Abstract
DNA repair takes place in the context of chromatin. Recently, it has become apparent that proteins that make up and modulate chromatin structure are involved in the detection and repair of DNA lesions. We previously demonstrated that Ser129 in the carboxyl-terminal tail of yeast histone H2A is important for double-strand-break responses. By undertaking a systematic site-directed mutagenesis approach, we identified another histone H2A serine residue (Ser122) that is important for survival in the presence of DNA-damaging agents. We show that mutation of this residue does not affect DNA damage-dependent Rad53 phosphorylation or G2/M checkpoint responses. Interestingly, we find that yeast lacking H2A S122 are defective in their ability to sporulate. Finally, we demonstrate that H2A S122 provides a function distinct from that of H2A S129. These data demonstrate a role for H2A S122 in facilitating survival in the presence of DNA damage and suggest a potential role in mediating homologous recombination. The distinct roles of H2A S122 and S129 in mediating these responses suggest that chromatin components can provide specialized functions for distinct DNA repair and survival mechanisms and point toward the possibility of a complex DNA damage responsive histone code.
THE consequences of inaccurately or inefficiently repaired DNA double-strand breaks (DSBs) include genomic instability, cell death, and, in higher eukaryotes, tumorigenesis. It is therefore not surprising that cells have rigorous mechanisms to detect and repair DNA damage, including DSBs. Members of the phosphatidylinositol-3 kinase-related kinase (PIKK) family play key roles in DNA damage detection and signaling in eukaryotes. This family of kinases includes the yeast proteins Mec1 and Tel1 and their human homologs, ATR and ATM, respectively. In response to DNA damage, Mec1 localizes to the sites of DNA lesions and activates a signal transduction cascade. This results in hyperphosphorylation and activation of the Rad53 protein kinase, and, ultimately, in the induction of repair mechanisms and the arrest of progression through the cell cycle (Weinert 1998; Rouse and Jackson 2002). In addition to detecting DNA DSBs, cells must repair the damaged DNA. In eukaryotes, DNA DSBs are repaired by two primary mechanisms: nonhomologous end-joining (NHEJ) and homologous recombination (HR). NHEJ results in the direct religation of the broken DNA ends (for review, see Lieber et al. 2003), while HR relies on the presence of a homologous template to repair the DNA DSB (for review, see West 2003). In both cases, the repair proteins must function in the context of chromatin structures present at the DNA lesion and also, in the case of HR, at the homologous template.
At its most basic level, chromatin is composed of DNA wrapped around an octamer of histone proteins, made up of an (H3-H4)2 tetramer and two H2A-H2B dimers, to form the nucleosome. Beyond this level of organization, chromatin can form more compact, higher-order structures, and this is mediated in part by the presence of linker histones. The compaction of DNA in this manner has been shown to be generally inhibitory to its manipulation and accessibility. By modulating chromatin structure, cells can achieve precise regulation of DNA-dependent functions such as transcription. Chromatin structure is modulated by two primary mechanisms: ATP-dependent chromatin remodeling and covalent modification of histone tails. Recent evidence suggests that both of these mechanisms are utilized by cells to facilitate DNA DSB repair. For example, yeast strains with mutations in either Ino80C or Swr-C ATP-dependent chromatin-remodeling complexes are sensitive to DNA-damaging agents (Shen et al. 2000; Mizuguchi et al. 2004). Additionally, covalent modification of the histone H4 N-terminal tail by the NuA4 histone acetyl transferase (HAT) complex has been implicated in DNA DSB repair (Bird et al. 2002). Consistent with this, expression of an enzymatically inactive subunit of a mammalian HAT complex that is homologous to the catalytic subunit of NuA4 results in slower double-strand-break repair (Ikura et al. 2000). Additionally, histone deacetylase activities have recently been implicated in DNA DSB repair (Fernandez-Capetillo and Nussenzweig 2004), raising the possibility that a complex series of chromatin-mediated events takes place after DNA damage. It is possible that these activities indirectly affect survival under these conditions by compromising the ability of cells to appropriately regulate transcription of genes necessary for normal DNA damage responses. Recently, however, subunits of the NuA4, Ino80-C, and Swr1C complexes have been found to be present at the site of DNA DSBs by chromatin immunoprecipitation (ChIP) assays (Bird et al. 2002; Downs et al. 2004; Morrison et al. 2004; van Attikum et al. 2004), suggesting that there is a direct role for one or more of these complexes in DNA repair.
One covalent chromatin modification that has been found to be directly involved in DNA DSB responses is the phosphorylation of the mammalian histone variant H2A-X on the carboxyl-terminal tail at position S139 (Rogakou et al. 1998). This has been shown to occur in proximity to DNA lesions (Rogakou et al. 1999) and is mediated by the PIKK family members ATM, ATR, and DNA-PK (Paull et al. 2000; Burma et al. 2001; Ward and Chen 2001). In yeast, this serine residue exists on the main H2A species (S129) and is phosphorylated in response to DNA damage by the PIKK family members Mec1 and Tel1 (Downs et al. 2000; Redon et al. 2003). Consistent with data generated in higher eukaryotes, we and others recently found that S129 is phosphorylated in the vicinity of DNA DSBs by ChIP analysis (Downs et al. 2004; Morrison et al. 2004; Shroff et al. 2004; van Attikum et al. 2004), suggesting that this covalent modification directly facilitates repair at the site of the DNA lesion. In addition to residues that are targets for covalent modifications or chromatin-remodeling activities, residues that contribute directly to chromatin structure are important for the ability of cells to appropriately access and manipulate DNA. For example, residues in histone H2B that are unlikely to be modified have been shown to be important for mediating DNA repair by the postreplicative pathway (Martini et al. 2002).
To determine whether there was a contribution to DNA damage responses being provided by residues in the H2A C-terminal tail in addition to that of S129, we examined the histone H2A C-terminal tail by systematic site-directed mutagenesis. We find that one residue, S122, is important for survival in the presence of DNA damage. Furthermore, we demonstrate that H2A S122 is not required for the Mec1-dependent DNA damage signal transduction cascade, suggesting a more direct role in the repair event itself. We also find that H2A S122A mutant yeast are drastically impaired in their ability to sporulate, pointing to a potential role in homologous recombination. In support of this hypothesis, continual expression of the homothallic (HO) endonuclease results in decreased survival in the H2A S122A mutant yeast strain. Finally, we show that this residue functions independently from H2A S129, suggesting the presence of a complex DNA DSB repair histone code.
MATERIALS AND METHODS
Yeast strains:
Yeast strains are listed in Table 1. Plasmids bearing mutations in the HTA1 gene were created by site-directed mutagenesis following the manufacturer's protocols (Stratagene, La Jolla, CA). Plasmids were then transformed into FY406 (Hirschhorn et al. 1995) and the transformants were grown on media containing 5-fluoroorotic acid (5-FOA) to select for cells that had lost the wild-type HTA1-containing plasmid. JDY94 was created by crossing a strain bearing the genomic hta1-S122A mutation (in W303α) with FY406 and sporulating. The resulting haploid strains were selected for loss of pAB6 by growth on 5-FOA-containing media and were subsequently analyzed for the presence of hta1-S122A by PCR and for hta2-htb2::TRP1 disruption by phenotypic analysis.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Plasmid (relevant sequences) | Reference |
|---|---|---|---|
| FY406 |
MATa (hta1-htb1)Δ::LEU2 (hta2-htb2)Δ:: TRP1 ura3-52,1 leu2Δ1 lys2Δ1 lys2-128Δ his3Δ200 trp1Δ63 |
pAB6 (HTA1-HTB1, URA3) | Hirschhorn et al. (1995) |
| FHY2 | As FY406 | pJD150 (HTA1-HTB1, HIS3) | Downs et al. (2000) |
| FHY3 | As FY406 | pJD151 (hta1-S129A-HTB1, HIS3) | |
| FHY44 | As FY406 | pJD187 (hta1-T103A-HTB1, HIS3) | This study |
| FHY45 | As FY406 | pJD188 (hta1-K120A-HTB1, HIS3) | This study |
| FHY46 | As FY406 | pJD189 (hta1-K121A-HTB1, HIS3) | This study |
| FHY47 | As FY406 | pJD190 (hta1-S122A-HTB1, HIS3) | This study |
| FHY48 | As FY406 | pJD191 (hta1-K124A-HTB1, HIS3) | This study |
| FHY49 | As FY406 | pJD192 (hta1-T126A-HTB1, HIS3) | This study |
| FHY50 | As FY406 | pJD193 (hta1-K127A-HTB1, HIS3) | This study |
| FHY54 | As FY406 | pJD197 (hta1-S122A/S129A-HTB1, HIS3) | This study |
| FHY58 | As FY406 | pJD210 (hta1-S122E-HTB1, HIS3) | This study |
| FHY12 | Diploid of FHY2 | pJD150 (HTA1-HTB1, HIS3) | This study |
| FHY69 | Diploid of FHY47 | pJD190 (hta1-S122A-HTB1, HIS3) | This study |
| JDY1 |
MATaura3-52 trp1Δ leu2-3, 112 his3Δ200 Δrad52::TRP1 |
Downs et al. (2000) | |
| Y865 | MATaura3-52 trp1-289 his3Δ1 leu2-3,112 gal2 gal10 | Costigan et al. (1994) | |
| Y869 | MATaura3-52 trp1-289 his3Δ1 leu2-3,112 gal2 gal10 nhp6A-Δ2::URA3 nhp6B-Δ3::HIS3 | Costigan et al. (1994) | |
| JDY94 | hta1-S122A; (hta2-htb2)Δ::TRP1 | This study |
Sensitivity assays:
Midlog cultures were diluted to a density corresponding to an absorbance of 0.3 at 600 nm and fivefold serial dilutions were plated onto medium containing the indicated concentration of DNA-damage- or cellular-stress-inducing agents. All assays were done using rich medium [yeast extract, peptone, adenine sulfate, dextrose (YPAD); Formedium] with the exception of the complementation experiment (Figure 6E), which was done on synthetic dropout medium (Formedium) lacking histidine and uracil. Plates were incubated for 3–4 days at 30°.
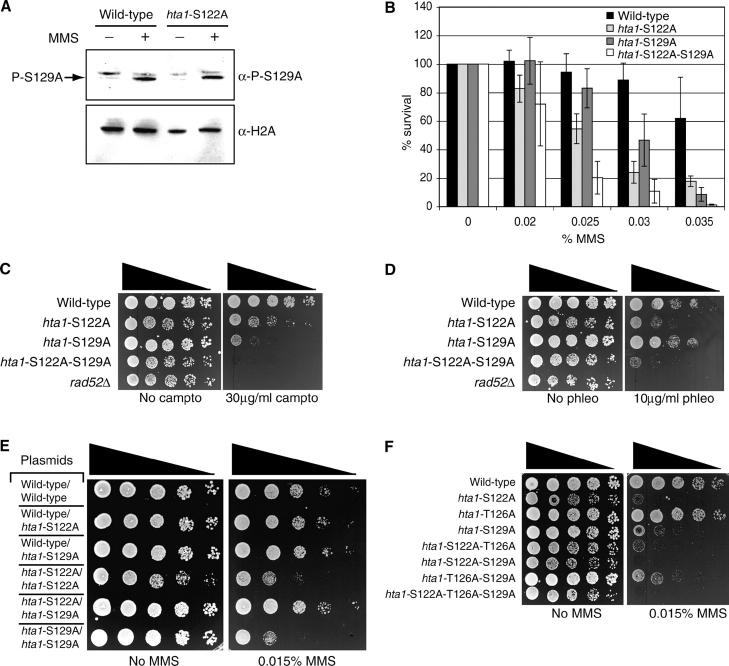
Figure 6.—
Histones H2A S122 and S129 provide distinct functions. (A) Western blot of wild-type (FHY2) and hta1-S122A mutant lysates prepared from cultures grown with or without 0.1% MMS. (Top) Lysates analyzed with antibody against histone H2A phospho-S129. (Bottom) Lysates analyzed using antihistone H2A antiserum. (B) Survival of indicated hta1-mutant strains and their isogenic wild-type control on medium containing indicated concentrations of MMS. (C and D) Serial dilutions of midlog cultures of the indicated strains plated onto medium containing the indicated agents. (E) Serial dilutions of midlog cultures of haploid strains harboring two H2A-encoding plasmids were plated onto selective medium containing the indicated concentration of MMS. (F) Serial dilutions of midlog cultures of the indicated strains grown in the presence or absence of MMS.
To quantitatively measure survival in the presence of methyl methanesulfonate (MMS), midlog cultures were diluted to a density corresponding to an absorbance of 0.002 at 600 nm and plated onto medium containing the indicated concentration of MMS. Plates were incubated at 30° for 3 days and then at 23° for 2 days. The number of surviving colonies at each concentration was counted and plotted as a percentage relative to the number of surviving colonies on medium lacking MMS.
To measure sensitivity to the expression of the EcoRI endonuclease, equal numbers of midlog yeast cells containing either the endonuclease gene under the control of the GAL1-10 promoter or an empty vector were grown in media containing 2% galactose as the sole carbon source. Surviving colonies were then counted after 4 days and are presented as the percentage of survival of the strains containing the endonuclease relative to the survival of the strains containing the empty vector. To measure sensitivity to the expression of the HO endonuclease, equal numbers of midlog yeast cells containing the HO endonuclease under the control of the GAL1-10 promoter were plated onto media containing either glucose or galactose as the sole carbon source, and surviving colonies were counted after 4 days. Survival is presented as the percentage of colonies on galactose-containing media relative to colonies on glucose-containing media. These assay were performed using strain JDY94 with or without complementation with wild-type HTA1.
Western blot analysis:
For preparation of whole-cell extracts for Western blot analysis, midlog cultures were either treated with 0.1% MMS or left untreated for 1 hr at 30° and then lysed using glass bead disruption into 20% trichloracetic acid. Lysates were electrophoresed on 18% (for H2A analysis) or 7.5% (for Rad53 analysis) SDS polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked using 5% (w/v) milk (Marvel) in Tris-buffered-saline containing 0.1% (v/v) Tween 20 (TBS-T) and then incubated with the indicated polyclonal primary antibody for 2 hr (for H2A and Tip49a analysis) or overnight (for Rad53 analysis) diluted in TBS-T. To detect H2A-S129 phosphorylation, an affinity-purified antibody specific for this modification (Downs et al. 2000) was used at a 1:1000 dilution. To detect H2A protein levels, antiserum that recognizes H2A regardless of S129 phosphorylation status (Downs et al. 2000) was used at a 1:4000 dilution in 5% milk in TBS-T. To detect Rad53, an antibody that recognizes unphosphorylated and hyperphosphorylated forms of the protein (gift of N. Lowndes) was used at 1:10,000 dilution. To detect Tip49a, an antibody raised against recombinant Tip49a was used at 1:1000 in TBS-T (gift of J. Cote). Signal was detected using horseradish-peroxidase-coupled anti-rabbit secondary antibody (Pierce, Rockford, IL) and enhanced chemiluminescence (Pierce).
Micrococcal nuclease digestion:
Spheroplasts were prepared according to a protocol developed by Kent and Mellor (1995). Briefly, 100-ml midlog cultures were harvested and normalized by measuring the OD600nm. Cell pellets were washed once in water, resuspended in 950 μl of freshly made YLE buffer [10 mg/ml zymolyase, 20,000 units/g (ICN), 1 m sorbitol, and 5 mm β-mercaptoethanol) and then incubated for 15 min at room temperature. The resulting spheroplasts were collected by centrifugation, gently washed twice in 950 μl of 1 m sorbitol, and resuspended in 1.2 ml of spheroplast digestion buffer (1 m sorbitol, 50 mm NaCl, 10 mm Tris-HCl pH 7.5, 5 mm MgCl2, 1 mm β-mercaptoethanol, 0.5 mm spermidine, and 0.075% nonidet P40). To this, 15 μl of 2.86 units/ml micrococcal nuclease (MNase; Sigma, St. Louis) was added and the reaction was incubated at 37°. At the indicated time points, 200-μl aliquots were removed and added to fresh microfuge tubes containing 20 μl of stop solution (5% SDS, 250 mm EDTA). The DNA was then purified by phenol/chloroform extraction and ethanol precipitation, analyzed on a 1% agar gel, and visualized with ethidium bromide.
Sporulation analysis:
Strains were grown overnight in YPAD, diluted 1:50 into YPA, and grown another 24 hr at 30°. The cultures were then washed two times in water and resuspended in sporulation media. After 5 days in sporulation media, cells were fixed in formaldehyde and examined microscopically for the presence of spores. Minimally, 300 cells were counted for each culture, and three independent sporulation cultures were analyzed per strain.
DNA damage checkpoint analysis:
Yeast cultures were grown to midlog phase and treated with 15 μg/ml nocodazole (Sigma) for 2.5 hr at 30°. One hour prior to release from nocodazole, MMS was added to one set of cultures to a final concentration of 0.10%. The cultures were then washed once and resuspended in fresh medium. Aliquots were removed at the indicated times, fixed with 5% formaldehyde, and sonicated. Samples were analyzed microscopically for the presence of large-budded cells (defined as cells with buds >70% the size of the mother cell).
RESULTS
H2A S122 is important for normal growth and for survival in the presence of MMS:
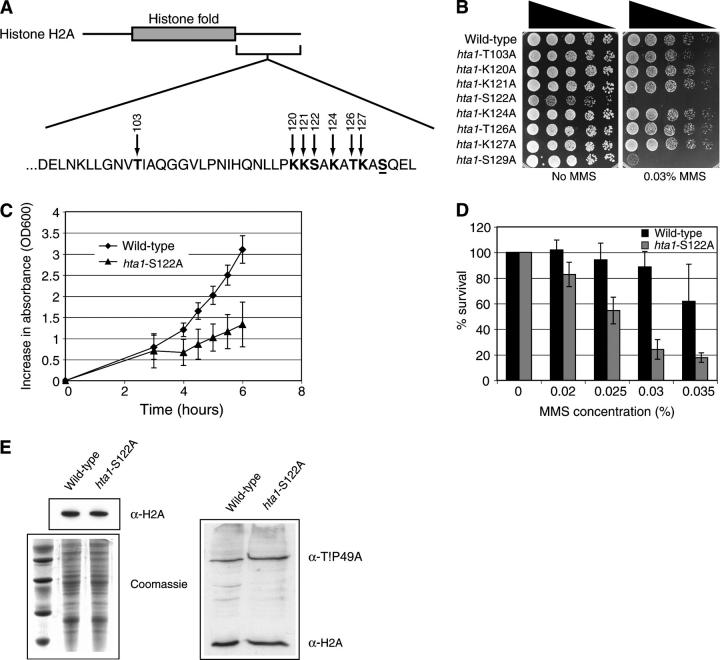
To determine whether residues in the H2A C-terminal tail, other than S129, were important for DNA repair, we examined all residues that have the potential to be covalently modified by using site-directed mutagenesis to change them to alanine residues. We did this using a plasmid-borne copy of wild-type H2A and H2B (HTA1-HTB1) in a haploid yeast strain in which both copies of the chromosomal H2A and H2B loci (HTA1-HTB1 and HTA2-HTB2) had been disrupted (Hirschhorn et al. 1995). An illustration of the mutagenized residues is shown in Figure 1A. While some of the residues that were mutagenized are part of the solved structure of the nucleosome, they are all downstream of the histone fold (Figure 1A; White et al. 2001). Strains harboring the mutagenized H2A constructs were analyzed for their ability to survive in the presence of the DNA-damage-inducing agent MMS. Notably, one strain, hta1-S122A, showed detectable sensitivity to MMS relative to the wild-type strain (Figure 1B).
Figure 1.—
Histone H2A S122 is important for survival in the presence of MMS. (A) Schematic showing the amino acid sequence of the S. cerevisiae histone H2A C-terminal tail. Residues that were changed to alanine are indicated by arrows. Serine 129 is underlined. (B) Serial dilutions of hta1-mutant yeast strains and their isogenic wild-type control (FHY2) grown with or without 0.03% MMS. (C) Growth of wild-type (diamonds) and hta1-S122A mutant (triangles) cultures grown at 30° was monitored by changes in OD600nm. (D) Survival of wild-type and hta1-S122A mutant yeast on MMS-containing plates. (E) Western blot analysis with antihistone H2A antiserum (top left) and Coomassie-stained SDS polyacrylamide gel (bottom left) of yeast lysates prepared from wild-type (lane 2) and hta1-S122A mutant strains (lane 3). Lane 1 contains molecular weight markers. Western blot analysis with anti-H2A and anti-Tip49a antibodies (right).
In performing these experiments, we observed that the hta1-S122A strain grew more slowly than the wild-type strain even in the absence of DNA damage (Figure 1B, left). We therefore examined the growth rates of wild-type and hta1-S122A yeast during exponential phase and confirmed that the hta1-S122A population has a significantly longer doubling time than the wild-type strain (Figure 1C). This phenotype makes it difficult to determine precisely how sensitive the hta1-S122A strain is to DNA damage compared to wild-type when using the qualitative spot tests. We therefore examined the ability of the strain to survive in the presence of MMS using a more quantitative approach. Yeast cultures were grown into midlog phase and equal numbers of cells were plated onto media containing variable amounts of MMS. The plates were then incubated for up to 5 days to facilitate detection of even very-slow-growing cells before colonies were counted. Doing this, we found that the hta1-S122A strain is significantly and reproducibly more sensitive to MMS than the wild-type strain (Figure 1D), indicating that the sensitivity to MMS seen in the spot tests relative to wild-type yeast is not due to the slow growth of the hta1-S122A strain. One possible mechanism by which mutation in a core histone might sensitize cells to DNA damage is by causing histone protein instability. We therefore examined the level of H2A by Western blot analysis in wild-type and hta1-S122A mutant strains and compared this to overall protein levels as well as to Tip49a protein levels and found no detectable difference (Figure 1E). Therefore, the S122A mutation does not markedly affect H2A protein stability.
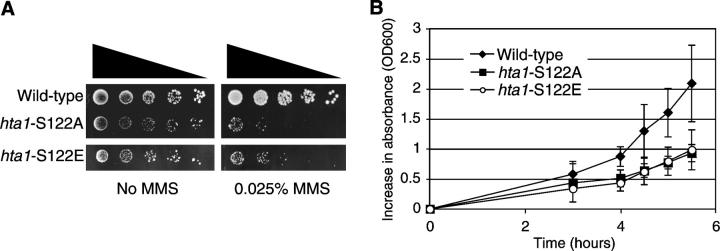
Interestingly, it has already been shown that H2A S122 is phosphorylated in vivo (Wyatt et al. 2003), although the timing and location of phosphorylation as well as the biological role of this event are as yet unknown. To investigate the possibility that phosphorylation of H2A S122 is important for survival in the presence of DNA damage, we created a strain in which S122 was replaced with a glutamic acid residue, which can, in some cases, mimic phosphorylation. This strain, however, appeared to be as sensitive to DNA damage as the hta1-S122A strain (Figure 2A), and exhibited the same slow growth as the hta1-S122A strain (Figure 2B). We next raised an antiserum against a peptide containing a phosphoserine residue corresponding to S122. While this antiserum was able to recognize the phospho-peptide, we were unable to detect phosphorylation of H2A S122 by Western blot analysis of both whole-cell extracts and histone preparations prepared from wild-type yeast under a variety of conditions, including treatment with MMS (data not shown). Therefore, while it is clear that H2A S122 is important for survival in the presence of MMS, we are unable to determine whether phosphorylation of this residue is important for this function.
Figure 2.—
An H2A S122E mutant strain has a phenotype that is indistinguishable from the H2A S122A mutant strain. (A) Serial dilutions of midlog cultures of hta1-mutant strains and their isogenic wild-type control (FHY2) grown with or without 0.025% MMS. (B) Growth of wild-type (diamonds), hta1-S122A mutant (squares), and hta1-S122E mutant (circles) cultures grown at 30° was monitored by changes in OD600nm.
DNA-damage-induced checkpoint responses appear to be normal in hta1-S122A mutant yeast:
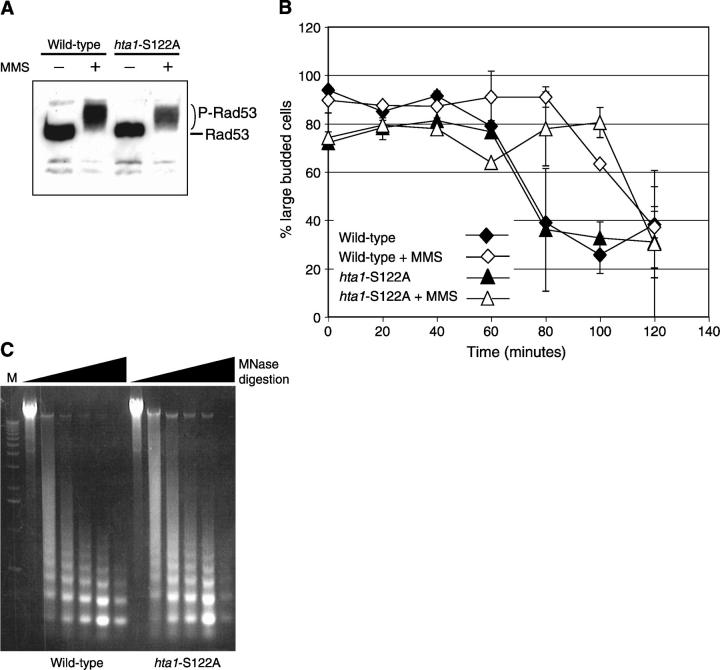
To more fully understand the role that H2A S122 plays in facilitating survival in the presence of MMS, we examined the ability of hta1-S122A mutant cells to respond appropriately to DNA damage. It has been shown that exposure of cells to MMS results in the rapid activation of the Mec1 kinase leading to hyperphosphorylation of Rad53 (Sanchez et al. 1996). These events are necessary for the ability of cells to appropriately upregulate DNA-damage-responsive transcription and to mediate cell cycle arrest (Weinert 1998). After treatment with MMS, we found that Rad53 is hyperphosphorylated in hta1-S122A mutant cells to a degree similar to that detected in wild-type cells (Figure 3A), indicating that the ability of Mec1 to detect damaged DNA and to initiate the signal transduction cascade is intact in the hta1-S122A mutant strain. Additionally, we examined the ability of the hta1-S122A mutant strain to arrest progression through the cell cycle in response to DNA damage. To do this, we first arrested midlog cultures in the G2/M phase with nocodazole and then treated one set of cultures with MMS for 1 hr. The MMS and nocodazole were then washed out, and the cells were examined microscopically to monitor their cell cycle progression. We found that the MMS-treated hta1-S122A mutant cells remained arrested as large-budded cells for ∼40 min longer than untreated cells (Figure 3B). This was in good agreement with the MMS-induced arrest seen in the wild-type strain (Figure 3B), indicating that the G2/M DNA damage checkpoint is intact in the hta1-S122A mutant strain.
Figure 3.—
Histone H2A S122 is not required for DNA damage checkpoint responses or gross chromatin structure. (A) Western blot analysis of wild-type (FHY2) and hta1-S122A lysates prepared from cultures grown with or without 0.1% MMS and analyzed using anti-Rad53 antiserum. (B) Wild-type (diamonds) and hta1-S122A mutant (triangles) cultures were arrested in G2/M using nocodazole and incubated with (open symbols) or without (solid symbols) 0.1% MMS. Cultures were released and the percentage of large-budded cells was calculated at the indicated time points. (C) An ethidium-bromide-stained gel of chromatin isolated from wild-type or hta1-S122A mutant yeast that was digested with MNase for various periods of time (0, 1, 2, 5, 10, or 20 min).
H2A S122 is not important for establishing normal global chromatin structure:
DNA lesions can occur nonrandomly in DNA, depending on the chromatin context. Consequently, one possible way in which a mutation in a core histone results in decreased survival after DNA damage is by altering the accessibility of DNA to the damaging agent. We therefore examined global chromatin structure by determining its sensitivity over time to digestion with MNase. Because MNase preferentially digests DNA in linker regions between nucleosomes, the rate of digestion can be used to determine whether there are any gross changes in chromatin structure. We found that the MNase digestion profile of chromatin from hta1-S122A mutant yeast showed no significant or reproducible differences from that of chromatin isolated from wild-type yeast (Figure 3C), suggesting that the sensitivity seen in this strain is unlikely to be due to increased accessibility of the DNA to the DNA-damaging agent.
Survival in the presence of DNA double-strand breaks is impaired in the hta1-S122A mutant strain:
Next, we tested our panel of histone H2A mutants for their ability to survive in the presence of a range of DNA-damaging agents that cause different DNA lesions. With the exception of hta1-K121A, which showed a weak sensitivity to camptothecin (Figure 4B), strains with mutations in other C-terminal tail residues of H2A did not show any significant phenotypes relative to the wild-type control in the presence of the drugs tested (Figure 4, A–F), suggesting that these residues are not individually important in mediating DNA damage responses in vivo. These results are in contrast to a previous report demonstrating that a strain with a T126A mutation in H2A is sensitive to bleomycin (Wyatt et al. 2003). There are numerous possible explanations for this discrepancy, including differences in the drugs tested, the assay conditions, and the yeast strains used in the two studies.
Figure 4.—
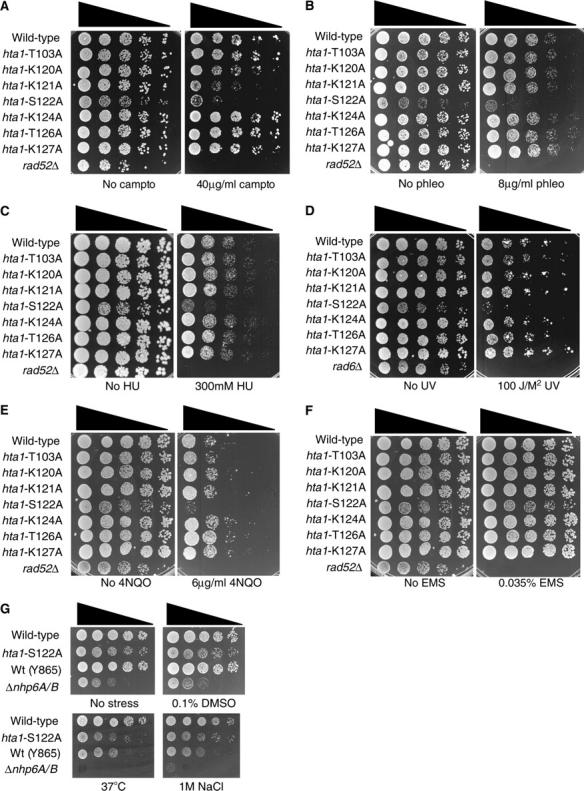
Histone H2A S122 is important for survival in the presence of DNA DSB-inducing agents. (A–G) Serial dilutions of midlog cultures of the indicated strains were plated onto medium containing the indicated concentrations of the specified agents.
When analyzing the agents to which these strains were sensitive, we found that the hta1-S122A mutant strain is sensitive to the topoisomerase inhibitor camptothecin, the radio-mimetic drug phleomycin, and, more weakly, to the dNTP synthesis inhibitor hydroxyurea, ultraviolet (UV) light, and the UV-mimetic drug 4-NQO (Figure 4, A–E). In contrast, we did not detect significant or reproducible sensitivity of the hta1-S122A mutant strain to the alkylating agent ethyl methanesulfonate (EMS) (Figure 4F). Furthermore, we tested the survival of the hta1-S122A mutant strain in the presence of other types of cellular stress. As a control, we compared the behavior of the hta1-S122A mutant strain to a strain lacking the HMG box-encoding genes NHP6-A and -B (Costigan et al. 1994). In contrast to this strain, the hta1-S122A mutant strain is not detectably sensitive to the presence of high salt, high temperature, low temperature, or DMSO (Figure 4G and data not shown), suggesting that there is no global defect in cellular stress responses. Taken together, these data suggest that H2A S122 is specifically important for DNA damage responses. Because the DNA-damaging agents to which the hta1-S122A strain is sensitive are capable of causing DNA double-strand breaks, either directly or indirectly, these data raise the possibility that H2A S122 facilitates DNA DSB repair.
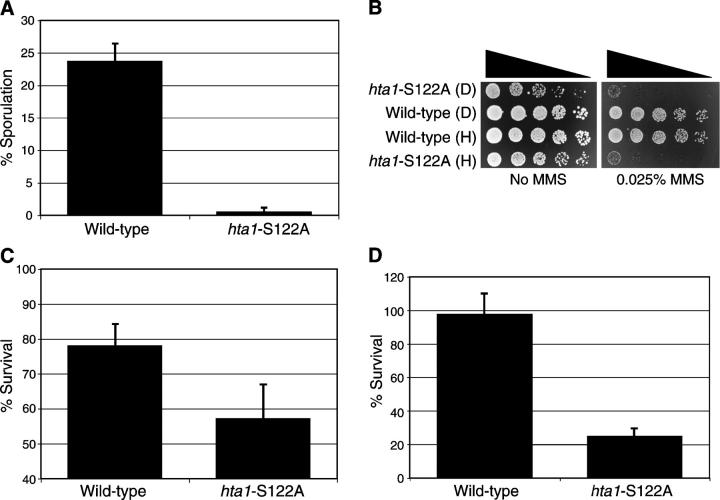
As previously mentioned, DNA DSB repair is performed by two major pathways in eukaryotes: HR and NHEJ. The repair of DNA DSBs by HR is an integral part of meiosis and strains lacking DNA damage signaling and repair genes are often defective in their ability to sporulate. We therefore analyzed the ability of the hta1-S122A strain to sporulate. Interestingly, after 5 days in sporulation media, the hta1-S122A mutant strain had ∼55-fold fewer spores than the wild-type strain (Figure 5A; 0.426 vs. 23.63%), demonstrating a defect in the ability to sporulate. It has previously been shown that NHEJ is downregulated in diploid cells when HR is the preferred pathway for DNA DSB repair (Valencia et al. 2001). We therefore examined the role of S122 in diploid strains in parallel with our haploid strains to see whether a difference in survival in the presence of DNA damage was apparent. In response to the presence of MMS, the diploid hta1-S122A mutant strain showed the same phenotype as the haploid hta1-S122A mutant strain (Figure 5B), indicating that the role of H2A S122 in mediating survival is not exclusive to haploid yeast. Taken together, these data are suggestive of a role in mediating HR responses. To further investigate this possibility, we examined the ability of hta1-S122A mutant yeast to survive in the presence of continual HO endonuclease expression. Survival under these conditions is severely compromised in strains lacking HR activity. In doing so, we found a modest, but reproducible decrease in survival when compared with the wild-type strain (Figure 5C). We additionally examined the ability of cells to survive in the presence of EcoRI expression, which severely compromises the ability of NHEJ-defective yeast to survive (Lewis et al. 1998). Interestingly, we found that survival in this assay was also reproducibly reduced in the hta1-S122A mutant yeast (Figure 5D). While this result is suggestive of a role for H2A S122 in NHEJ-mediated DNA repair responses, we note that the phenotype detected in the hta1-S122A mutant yeast strain is significantly less severe than that detected in strains lacking NHEJ components. Moreover, strains lacking genes required for HR are mildly sensitive to the overexpression of EcoRI (Lewis et al. 1998), making it difficult to definitively place H2A S122 on either DNA DSB repair pathways. Nevertheless, these assays clearly demonstrate that survival in the presence of DNA double-strand breaks is defective in the hta1-S122A mutant strain.
Figure 5.—
Histone H2A S122 is important for sporulation and survival in the presence of DNA double-strand breaks. (A) Sporulation levels of wild-type and hta1-S122A mutant cells after 5 days in sporulation media. (B) Serial dilutions of haploid (H) or diploid (D) wild-type and hta1-S122A mutant yeast grown in the presence or absence of MMS. (C) Survival of JDY94 complemented with wild-type HTA1 (wild type) or an empty vector (hta1-S122A) in the presence of continual HO endonuclease expression. Survival is presented as a percentage of colonies grown on galactose-containing media compared with strains grown on glucose-containing media. (D) Survival of JDY94 complemented with wild-type HTA1 (wild type) or an empty vector (hta1-S122A) in the presence of EcoRI expression. Survival is presented as a percentage compared with strains lacking the EcoRI endonuclease construct.
H2A S122 and S129 provide distinct functions:
It has previously been shown that H2A S129 is phosphorylated in response to DNA damage and that this event is important for the ability of yeast to survive in the presence of MMS (Downs et al. 2000). Because of the proximity of S122 to S129, we considered the possibility that the MMS sensitivity conferred by the S122A mutation was a result of impaired S129 phosphorylation or S129-dependent downstream events. However, by using an antibody specific for H2A phosphorylated at S129, we found no detectable loss of MMS-dependent S129 phosphorylation in the hta1-S122A mutant strain when compared with the wild-type strain (Figure 6A). Next, we made a strain in which both serine residues were replaced with alanine (hta1-S122A/S129A) and examined its survival in the presence of MMS compared with strains harboring each individual mutation (hta1-S122A or hta1-S129A). We found that the hta1-S122A mutant strain appears to be slightly more sensitive to MMS than the hta1-S129A mutant strain (Figure 6B). Importantly, the hta1-S122A/S129A strain is more sensitive than either single-mutant strain (Figure 6B), suggesting that the two residues provide distinct functions in response to DNA damage. We then tested the ability of the hta1-S122A, hta1-S129A, and hta1-S122A/S129A strains to survive in the presence of other types of DNA damage. We find that, as with MMS, the hta1-S122A mutant strain appears slightly more sensitive to phleomycin than the hta1-S129A mutant strain, but interestingly, the hta1-S122A mutant strain appears less sensitive to camptothecin than the hta1-S129A mutant strain (Figure 6, C and D). This change in sensitivity profile suggests that the two residues may perform functions that are of different importance, depending on the nature of the DNA lesion. In agreement with the postulation that these two residues have individual roles, we find that the hta1-S122A/S129A double mutant is significantly more sensitive to these agents than either single mutant (Figure 6, C and D). Furthermore, we find no defect in the ability of hta1-S129A yeast to sporulate (data not shown), again pointing toward separate cellular roles.
While the DNA damage sensitivity profiles of the hta1-S122A, hta1-S129A, and hta1-S122A/S129A mutant strains imply that the two residues work independently to facilitate DNA repair, there are other viable interpretations, particularly since these residues are in such close proximity on the same molecule. For instance, if both residues are important for binding by a common downstream factor and each single mutation reduces, but does not abrogate, binding, then the double-mutant strain in which binding is further impaired would have a more severe phenotype than either single-mutant strain. To discriminate between these possibilities, we performed a complementation experiment in which we supplied the yeast with two H2A-expressing plasmids. As expected, a strain containing two hta1-S122A mutant constructs was significantly more sensitive to MMS than a strain containing two wild-type constructs (Figure 6E; compare the top row to the the fourth row). This is also the case for a strain containing two hta1-S129A mutant constructs (Figure 6E; compare the top and bottom rows). Importantly, strains containing one wild-type H2A construct and one mutant H2A construct are no more sensitive to MMS than the wild-type strain is, demonstrating that the hta1-S122A and hta1-S129A mutations are both recessive (Figure 6E; compare the top row to the second and third rows). Importantly, however, if cells contain one hta1-S122A mutant construct and one hta1-S129A mutant construct, the strain is indistinguishable from the wild-type strain in the presence of DNA damage (Figure 6E; compare the top row to the fifth row), demonstrating that both mutant phenotypes can be complemented by the presence of the other mutant H2A. These data indicate that the two residues provide independent functions and that if both serines are present, regardless of whether they are on the same histone molecule, this is sufficient for the cell to respond appropriately and to survive in the presence of DNA damage.
As previously mentioned, it has been demonstrated that H2A S122 and S129 are phosphorylated (Downs et al. 2000; Wyatt et al. 2003). Interestingly, H2A T126 was also found to be phosphorylated, and in an hta1-S122P mutant strain, the amount of phosphorylation at other sites was increased (Wyatt et al. 2003). Therefore, one possible mechanism by which loss of H2A S122 affects DNA damage responses is via increased H2A T126 phosphorylation, which indirectly inhibits normal H2A S129 responses. The different profiles of hta1-S122A and hta1-S129A phenotypes and DNA damage sensitivities make this an unlikely explanation. Nevertheless, to test this hypothesis, we made an hta1-S122A/T126A double-mutant strain to remove the possibility that T126 was phosphorylated inappropriately. If H2A S122 contributes to DNA damage responses only by preventing inappropriate H2A T126 phosphorylation, then there should be no DNA damage sensitivity when both residues are changed to alanine residues. In contrast to this hypothesis, we find that the hta1-S122A/T126A double mutant is still sensitive to the presence of DNA damage (Figure 6F). Additionally, an hta1-T126A/S129A mutant strain is no more sensitive than an hta1-S129A mutant strain, and an hta1-S122A/T126A/S129A mutant strain is no more sensitive to DNA damage than an hta1-S122A/S129A mutant strain (Figure 6F), suggesting that H2A T126 plays no significant role in DNA damage responses in these assays. Importantly, these data indicate that H2A S122 provides a function in mediating DNA damage responses that is independent of H2A T126 and further supports the conclusion that the function is also independent of H2A S129.
DISCUSSION
Here we have demonstrated a role for H2A S122 in DNA DSB repair in yeast. While it has been shown that H2A S122 is phosphorylated (Wyatt et al. 2003), we were unable to determine whether phosphorylation of this serine is important for facilitating DNA damage responses. The surrounding sequence of S122 weakly agrees with the protein kinase C (PKC) consensus site, and interestingly, PKC in higher eukaryotes has been implicated in the DNA damage response (Yoshida et al. 2003). Furthermore, the analogous residue in Drosophila H2A (T119) has recently been found to be phosphorylated during mitosis (Aihara et al. 2004). A role for this residue during the cell cycle, even in the absence of exogenous DNA damage, may explain why our hta1-S122A mutant strain grows more slowly than wild type (Figure 1). Aihara et al. (2004) identified a kinase from the Drosophila extract, NHK1, which phosphorylates DmH2A T119 in vitro. Moreover, they demonstrated the existence of an activity in yeast extract that is able to phosphorylate the Drosophila residue, suggesting that this activity may also phosphorylate yeast H2A S122 and be responsible for the phosphorylation detected in the study by Wyatt et al. (2003). While we were unable to determine whether phosphorylation of H2A S122 is important for DNA repair responses, it seems reasonable to speculate that it is.
Regardless of whether H2A S122 is phosphorylated, these studies demonstrate that it is important in yeast for DNA damage responses and show that this residue provides a function independent from the known DNA-damage-responsive residue H2A S129. The role of histone H2A S129 in response to DNA damage has been shown to be conserved in a number of eukaryotes (Rogakou et al. 1999). As previously mentioned, the motif surrounding S129 exists on the major H2A species in some lower eukaryotes, such as Saccharomyces cerevisiae, but is found on histone variants in higher eukaryotes, such as H2AX in humans, and not in the major H2A species. We investigated the conservation of sequences homologous to S. cerevisiae H2A S122 and found that, while the sequence downstream of S122 is not well conserved, the sequence immediately upstream of S122 is highly conserved and is present in the eukaryotic organisms that we have examined thus far (Figure 7). The conserved upstream sequence is present in the major histone H2A encoding genes as well as in some H2AX and H2AF/Z variants. Moreover, S122 itself is reasonably highly conserved, and the analogous residue is a serine or threonine, again pointing to the strong possibility that phosphorylation will play a role in its cellular function. The degree of conservation of this region of histone H2A is suggestive of a conserved function, and it will be of great interest to determine whether the analogous residue in other eukaryotes is also important in DNA damage responses.
Figure 7.—
Lineup of C-terminal tails of histone H2A proteins from a range of eukaryotic species. S. cerevisiae S122 and homologous residues in the lineup are indicated with an arrow.
There are a number of potential mechanisms by which S122 could facilitate survival in the presence of DNA damage. One possibility is that S122 is important for the transcriptional response to DNA damage. Indeed, the fact that the hta1-S122A mutant strain has difficulty growing in the absence of exogenously added DNA damage may indicate that H2A S122 plays a role in transcriptional responses regardless of whether this is the causative mechanism of the DNA repair defect. Alternatively, H2A S122 may be important for providing binding sites in the vicinity of DNA lesions for repair factors. The regulation of any putative interactions could be modulated by covalent modification of the histone tail or of the binding partners, or both. The proximity of S122 to S129 suggests that both residues would be important for mediating interactions with binding partners. However, because we demonstrated that S122 and S129 provide independent functions and are able to provide complementary functions on separate histone proteins, this is unlikely. Instead, an attractive hypothesis is that S122 and S129 are important for binding to their respective protein partners in either a physically or a temporally distinct manner, such as in response to specific lesions or at particular points in the cell cycle.
Additionally, S122 might be necessary to create a chromatin structure that is compatible with the manipulation required for DNA repair. While the gross chromatin structure in an hta1-S122A-containing strain is indistinguishable from wild-type chromatin, it is possible that subtle differences in chromatin structure that would be undetectable in our assays could have profound effects on the ability of DNA in the vicinity of lesions to be appropriately manipulated. These possibilities are currently being investigated. From the data presented here, it is clear that multiple histone residues are important for mediating DNA repair and, with the emerging studies of chromatin-modulating activities in DNA repair, suggest the existence of a complex DNA-damage-dependent histone code.
Acknowledgments
We thank Mike Snyder, Noel Lowndes, Jacques Cote, and Fred Winston for strains and reagents and Steve Bell, Daniel Easton, and members of the J.A.D. lab for helpful discussions. This work was supported by Cancer Research UK grant SP2143/0403, and J.A.D. is a Jenner Fellow of the Lister Institute.
References
- Aihara, H., T. Nakagawa, K. Yasui, T. Ohta, S. Hirose et al., 2004. Nucleosomal histone kinase-1 phosphorylates H2A Thr119 during mitosis in the early Drosophila embryo. Genes Dev. 18: 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon et al., 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. [DOI] [PubMed] [Google Scholar]
- Burma, S., B. P. Chen, M. Murphy, A. Kurimasa and D. J. Chen, 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276: 42462–42467. [DOI] [PubMed] [Google Scholar]
- Costigan, C., D. Kolodrubetz and M. Snyder, 1994. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 14: 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs, J. A., N. F. Lowndes and S. P. Jackson, 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger et al., 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 17: 979–990. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo, O., and A. Nussenzweig, 2004. Linking histone deacetylation with the repair of DNA breaks. Proc. Natl. Acad. Sci. USA 101: 1427–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse and F. Winston, 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15: 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang et al., 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102: 463–473. [DOI] [PubMed] [Google Scholar]
- Kent, N. A., and J. Mellor, 1995. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 23: 3786–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, L. K., J. M. Kirchner and M. A. Resnick, 1998. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol. Cell. Biol. 18: 1891–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber, M. R., Y. Ma, U. Pannicke and K. Schwarz, 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4: 712–720. [DOI] [PubMed] [Google Scholar]
- Martini, E. M. D., S. Keeney and M. A. Osley, 2002. A role for histone H2B during repair of UV-induced DNA damage in Saccharomyces cerevisiae. Genetics 160: 1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W.-H. Wu, S. Sen et al., 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt et al., 2004. INO80 and g-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775. [DOI] [PubMed] [Google Scholar]
- Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert et al., 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10: 886–895. [DOI] [PubMed] [Google Scholar]
- Redon, C., D. R. Pilch, E. Rogakou, A. H. Orr, N. F. Lowndes et al., 2003. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova and W. M. Bonner, 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Rogakou, E. P., C. Boon, C. Redon and W. M. Bonner, 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146: 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, J., and S. P. Jackson, 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297: 547–551. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang et al., 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360. [DOI] [PubMed] [Google Scholar]
- Shen, X., G. Mizuguchi, A. Hamich and C. Wu, 2000. A chromatin remodeling complex involved in transcription and DNA processing. Nature 406: 541–544. [DOI] [PubMed] [Google Scholar]
- Shroff, R., A. Arbel-Eden, D. Pilch, G. Ira, W. M. Bonner et al., 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14: 1703–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus et al., 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 416: 666–669. [DOI] [PubMed] [Google Scholar]
- van Attikum, H., O. Fritsch, B. Hohn and S. M. Gasser, 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788. [DOI] [PubMed] [Google Scholar]
- Ward, I. M., and J. Chen, 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276: 47759–47762. [DOI] [PubMed] [Google Scholar]
- Weinert, T., 1998. DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell 94: 555–558. [DOI] [PubMed] [Google Scholar]
- West, S. C., 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4: 1–11. [DOI] [PubMed] [Google Scholar]
- White, C. L., R. K. Suto and K. Luger, 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20: 5207–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt, H. R., H. Liaw, G. R. Green and A. J. Lustig, 2003. Multiple roles for Saccharomyces cerevisiae histone H2A in telomere position effect, Spt phenotypes and double-strand-break repair. Genetics 164: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K., H.-G. Wang, Y. Miki and D. Kufe, 2003. Protein kinase Cδ is responsible for constitutive and DNA damage-induced phosphorylation of Rad9. EMBO J. 22: 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]