 | 50 | 50 |
| Mean (days) | 35.0 | 27.1 | 30.0 | 23.6 |
| Standard deviation (days) | 8.0 | 7.5 | 7.2 | 5.9 |
| Minimum (days) | 15.8 | 10.5 | 15.6 | 10.4 |
| Maximum (days) | 64.6 | 43.9 | 48.9 | 36.3 |
| No. of flies | 14,979 | 16,336 | 10,290 | 10,173 |
The numbers of lines with assayed genotypes were 128 and 48; correspondingly, only those lines were retained for subsequent analyses.
Sex and age effects on survival:
As shown in Table 1, the mean life span of females was lower than that of males by ∼13% (P < 0.0001), as is typical in mixed-sex populations (Curtsinger et al. 1998; Resler et al. 1998; Khazaeli and Curtsinger 2000; Curtsinger and Khazaeli 2002; Reiwitch and Nuzhdin 2002). Age-specific survivorship and mortality rates for data pooled over lines for block 1 are shown in Figure 1 (data are similar for block 2; not shown). Males exhibit higher survivorship and lower mortality than females at all ages up to ∼50 days after emergence. As observed in previous studies of Drosophila and other organisms (Curtsinger et al. 1992, 2005; Fukui et al. 1993, 1996; Promislow et al. 1996; Pletcher and Curtsinger 1998; Vaupel et al. 1998), age-specific mortality rates increase approximately exponentially early in adult life and then decelerate at later ages (Figure 1B).
Figure 1.—

Survival (A) and mortality (B) curves for males (blue) and females (red) in block 1.
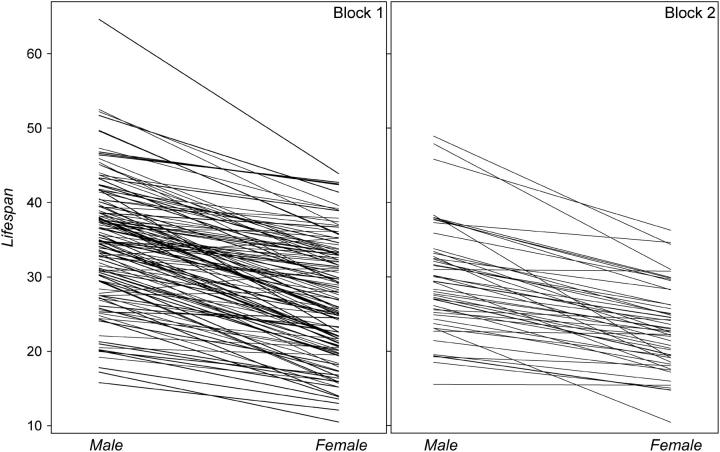
Line mean longevities for males and females varied more than twofold between genotypes (Figure 2) and were largely concordant between sexes and blocks. Both line and sex components of variance were highly significant (P < 0.0001, Table 2). As we (Curtsinger and Khazaeli 2002; Reiwitch and Nuzhdin 2002) previously observed for mated flies, the sex × line interaction term was not significantly different from zero (Table 2). We have also redone the analyses with the procedure MIXED (SAS Institute 1988) that treats random and fixed effects in a more sensible way than the procedure GLM. We used the model with sex (fixed), line, and line × sex (random). Sex was a significant effect as before (F = 195.06, P < 0.0001). Comparing the log likelihoods of the full and reduced models—with line and line × sex effects omitted—established significance of the former term 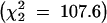 , but not of the latter.
, but not of the latter.
Figure 2.—
Mean line life span in males and females of blocks 1 and 2.
TABLE 2.
Analysis of variance in life span
| Source | d.f. | Mean square | F-value | Significance |
|---|---|---|---|---|
| Line | 128 | 0.0291 | 9.20 | <0.0001 |
| Sex | 1 | 1.1444 | 291.20 | <0.0001 |
| Sex × line | 128 | 0.0032 | 0.30 | 1.0000 |
| Error | 94 | 0.0106 |
The correlation between line means for male and female life spans was estimated as 0.81 within block 1 (P < 0.0001; Table 3) and 0.82 within block 2 (P < 0.0001). The high correlation of life spans in males and females within experimental blocks is inflated by shared microenvironmental variations, since males and females of each line share the same population cage. This problem can be overcome by examining correlations between the sexes among blocks. Line means for males in block 1 are significantly correlated with line means for females of the same genotype in block 2 (r = 0.50, P < 0.001), and the reciprocal correlation is also significant (r = 0.39, P < 0.01). Note that the phenotypic correlation across blocks estimates the genetic correlation of life span between the two sexes.
TABLE 3.
Correlations between life span and age-specific mortality
| Tr.a | Lm | m1 | m2 | m3 | m4 | M5 | m6 | m7 | m8 | m9 | Lf | f1 | f2 | f3 | f4 | f5 | f6 | f7 | f8 | f9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lm | — | −0.260 | −0.606 | −0.711 | −0.818 | −0.835 | −0.716 | −0.618 | −0.478 | −0.416 | 0.809 | −0.367 | −0.554 | −0.711 | −0.601 | −0.559 | −0.395 | 0.062 | −0.346 | −0.528 |
| m1 | 0.048 | — | 0.404 | 0.339 | 0.212 | 0.168 | 0.104 | 0.072 | −0.133 | 0.030 | −0.253 | 0.469 | 0.291 | 0.293 | 0.054 | −0.006 | −0.123 | 0.132 | 0.428 | 0.785 |
| m2 | <10−4 | 0.004 | — | 0.648 | 0.596 | 0.546 | 0.193 | 0.077 | 0.000 | 0.348 | −0.601 | 0.406 | 0.712 | 0.644 | 0.424 | 0.222 | −0.174 | −0.057 | −0.161 | −0.780 |
| m3 | <10−4 | 0.012 | <10−4 | — | 0.762 | 0.608 | 0.348 | 0.150 | −0.152 | −0.222 | −0.720 | 0.340 | 0.698 | 0.820 | 0.543 | 0.233 | −0.040 | −0.227 | −0.340 | −0.125 |
| m4 | <10−4 | NS | <10−4 | <10−4 | — | 0.802 | 0.505 | 0.288 | 0.132 | −0.024 | 0.755 | 0.337 | 0.605 | 0.758 | 0.698 | 0.472 | 0.051 | −0.257 | −0.107 | −0.147 |
| m5 | <10−4 | NS | <10−4 | <10−4 | <10−4 | — | 0.690 | 0.524 | 0.230 | 0.077 | −0.702 | 0.252 | 0.482 | 0.669 | 0.618 | 0.624 | 0.361 | −0.103 | −0.064 | 0.130 |
| m6 | <10−4 | NS | NS | 0.0003 | <10−4 | <10−4 | — | 0.611 | 0.423 | 0.361 | −0.506 | 0.192 | 0.148 | 0.415 | 0.438 | 0.508 | 0.633 | 0.056 | 0.245 | 0.319 |
| m7 | <10−4 | NS | NS | NS | 0.003 | <10−4 | <10−4 | — | 0.544 | 0.571 | −0.408 | 0.162 | 0.122 | 0.244 | 0.248 | 0.388 | 0.548 | 0.113 | 0.474 | 0.852 |
| m8 | <10−4 | NS | NS | NS | NS | NS | 0.0003 | <10−4 | — | 0.692 | −0.225 | 0.204 | −0.082 | 0.011 | 0.025 | 0.177 | 0.367 | 0.054 | 0.678 | 0.924 |
| m9 | <10−4 | NS | NS | NS | NS | NS | 0.013 | <10−4 | <10−4 | — | −0.239 | 0.062 | −0.177 | −0.102 | −0.005 | 0.201 | 0.546 | 0.068 | 0.618 | 0.804 |
| Lf | <10−4 | NS | <10−4 | <10−4 | <10−4 | <10−4 | <10−4 | <10−4 | NS | NS | — | 0.500 | −0.725 | −0.879 | −0.832 | −0.732 | 0.297 | 0.227 | 0.060 | −0.043 |
| f1 | 0.0009 | 0.0006 | 0.0009 | 0.003 | 0.003 | 0.025 | NS | NS | NS | NS | <10−4 | — | 0.505 | 0.508 | 0.198 | 0.096 | 0.030 | 0.022 | 0.302 | 0.350 |
| f2 | <10−4 | 0.032 | <10−4 | <10−4 | <10−4 | <10−4 | NS | NS | NS | NS | <10−4 | <10−4 | — | 0.744 | 0.468 | 0.212 | −0.120 | −0.150 | −0.141 | −0.537 |
| f3 | <10−4 | 0.024 | <10−4 | <10−4 | <10−4 | <10−4 | <10−4 | 0.014 | NS | NS | <10−4 | <10−4 | <10−4 | — | 0.740 | 0.526 | 0.050 | −0.076 | −0.324 | −0.299 |
| f4 | <10−4 | NS | <10−4 | <10−4 | <10−4 | <10−4 | <10−4 | 0.011 | NS | NS | <10−4 | NS | <10−4 | <10−4 | — | 0.722 | 0.125 | −0.246 | −0.377 | −0.484 |
| f5 | <10−4 | NS | NS | 0.020 | <10−4 | <10−4 | <10−4 | <10−4 | NS | NS | <10−4 | NS | 0.040 | <10−4 | <10−4 | — | 0.353 | −0.155 | −0.359 | −0.353 |
| f6 | 0.0002 | NS | NS | NS | NS | 0.0006 | <10−4 | <10−4 | 0.004 | 0.0002 | 0.005 | NS | NS | NS | NS | 0.0008 | — | 0.244 | 0.341 | 0.534 |
| f7 | NS | NS | NS | NS | 0.046 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | — | 0.145 | −0.508 |
| f8 | NS | NS | NS | NS | NS | NS | NS | 0.015 | 0.0002 | 0.002 | NS | NS | NS | NS | NS | NS | NS | NS | — | 0.737 |
| f9 | NS | NS | NS | NS | NS | NS | NS | 0.002 | 0.0004 | 0.009 | NS | NS | NS | NS | NS | NS | NS | NS | 0.015 | — |
Traits (Tr.) are Lm, life span of males; Lf, life span of females; m1–m9, male mortality; and f1–f9, female mortality in weeks 1–9, respectively. Estimates are above (underlined if significant after correction for multiple testing and italicized if significant at the 0.05 level) and significances are below the diagonal.
As expected, life span was strongly negatively correlated with age-specific mortality (Table 3). The correlations appeared to be the strongest at weeks 4 and 5 for both sexes, when much of the death occurs. Life span is weakly though significantly correlated with mortality rate during the first and final weeks of life (Table 3). Age-specific mortality rates in males and females are highly correlated, especially at midlife and the oldest ages. A subset of RILs was previously used in a smaller experiment measuring life span and survival in the absence of food (Wang et al. 2004). Line mean life spans are positively though not significantly correlated with the present results (Table 4). Interestingly, the survival data of Wang et al. (2004) are strongly correlated with both independent measurements of life span. Weak correlation of the flies' life spans measured in different environments is not surprising and has been carefully documented (Vieira et al. 2000).
TABLE 4.
Correlation of line means across experiments and traits
|
Wang et al. (2004)
|
||||||
|---|---|---|---|---|---|---|
| Life span
|
Survival
|
|||||
| Trait | Males | Females | Males | Females | Males | Females |
| LM | — | 0.809 | 0.100 | 0.100 | 0.385 | 0.245 |
| LF | <10−4 | — | 0.077 | 0.146 | 0.397 | 0.263 |
| LMW | NS | NS | — | 0.855 | 0.248 | 0.192 |
| LFW | NS | NS | <10−4 | — | 0.209 | 0.198 |
| SMW | <10−4 | <10−4 | 0.014 | 0.039 | — | 0.587 |
| SFW | 0.008 | 0.005 | NS | 0.048 | <10−4 | — |
Note that the estimates are in very close agreement with those in Table 1 of Wang et al. (2004) but they deviate slightly as we applied a different transformation of the raw data. Estimates are above (underlined if significant at the 0.001 level and italicized if significant at the 0.05 level) and significances are below the diagonal.
Marker-specific survival analysis:
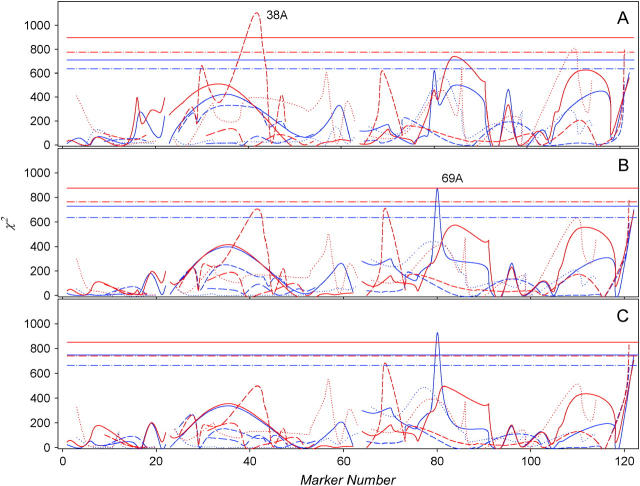
The genotype of every RIL is known at 123 marker loci. We pooled data over lines and then stratified according to marker locus genotypes for each sex and block. The stratified data were used to test the hypothesis that allelic states are associated with differential survival of flies by using nonparametric Wilcoxon (Figure 3A) and log-rank (Figure 3B) tests. We also used a parametric test of deviation from the Weibull distribution, the null hypothesis being justified by the observation that −log (survivorship) increases approximately linearly with age on the log-log scale (data not shown). Significance of the survival differences assuming this distribution was evaluated with χ2 (SAS procedure LIFEREG). Each of the three tests was performed for every marker locus, and significance was evaluated by permutations. The results are largely concordant between tests (see Figure 3). Marker 38A in the third haplotype of the second chromosome is significantly associated with female survival, and marker 69A of the first haplotype of chromosome 3 is associated with male survival. Females with the roo marker present at chromosomal location 38A die faster than those carrying the alternative allele at the beginning of life, but appear to survive better at more advanced ages. However, the QTL analysis described below does not support this result. Flies carrying the roo marker at chromosomal position 69A have lower survival across all ages compared to the flies with the alternative allele.
Figure 3.—
Significance of association between marker alleles and survival of males (blue) and females (red) for Wilcoxon (A) and log-rank (B) nonparametric tests and (C) a parametric test assuming Weibull mortality distribution. Lines represent the test statistics for markers sorted by haplotypes. Horizontal lines are corresponding significance thresholds.
QTL maps:
As a second method of analyzing the data, we mapped QTL affecting mean adult life spans and weekly mortality rates in each sex and block. Results for correlations between log-transformed life span and age-specific mortality are shown in Table 5.
TABLE 5.
QTL effects on life span and mortality
| Life span
|
Mortality
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome: haplotype |
QTL | Sex | Block 1 |
Block 2 |
Wang et al. (2004)a | Wk 1 | Wk 2 | Wk 3 | Wk 4 | Wk 5 | Wk 6 | Wk 7 | Wk 8 | Wk 9 |
| X-1: 1 | 3A | m | 0.0127 | 0.0101 | 0.0110 | −0.032 | −0.023 | −0.070 | −0.048 | −0.012 | 0.012 | 0.123 | 0.130 | −0.411 |
| f | 0.0147 | 0.0282 | 0.0131 | −0.057 | −0.081 | 0.028 | −0.016 | 0.011 | −0.130 | 9.2e−3 | −0.066 | −0.239 | ||
| X-1: 2 | 12C | m | −9.17e−3 | 8.89e−3 | −3.70e−3 | 0.011 | 0.174 | 0.217 | 0.079 | 0.036 | −0.083 | −0.050 | −0.102 | −0.152 |
| f | −.0210 | −2.50e−3 | −8.55e−3 | 0.082 | 0.120 | 0.197 | 0.063 | −0.082 | −0.142 | 7.1e−3 | −0.104 | −0.113 | ||
| X-2: 1 | 1E | m | 0.0161 | 0.0244 | −5.97e−3 | 2.8e−3 | −0.032 | −0.114 | −0.136 | −0.107 | −0.072 | −8.0e−3 | 0.025 | 9.7e−3 |
| f | 0.0251 | 0.0304 | 1.88e−3 | −0.042 | −0.054 | −0.193 | −0.121 | −0.047 | 5.5e−3 | 0.089 | −3.5e−3 | −0.051 | ||
| 2-1: 1 | 22C | m | 0.0344 | 0.0661 | 9.72e−3 | −0.030 | −0.092 | −0.110 | −0.238 | −0.110 | −0.059 | −0.130 | −0.076 | −0.017 |
| f | 0.0375 | 0.0544 | −1.74e−3 | −0.057 | −0.107 | −0.179 | −0.126 | −0.084 | 0.019 | −0.012 | −0.092 | −1.5e−3 | ||
| 2-1: 2 | 33F | m | 0.0364 | 0.0648 | 0.0715 | −0.047 | −0.133 | −0.123 | −0.132 | −0.168 | −0.262 | −0.129 | −0.028 | 3.4e−3 |
| f | 0.0381 | 0.0625 | 0.0451 | −0.151 | −0.050 | −0.249 | −0.035 | −0.160 | −0.134 | −0.047 | −0.040 | 0.022 | ||
| 2-2: 1 | 31F | m | 0.0172 | −5.01e−3 | 0.0159 | −0.047 | −0.101 | −0.104 | −0.107 | −0.032 | 0.025 | −0.021 | 0.100 | 0.068 |
| f | 0.0231 | 0.0160 | 0.0314 | −0.018 | −0.034 | −0.127 | −0.160 | −0.097 | 0.109 | 0.056 | 0.239 | 0.088 | ||
| 2-2: 2 | 54C | m | 0.0222 | 3.90e−3 | −0.0119 | −0.062 | −0.118 | −0.108 | −0.121 | −0.089 | −0.025 | −0.037 | −3.4e−3 | −0.045 |
| f | 0.0359 | 0.0180 | −3.08e−3 | −0.055 | −0.120 | −0.161 | −0.121 | −0.133 | 0.063 | 0.018 | 1.7e−3 | −9.8e−3 | ||
| 2-3: 1 | 24C–D | m | −0.0312 | −0.0166 | 3.76e−3 | 0.018 | 0.154 | 0.100 | 0.168 | 0.093 | 0.041 | 0.044 | 0.116 | 0.106 |
| f | −0.0238 | −0.0343 | −7.45e−4 | 5.7e−3 | 0.063 | 0.086 | 0.033 | 0.067 | 0.060 | 5.7e−4 | 0.153 | 0.546 | ||
| 2-3: 2 | 28C | m | −0.0509 | −0.0343 | −0.0202 | 0.062 | 0.270 | 0.210 | 0.262 | 0.239 | 0.085 | −0.035 | 4.0e−3 | −0.031 |
| f | −0.0628 | −0.0599 | −0.0259 | 0.111 | 0.304 | 0.335 | 0.213 | 0.125 | −5.3e−3 | 1.6e−3 | 0.050 | −0.138 | ||
| 2-3: 3 | 38A | m | −0.0278 | −0.0585 | −0.0496 | 0.086 | 0.115 | 0.151 | 0.151 | 0.151 | 0.167 | 0.030 | −0.039 | 7.3e−3 |
| f | −0.0354 | −0.0539 | −0.0559 | 0.072 | 0.102 | 0.265 | 0.162 | 0.116 | 0.041 | 8.8e−3 | −0.167 | −0.115 | ||
| 2-3: 4 | 41F | m | −0.0634 | −0.0645 | −0.1134 | 0.222 | 0.249 | 0.330 | 0.390 | 0.242 | 0.228 | 0.118 | −0.054 | 0.074 |
| f | −0.1016 | −0.0537 | −0.0869 | 0.283 | 0.350 | 0.444 | 0.446 | 0.168 | −0.021 | 0.055 | −0.135 | −0.131 | ||
| 2-3: 5 | 41F | m | 0.0617 | 0.0625 | 0.1119 | −0.226 | −0.258 | −0.328 | −0.389 | −0.251 | −0.237 | −0.105 | 0.049 | −0.068 |
| f | 0.1009 | 0.0494 | 0.0827 | −0.297 | −0.380 | −0.463 | −0.458 | −0.177 | 0.052 | −0.052 | 0.122 | 0.132 | ||
| 2-4: 1 | 49B | m | 0.0129 | 0.0196 | 0.0693 | −0.103 | 0.095 | 0.208 | 0.054 | −0.082 | −0.114 | −0.056 | 0.030 | 0.052 |
| f | 0.0316 | 0.0235 | 0.0799 | −0.187 | −0.098 | −0.152 | −0.117 | −0.338 | −0.061 | −0.203 | 8.7e−3 | 0.216 | ||
| 3-1: 1 | 69A | m | −0.0346 | −0.0449 | −1.72e−3 | −0.055 | 0.065 | 0.128 | 0.315 | 0.124 | 0.076 | 0.064 | 0.139 | 0.095 |
| f | −0.0284 | −0.0381 | 0.0156 | 1.2e−3 | 0.097 | 0.217 | 0.113 | −0.030 | 0.072 | −0.022 | 0.119 | 0.330 | ||
| 3-1: 2 | 76B | m | −0.0174 | 7.54e−3 | 0.0118 | −0.039 | 0.076 | 0.126 | 0.185 | 0.151 | 0.016 | 0.017 | 0.056 | 0.077 |
| f | −0.0381 | −0.0456 | 1.45e−3 | −0.168 | 0.099 | 0.334 | 0.139 | −6.7e−3 | 0.040 | 0.055 | −0.086 | −0.194 | ||
| 3-1: 3 | 83D | m | −0.0206 | 2.61e−3 | 0.0239 | −0.040 | 0.068 | 0.115 | 0.118 | 0.173 | 0.011 | −0.023 | 0.073 | 0.185 |
| f | −0.0166 | −0.0339 | −6.34e−3 | −0.165 | 0.115 | 0.278 | 0.089 | −0.145 | −0.016 | 0.083 | −0.059 | −0.173 | ||
| 3-1: 4 | 87B | m | 0.0655 | 0.0196 | 9.71e−3 | −0.131 | −0.138 | −0.144 | −0.108 | −0.140 | −0.120 | −0.046 | −0.054 | −0.066 |
| f | 0.0195 | 0.0311 | 8.41e−3 | −0.051 | 6.9e−3 | −0.451 | −0.442 | −0.175 | 0.011 | −0.026 | 4.0e−3 | −0.016 | ||
| 3-1: 5 | 89A–B | m | −8.32e-3 | −0.0188 | −3.20e−3 | 0.066 | 0.051 | 0.193 | 0.120 | 0.013 | −0.018 | −0.064 | −0.028 | −0.102 |
| f | −0.0614 | −0.0429 | −9.82e−3 | 0.173 | 0.188 | 0.129 | 0.229 | 0.097 | 4.6e−3 | −0.026 | 0.047 | −0.181 | ||
| 3-1: 6 | 97B | m | −0.0167 | 0.0217 | 0.0286 | 3.7e−3 | 0.100 | 0.233 | 0.081 | 0.100 | −0.018 | −1.1e−3 | −0.036 | 5.7e−3 |
| f | −0.0603 | −3.75e−3 | 4.47e−3 | 0.081 | 0.168 | 0.236 | 0.304 | 0.106 | 0.075 | −0.062 | 2.6e−5 | 0.138 | ||
| 3-2/3: 1 | 67C | m | 0.0506 | 0.0521 | −0.0108 | 0.063 | −9.1e−3 | −0.100 | −0.198 | −0.179 | −0.135 | −0.171 | −0.182 | −0.098 |
| f | −0.0328 | 0.0454 | 8.29e−3 | −0.075 | −0.022 | −0.068 | −0.096 | −0.055 | −0.120 | 0.019 | −0.182 | −0.250 | ||
| 3-2/3: 2 | 77B | m | 0.0234 | 0.0424 | 5.83e−4 | 0.023 | −0.103 | −0.108 | −0.127 | −0.102 | −0.063 | 0.012 | −0.011 | −0.048 |
| f | 0.0420 | 8.436e−3 | −0.0191 | −0.054 | −0.146 | −0.157 | −0.037 | −0.068 | 0.076 | 0.026 | 6.2e−3 | 0.469 | ||
| 3-2/3: 3 | 84F | m | 0.0231 | 0.0499 | −5.85e−3 | 0.024 | −0.114 | −0.140 | −0.105 | −0.138 | −0.128 | 4.1e−3 | −0.028 | −0.130 |
| f | 0.0450 | 0.0144 | −0.0323 | −0.047 | −0.183 | −0.284 | −0.187 | −0.184 | 0.046 | −4.2e−3 | 0.010 | 0.376 | ||
| 3-2/3: 4 | 88E | m | 0.0124 | 0.0392 | 6.599e−3 | −0.065 | −0.089 | −0.145 | −0.146 | −0.127 | −0.120 | −1.9e−3 | 0.012 | −0.017 |
| f | 0.0395 | 0.0203 | −0.0168 | −0.057 | −0.160 | −0.250 | −0.165 | −0.120 | −3.6e−3 | 0.042 | 0.052 | 0.469 | ||
| 3-2/3: 5 | 90E–F | m | 0.0184 | 0.0219 | 0.0244 | −0.053 | −0.049 | −0.030 | −0.102 | −0.133 | −0.102 | −0.030 | −0.040 | 4.6e−3 |
| f | 0.0455 | 2.064e−3 | 9.257e−3 | −0.055 | −0.148 | −0.260 | −0.204 | −0.177 | 0.018 | 3.6e−3 | −0.024 | 0.147 | ||
| 3-4: 1 | 65C | m | −0.1032 | −0.0433 | −0.0428 | 0.303 | 0.161 | 0.408 | 0.465 | 0.361 | 0.213 | 6.2e−3 | 0.075 | 0.090 |
| f | −0.0746 | −0.1047 | −0.0149 | 0.197 | 0.317 | 0.420 | 0.161 | 0.235 | 0.111 | 0.117 | 0.000 | 0.000 | ||
| 3-4: 2 | 98F | m | 0.0377 | 0.0258 | 9.407e−4 | 1.6e−3 | −0.105 | −0.241 | −0.168 | −0.153 | −0.085 | −0.070 | −0.082 | −0.059 |
| f | 0.0518 | 0.0385 | 4.440e−3 | −0.037 | −0.177 | −0.316 | −0.195 | −0.154 | −0.040 | 0.041 | 0.012 | −0.145 | ||
Note that the estimate is at the position most significant in this study. See Wang et al. (2004) for the estimates at the most significant positions in their experiment. Underlined estimates are significant at the 0.05 level.
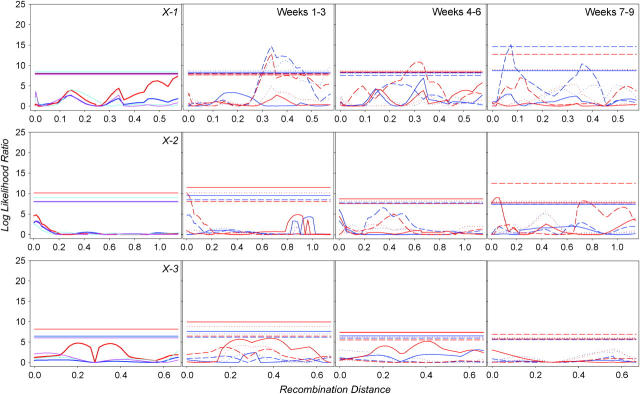
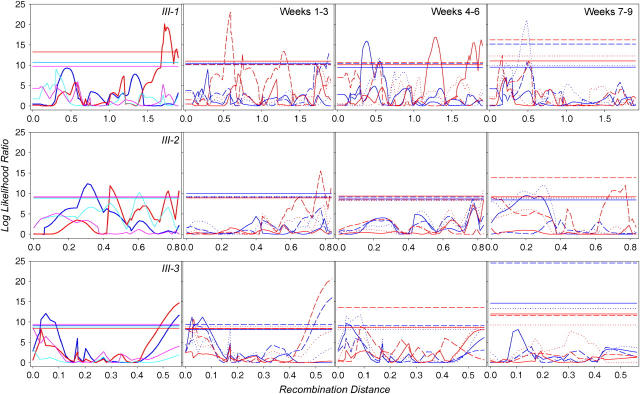
Log-likelihood-ratio plots are shown in Figure 4 for the 3 haplotypes of the X chromosome, in Figure 5 for the 4 haplotypes of the second chromosome, and in Figure 6 for the three haplotypes of the third chromosome. For every haplotype and trait, we also performed 1000 permutations to define an appropriate 5% significance threshold. Note that for 10 total haplotypes and 20 traits, we performed 200 analyses altogether, and thus 10 QTL significant at the 0.05 level were expected by chance alone. For block 1, we found 80 significant QTL in 25 genomic positions—significantly more than expected under the null hypothesis of no trait-genotype association. A false discovery rate might be approximated as 10/80 = 12.5%. Note that composite interval mapping detects as significant both QTL 38A and 69A, which were identified by survival analysis.
Figure 4.—
Quantitative trait loci affecting male (blue for block 1, light blue for block 2) and female (red for block 1, pink for block 2) life span (far left column) on haplotypes 1, 2, and 3 of the X chromosome. QTL for age-specific mortality rate for males and females are in the right columns, with solid lines corresponding to earlier, dotted lines to intermediate, and dashed lines to later weeks. Horizontal lines are corresponding significance thresholds, some of which overlap.
Figure 5.—
QTL of the second chromosome affecting life span and age-specific mortality. Line designations are analogous to those in Figure 4.
Figure 6.—
QTL of the third chromosome affecting life span and age-specific mortality. Line designations are analogous to those in Figure 4.
A summary of QTL positions and estimates of their effects on life span and age-specific mortality is presented in Table 5. For each QTL position detected in block 1 (having the most power of QTL detection), we also provide the estimated effects for block 2, as well as those for the study of Wang et al. (2004) using the same RILs. Even though a small number of RILs (48) were scored for block 2, diminishing the power to detect QTL, the estimated effects are highly concordant at 0.82 for males and 0.85 for females. Unfortunately, the significance of these correlations is difficult to evaluate, because the estimated QTL effects are not independent. Even more surprising is the marked similarity of estimates between the current experiment and the data of Wang et al. (2004). While the line means are only weakly positively associated, the QTL effects are remarkably similar, with a correlation of 0.58 for both males and females. Of six QTL detected for life span by Wang et al. (2004), four are detected here at the same positions and with very similar magnitudes. The statistical nature of the discrepancy between highly correlated QTL effects and loosely correlated line means merits further study.
DISCUSSION
What are the genetic factors that underlie age-specific mortality in Drosophila? An answer to this question has been available only for QTL alleles that were fixed due to selection for an extended reproduction period (Luckinbill and Clare 1985). The effects of those QTL alleles were consistent across ages and confer resistance to oxidative stress (Curtsinger and Khazaeli 2002). Are these patterns common for alleles segregating in natural populations? To answer this question, we generated and analyzed a panel of recombinant inbred lines derived from flies caught in nature (Kopp et al. 2003). Using moderate sample sizes, we previously established genetic variation in life span of these lines in a normal and a no food environment (Wang et al. 2004). Here, we report age-specific mortality QTL.
We used a combination of survival analysis and composite interval mapping—two powerful statistical approaches. Our analysis could have possibly benefited from employing other recently promoted techniques: a character process (Pletcher and Geyer 1999) and a logistic mixture model (Wu et al. 2002). The former technique is, unfortunately, not supported by the programs amenable to QTL mapping. In the spirit of the latter technique, we estimated line-specific survival parameters and used them for QTL mapping. No QTL were supported by permutations as parameter estimation was too noisy (data not shown). It appears that our sample sizes—∼100 flies per sex for most genotypes—were insufficient.
With conventional QTL analyses, the effects of QTL, while not statistically significant at every age, were largely concordant between ages and sexes (Table 5). Two QTL deviate from this pattern. The presence of roo at 12C is associated with increased mortality at age 3 weeks in both males and females. This same marker allele corresponds to slower death rate at age 9 weeks in males and at week 6 in females. Similarly, roo at 1E is associated with better survival of females early in life but poorer survival late in life. These data could be interpreted as age-limited effects of at least two (per QTL) mutations. Alternatively, they might represent long-sought-for QTL with antagonistic pleiotropic effects on mortality. Testing this interesting hypothesis might be challenging. Only a few life span QTL were further dissected to the effects of individual genes (Pasyukova et al. 2000; DeLuca et al. 2003). Every QTL appeared to be a composite effect of polymorphisms in several genes. We will have to split QTL support intervals with additional recombination breakpoints and test whether both antagonistic age-specific effects map to the same smaller interval.
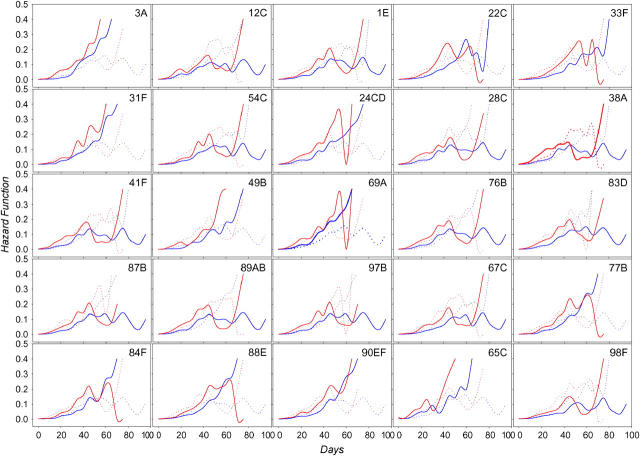
Hazard functions specific for two marker alleles at significant QTL are plotted in Figure 7. One should be cautious interpreting these figures at advanced ages as they are based on a small number of flies surviving in a few cages. Compare, for instance, the plots for markers 24CD, 69A, and 97B. After the age of ∼60 days, they appear nearly identical for females. Suffice it to say that at this age, only 11 RILs contributing to the estimation remain (data not shown). If very few RILs sharing a genotype at these markers can survive, the survival patterns for these markers will appear identical. This pattern can be explained by chance alone. Alternatively, interactions between markers might be required for the flies to be able to survive to older age. Careful estimation of intralocus and interlocus allelic interactions across ages might be a worthwhile direction for future research.
Figure 7.—
Hazard rates for males (blue) and females (red) for significant markers. Solid lines show the allele marked by the presence of roo; dotted lines show the absence of roo.
The genetic makeup of variation in age-specific mortality has been a subject of much debate. The interest comes from different predictions on the additive portion of age-specific mortality variance by alternative evolutionary aging theories (Charlesworth and Hughes 1996). While straightforward at first sight, the inferences are murky due to statistical difficulties of complex estimation procedures (Shaw et al. 1999). Enflaming this heated discussion is not the goal of this article. Rather, we point out that estimation of intralocus (dominance) and interloci interactions can be made in marker-based fashion—potentially providing new types of data for the discussion.
Our study provides estimates of life span QTL allelic effects in different ages. Further, it compares the effect of an allele to the “population mean”—constructed for our population of recombinant inbred lines from the mean of two other alleles for the X and third chromosomes—and of three other alleles for the second chromosome. This lets us test whether effects of lower-frequency alleles are primarily to increase mortality, as predicted by the mutational hypothesis of aging (Medawar 1952). Of 55 age-specific significant effects, 35 increase mortality and 20 decrease it. Effects increasing mortality are more frequent at the beginning of life (29 of 41 during weeks 1–5) in comparison with those later in life (6 of 14). One should be cautious taking these numbers at face value. For instance, marker 65C on haplotype 4 of chromosome 3 is associated with decreased life span, but marker 67C on haplotype 2/3 is associated with increased life span by a smaller amount. In fact, the latter estimate might be a shadow of the former as the estimates of allelic effects on different haplotypes are not independent (see materials and methods for explanations; note that when a QTL is a part of a mixed distribution, the power of detecting its effect is low). This explanation might account for lower-frequency QTL alleles decreasing mortality (another example is 88E on haplotype 2/3 of chromosome 3 vs. 89AB on haplotype 1). Note, however, that some positive effects (see, for instance, 54C on the second haplotype of chromosome 2) are not colocalized with poor alleles on other haplotypes. Overall, we conclude that lower-frequency alleles increasing mortality are more prevalent, especially with effects at the beginning of life.
Mutations in numerous genes appear to affect life span. QTL mapped between different parental stocks were thought to have, perhaps, low overlap. Somewhat surprisingly, it appears that there is substantial overlap. A number of QTL found here coincide with those found by Vieira et al. (2000). Pasyukova et al. (2000) precisely mapped life span QTL within two regions—33E–46C and 65D–85F. Both markers significant in survival analysis here, 38A and 69A, were identified as contributing to life span. Curtsinger and Khazaeli (2002) found three significant intervals: 22B–26D, 34F–45D, and 66A–68D—all colocated with QTL detected by Vieira et al. (2000) and found here. Those might be mere coincidences. Alternatively, Mackay (2002) and DeLuca et al. (2003) hypothesized that variation in some of the life span QTL is maintained due to balancing selection. Then, identical alleles might be segregating in different natural and mapping populations and independently picked by QTL analysis. Testing these hypotheses represents an interesting future task. Interestingly, the correspondence is also apparent in the data of whole-genome expression experiments of Pletcher et al. (2002) and Jin et al. (2001), both of which examined the differences in gene expression in flies between ages. The former study, for instance, implicated the region 66D10–66F6 on chromosome 3 as having significantly different levels of gene expression at different ages, potentially narrowing down the list of candidate genes for follow-up work.
Acknowledgments
Research of S.V.N. is supported by the National Institutes of Health (NIH 1R24 GM65513-01 and NIH 1R01 GM61773-01). J.W.C. is supported by NIH AG11722.
References
- Basten, C. J., B. S. Weir and Z-B. Zeng, 1999 QTL Cartographer: Version 1.13. Department of Statistics, North Carolina State University, Raleigh, NC.
- Carey, J. R., 2002 Longevity: The Biology and Demography of Life Span. Princeton University Press, Princeton, NJ.
- Charlesworth, B., 1994 Evolution in Age-Structured Populations, Ed. 2. Cambridge University Press, Cambridge, UK.
- Charlesworth, B., and K. A. Hughes, 1996. Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc. Natl. Acad. Sci. USA 93: 6140–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger, J. W., and A. A. Khazaeli, 2002. Life span, QTL's, age-specificity, and pleiotropy in Drosophila. Mech. Ageing Dev. 123: 81–93. [DOI] [PubMed] [Google Scholar]
- Curtsinger, J. W., H. Fukui, D. Townsend and J. W. Vaupel, 1992. Demography of genotypes: failure of the limited life-span paradigm in Drosophila melanogaster. Science 258: 461–463. [DOI] [PubMed] [Google Scholar]
- Curtsinger, J. W., H. H. Fukui, A. A. Khazaeli, A. Kirscher, S. D. Pletcher et al., 1995. Genetic variation and aging. Annu. Rev. Genet. 29: 553–575. [DOI] [PubMed] [Google Scholar]
- Curtsinger, J. W., H. H. Fukui, A. S. Resler, K. Kelly and A. A. Khazaeli, 1998. Genetic analysis of extended life span in Drosophila melanogaster. I. RAPD screen for genetic divergence between selected and control lines. Genetica 104: 21–32. [DOI] [PubMed] [Google Scholar]
- Curtsinger, J. W., N. S. Gavrilova and L. A. Gavrilov, 2005 Age-specific mortality in Drosophila melanogaster: investigations at the intersection of genetics and demography. Handb. Biol. Aging (in press).
- DeLuca, M. D., N. V. Roshina, G. L. Geiger-Thornsberry, R. F. Lyman, E. G. Pasyukova et al., 2003. Dopa decarboxilase (Ddc) affects variation in Drosophila longevity. Nat. Genet. 34: 429–433. [DOI] [PubMed] [Google Scholar]
- Doerge, R. W., Z-B. Zeng and B. S. Weir, 1997. Statistical issues in the search for genes affecting quantitative traits in experimental populations. Stat. Sci. 12: 195–219. [Google Scholar]
- Flatt, T., 2004. Assessing natural variation in genes affecting Drosophila lifespan. Mech. Ageing Dev. 125: 155–159. [DOI] [PubMed] [Google Scholar]
- Forbes, S. N., R. K. Valenzuela, P. Keim and P. M. Service, 2004. Quantitative trait loci affecting life span in replicated populations of Drosophila melanogaster. I. Composite interval mapping. Genetics 168: 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui, H. H., and A. W. Kirshner, 1993. Thanatometer II: a chamber designed for large mixed-sex populations of Drosophila melanogaster. Dros. Inf. Serv. 72: 72–73. [Google Scholar]
- Fukui, H., L. Xiu and J. W. Curtsinger, 1993. Slowing of age-specific mortality rates in Drosophila melanogaster. Exp. Gerontol. 28: 585–599. [DOI] [PubMed] [Google Scholar]
- Fukui, H. H., L. Ackert and J. W. Curtsinger, 1996. Deceleration of age-specific mortality rates in chromosomal homozygotes and heterozygotes of Drosophila melanogaster. Exp. Gerontol. 31: 517–531. [DOI] [PubMed] [Google Scholar]
- Geiger-Thornsberry, G. L., and T. F. C. Mackay, 2004. Quantitative trait loci affecting natural variation in Drosophila longevity. Mech. Ageing Dev. 125: 179–189. [DOI] [PubMed] [Google Scholar]
- Jin, W., R. M. Riley, R. D. Wolfinger, K. P. White, G. Passador-Gurgel et al., 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29: 389–395. [DOI] [PubMed] [Google Scholar]
- Khazaeli, A. A., and J. W. Curtsinger, 2000. Genetic analysis of extended life span in Drosophila melanogaster. III. On the relationship between artificially selected and wild stocks. Genetica 109: 245–253. [DOI] [PubMed] [Google Scholar]
- Khazaeli, A. A., W. Van Voorhies and J. W. Curtsinger, 2005. Longevity and metabolism in Drosophila melanogaster: genetic correlations between life span and age-specific metabolic rate in populations artificially selected for long life. Genetics 169: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp, A., R. M. Graze, S. Xu, S. B. Carroll and S. V. Nuzhdin, 2003. Quantitative trait loci responsible for variation in sexually dimorphic traits in Drosophila melanogaster. Genetics 163: 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips, J., and T. F. C. Mackay, 2000. Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics 155: 1773–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckinbill, L. S., and M. J. Clare, 1985. Selection for life span in Drosophila melanogaster. Heredity 55: 9–18. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., 2002. The nature of quantitative genetic variation for Drosophila longevity. Mech. Ageing Dev. 123: 95–104. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., and J. D. Fry, 1996. Polygenic mutation in Drosophila melanogaster: genetic interactions between selection lines and candidate quantitative trait loci. Genetics 144: 671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar, P. B., 1952 An Unsolved Problem of Biology. H. K. Lewis, London.
- Mezey, J. G., D. Houle and S. V. Nuzhdin, 2005. Naturally segregating QTL affecting wing shape of Drosophila melanogaster. Genetics 169: 2101–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin, S. V., E. G. Pasyukova, C. A. Dilda, Z-B. Zeng and T. F. C. Mackay, 1997. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94: 9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge, L., and D. Gems, 2002. Mechanisms of ageing: Public or private? Nat. Rev. Genet. 3: 165–175. [DOI] [PubMed] [Google Scholar]
- Pasyukova, E. G., C. Vieira and T. F. C. Mackay, 2000. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics 156: 1129–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher, S. D., and J. W. Curtsinger, 1998. Mortality plateaus and the evolution of senescence: Why are old-age mortality rates so low? Evolution 52: 454–464. [DOI] [PubMed] [Google Scholar]
- Pletcher, S. D., and C. J. Geyer, 1999. The genetic analysis of age-dependent traits: modeling the character process. Genetics 151: 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher, S. D., S. J. Macdonald, R. Marguerie, U. Certa, S. C. Stearns et al., 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12: 712–723. [DOI] [PubMed] [Google Scholar]
- Promislow, D. E. L., and S. D. Pletcher, 2002. Advise to an aging scientist. Mech. Ageing Dev. 123: 841–850. [DOI] [PubMed] [Google Scholar]
- Promislow, D. E. L., M. Tatar, A. A. Khazaeli and J. W. Curtsinger, 1996. Age-specific patterns of genetic variation in Drosophila melanogaster. I. Mortality. Genetics 143: 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiwitch, S. G., and S. V. Nuzhdin, 2002. Quantitative trait loci for lifespan of mated Drosophila melanogaster affect both sexes. Genet. Res. 80: 1–6. [DOI] [PubMed] [Google Scholar]
- Resler, A. S., K. Kelly, G. Cantor, A. A. Khazaeli, M. Tatar et al., 1998. Genetic analysis of extended life span in Drosophila melanogaster. II. Replication of the backcross test and molecular characterization of the N14 locus. Genetica 104: 33–39. [DOI] [PubMed] [Google Scholar]
- SAS Institute, 1988 SAS/STAT User's Guide. Release 6.03 Edition. SAS Institute, Cary, NC.
- Shaw, F. H., D. E. L. Promislow, M. Tatar, K. A. Hughes and C. J. Geyer, 1999. Toward reconciling inferences concerning genetic variation in senescence in Drosophila melanogaster. Genetics 152: 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimpton, A. E., E. A. Montgomery and C. H. Langley, 1986. Orc mutations in Drosophila ananassae are linked to insertions of a transposable element. Genetics 114: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, C. C., C. E. Howell, A. R. Wright and D. E. L. Promislow, 2003. Testing an ‘aging gene’ in long-lived Drosophila strains: increased longevity depends on sex and genetic background. Aging Cell 2: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahoe, N. M. A., A. M. Dean and J. W. Curtsinger, 2002. Nucleotide variations in the lxd region of Drosophila melanogaster: characterization of a candidate modifier of lifespan. Gene 297: 221–228. [DOI] [PubMed] [Google Scholar]
- Valenzuela, R. K., S. N. Forbes, P. Keim and P. M. Service, 2004. Quantitative trait loci affecting life span in replicated populations of Drosophila melanogaster. II. Response to selection. Genetics 168: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voorhies, W., A. A. Khazaeli and J. W. Curtsinger, 2003. Selected contribution: long-lived Drosophila melanogaster exhibit normal metabolic rates. J. Appl. Physiol. 95: 2605–2613. [DOI] [PubMed] [Google Scholar]
- Van Voorhies, W. W., A. A. Khazaeli and J. W. Curtsinger, 2004. a Testing the “rate of living” model: further evidence that longevity and metabolic rate are not inversely correlated in Drosophila melanogaster. J. Appl. Physiol. 97: 1915–1922. [DOI] [PubMed] [Google Scholar]
- Van Voorhies, W., A. A. Khazaeli and J. W. Curtsinger, 2004. b Lack of correlation between body mass and metabolic rate in Drosophila melanogaster. J. Insect Physiol. 50: 445–453. [DOI] [PubMed] [Google Scholar]
- Vaupel, J. W., J. R. Carey, K. Christiansen, T. E. Johnson, A. I. Yashin et al., 1998. Biodemographic trajectories of longevity. Science 280: 855–860. [DOI] [PubMed] [Google Scholar]
- Vieira, C., E. G. Pasyukova, Z-B. Zeng, J. B. Hackett, R. F. Lyman et al., 2000. Genotype-environment interaction for quantitative trait loci affecting life in Drosophila melanogaster. Genetics 154: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. H., O. Lazebny, L. G. Harshman and S. V. Nuzhdin, 2004. Environment dependent survival of Drosophila melanogaster: a quantitative genetic analysis. Aging Cell 3: 133–141. [DOI] [PubMed] [Google Scholar]
- Williams, G. C., 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11: 398–411. [Google Scholar]
- Wu, R., C.-X. Ma, M. Chang, R. C. Littell, S. S. Wu et al., 2002. A logistic mixture model for characterizing genetic determinants causing differentiation in growth trajectories. Genet. Res. 79: 235–245. [DOI] [PubMed] [Google Scholar]