Abstract
This study evaluates the potential of flow cytometry for chromosome sorting in durum wheat (Triticum turgidum Desf. var. durum, 2n = 4x = 28). Histograms of fluorescence intensity (flow karyotypes) obtained after the analysis of DAPI-stained chromosomes consisted of three peaks. Of these, one represented chromosome 3B, a small peak corresponded to chromosomes 1A and 6A, and a large peak represented the remaining 11 chromosomes. Chromosomes sorted onto microscope slides were identified after fluorescence in situ hybridization (FISH) with probes for GAA microsatellite, pSc119.2, and Afa repeats. Genomic distribution of these sequences was determined for the first time in durum wheat and a molecular karyotype has been developed for this crop. Flow karyotyping in double-ditelosomic lines of durum wheat revealed that the lines facilitated sorting of any arm of the wheat A- and B-genome chromosomes. Compared to hexaploid wheat, flow karyotype of durum wheat is less complex. This property results in better discrimination of telosomes and high purities in sorted fractions, ranging from 90 to 98%. We have demonstrated that large insert libraries can be created from DNA purified using flow cytometry. This study considerably expands the potential of flow cytogenetics for use in wheat genomics and opens the possibility of sequencing the genome of this important crop one chromosome arm at a time.
DURUM wheat (Triticum turgidum Desf. var. durum) is an allotetraploid species with 2n = 4x = 28 (AABB genome) that originated through intergeneric hybridization and polyploidization involving two diploid grass species: T. urartu (2n = 2x = 14, AA genome) and a B-genome diploid related to Aegilops speltoides (2n = 2x = 14, SS genome) (Kihara 1944; McFadden and Sears 1946). Molecular genetic data indicate that T. aestivum (the cultivated bread wheat) arose from hybridization of T. turgidum (durum or pasta wheat) and Ae. tauschii only 8000 years ago (Huang et al. 2002). Thus the A- and B-genome chromosomes in these two cultivated wheat species are >99% identical.
Durum wheat is an important cereal whose grain is used predominantly for food products such as pasta, couscous, and burghul. It is extensively cultivated not only in the Mediterranean Basin, but also in Argentina, Australia, Canada, India, the United States, and several other countries. The annual worldwide grain production of durum wheat is estimated at 27.5 million metric tons, >10% of the worldwide wheat production. Losses due to disease, pest, and environmental problems each year influence durum production. It is expected that the use of molecular techniques will speed the development of improved varieties and enable creation of novel germplasm that cannot be obtained by classical approaches. However, isolation of important genes in wheat is a major challenge and a prerequisite for the exploitation of such molecular techniques.
Recent analyses of the wheat genome organization indicate that genes are clustered in gene-rich islands where the gene density is similar to that found in small-genome species (Erayman et al. 2004). The presence of small gene-rich islands should facilitate gene cloning based on recombination mapping, originally considered difficult in polyploid species. Indeed, several laboratories reported successful map-based cloning in wheat (Faris et al. 2003; Huang et al. 2003; Yan et al. 2003). This success renewed interest in the construction of physical maps, which are based on large insert DNA libraries, through global fingerprinting and/or genetic anchoring to high-resolution maps (Lander et al. 2001; Luo et al. 2003). A bacterial artificial chromosome (BAC) library has already been prepared for durum wheat (Cenci et al. 2003). Because of the large genome size of durum wheat (1C 13,000 Mbp, Bennett and Leitch 1995), 516,096 clones were required to achieve a 5.1× genome coverage. While the library is a unique genomic resource, its size makes its maintenance and handling technically demanding.
Since the early 1990s, we have pioneered the use of flow cytogenetics for targeted analysis of plant genomes. The strategy relies on purification of specific chromosomes using flow cytometry and is especially attractive for species with large genomes (Doležel et al. 1994, 2004). Flow-sorted chromosomes have been used for physical mapping using fluorescence in situ hybridization (FISH) and PCR with specific primers (Lysák et al. 1999; Kejnovský et al. 2001; Valárik et al. 2004), integration of genetic and physical maps (Neumann et al. 2002; Vláčilová et al. 2002), and targeted isolation of molecular markers (Požárková et al. 2002; Román et al. 2004). Recently, chromosomes sorted from hexaploid wheat were used for preparation of the first subgenomic BAC library specific for chromosomes 1D, 4D, and 6D (Janda et al. 2004), a BAC library specific for chromosome 3B (Šafář et al. 2004), and a BAC library for the short arm of chromosome 1B (J. Janda, J. Šafář, M. Kubaláková, J. Bartoš, P. Kovářová, P. Suchánková, S. Pateyron, J. Číhalíková, P. Sourdille, H. Šimková, M. Bernard, A. Lukaszewski, B. Chalhoub and J. Doležel, unpublished results). These genomic resources should greatly facilitate physical mapping and gene cloning in hexaploid wheat.
Flow cytometric purification of single chromosomes and chromosome arms in barley, rye, and hexaploid wheat relies on the use of cytogenetic stocks, such as telosomic and chromosome addition lines (Kubaláková et al. 2002, 2003; P. Suchánková, M. Kubaláková, P. Kovářová, J. Bartoš, J. Číhalíková, T. Endo and J. Doležel, unpublished results). These were first created by Sears (1966), who demonstrated that in hexaploid wheat (2n = 6x = 42, AABBDD genome), a specific chromosome in each of the genomes (A, B, or D) could compensate for the loss of a homeologous chromosome in the other genomes. Sears' work also indicated the possibility of substituting a D-genome chromosome for its homeologue from the A- or B-genome of tetraploid durum wheat. Crosses of these aneuploid stocks with tetraploid durum wheat followed by repeated backcrossing and selection of appropriate progeny lead to the development of a complete set of D-genome disomic substitution lines (Joppa and Williams 1988; Joppa 1993). Joppa (1993) also generated a set of durum wheat lines carrying separate arms of each chromosome (double-ditelosomic lines; 2n = 26 + 2tL + 2tS) from crosses to hexaploid wheat aneuploids. An additional set of durum double-ditelosomic lines have also been produced independently (K. Nishikawa, unpublished data). In addition to the nullisomic-tetrasomic, D-genome chromosome substitution lines, and double-ditelosomic lines, a plethora of other aneuploids in durum and hexaploid wheat exists (see http://www.ksu.edu/wgrc/; Sears 1969; Joppa 1993).
In this study individual chromosomes were isolated from various cultivars of durum wheat, from a strain of T. turgidum Desf. var. melanopus with 2n = 30 and from a series of double-ditelosomic lines to explore the feasibility of sorting individual chromosomes and chromosome arms. A set of repetitive DNA probes identified in this study was used to develop the first molecular karyotype of durum wheat. These and the protocols developed provide an important step toward the development of BAC libraries for individual wheat chromosome arms.
MATERIALS AND METHODS
Plant material:
Seeds of T. turgidum Desf. var. durum (2n = 4x = 28) cv. Creso and cv. Langdon, double-ditelosomic (dDt) lines of cv. LD222 (dDt 1A, dDt 1B, dDt 2A, dDt 2B, dDt 3A, dDt 3B, dDt 7B) (K. Nishikawa, unpublished data) and cv. Langdon (dDt1A and dDt1B) (Joppa 1993), and seeds of a strain of T. turgidum Desf. var. melanopus (Al.) Körn with 2n = 30 (Tsunewaki 1963) were used in this study. The strain is believed to have originated spontaneously in Tibet and its karyotype consists of 13 metacentric and submetacentric chromosomes and 2 telocentric chromosomes whose origin was not clear (Tsunewaki 1963).
Cell cycle synchronization and accumulation of metaphases:
The protocol of Vrána et al. (2000) was used, with modifications. All incubations were performed in the dark at 25° ± 0.5°, and all solutions were aerated. Seeds were germinated on moistened filter paper in glass petri dishes for 2–3 days to achieve optimal root length of 2–3 cm. Seedlings were transferred to an open mesh basket positioned on a tray filled with Hoagland's nutrient solution (Gamborg and Wetter 1975) containing 1.25 mm hydroxyurea (HU) and incubated for 18 hr. Then the roots were washed in distilled water and cultured in HU-free Hoagland's solution. To accumulate cells at metaphase, the roots were treated with Hoagland's solution containing amiprophos-methyl (APM; 2.5 μm) 5 hr after recovery from HU and incubated overnight in ice water (1°–2°).
Preparation of chromosome suspensions:
Suspensions of intact chromosomes were prepared according to Vrána et al. (2000). Briefly, 30 roots were cut 1 cm from the root tip, rinsed in deionized water, and fixed in 2% (v/v) formaldehyde in Tris buffer at 5° for 20 min. After three washes in Tris buffer for 5 min at 5°, the meristem tips (∼1 mm) were excised and transferred to a 3.5-ml polystyrene tube containing 1 ml of LB01 buffer (Doležel et al. 1989) at pH 9. Metaphase chromosomes were released after homogenization with a Polytron PT1300 homogenizer (Kinematica AG, Littau, Switzerland) at 20,000 rpm for 13 sec. The suspension was passed through a 50-μm pore size nylon mesh to remove large cellular debris and stored on ice until analyzed on the same day.
Chromosome analysis and sorting:
The samples were analyzed using a FACSVantage SE flow cytometer (Becton Dickinson, San Jose, CA) equipped with an argon ion laser set to multiline UV and 300 mW output power. A solution of 50 mm NaCl was used as sheath fluid. Chromosome suspensions were stained with DAPI at a final concentration of 2 μg/ml and analyzed at rates of 200–400 particles/sec. DAPI fluorescence was acquired through a 424/44 band-pass filter. Approximately 30,000 chromosomes were analyzed from each sample and data were collected using Cell Quest software (Becton Dickinson). The results were displayed as histograms of relative fluorescence intensity (flow karyotypes). To verify chromosome content of individual peaks on the flow karyotype, 1000 chromosomes were sorted from each peak at rates of ∼5–10/sec onto a microscope slide into a 15-μl drop of PRINS buffer supplemented with 5% sucrose (Kubaláková et al. 1997), air dried, and used for FISH with probes for DNA repeats that give chromosome-specific fluorescent-labeling patterns.
FISH:
Probes for 5S rDNA labeled by biotin were prepared using PCR with a pair of specific primers (RICRGAC1, RICRGAC2), which amplify 303 bp in rice (Fukui et al. 1994) using rice genomic DNA as a template. A DNA clone Radka1, carrying part of the 45S rDNA of Musa (Valárik et al. 2002), was labeled by biotin using PCR with a pair of T3 and T7 primers. Digoxigenin-labeled probe for GAA microsatellites was prepared using PCR with (GAA)7 and (CCT)7 primers and rye genomic DNA as a template; a probe for telomeric repeats labeled by digoxigenin was prepared using PCR with (AGGGTTT)3 and (CCCTAAA)5 primers. A 260-bp fragment of the Afa family repeat was prepared using PCR with primers AS-A and AS-B on wheat genomic DNA (Nagaki et al. 1995). The product was separated by agarose gel electrophoresis and the 260-bp band was cut from the gel and the DNA was labeled by digoxigenin or biotin using PCR and the same primers. The DNA clone pSc119.2, which was originally isolated from rye and contains highly repeated sequences from a 120-bp repeat family (Bedbrook et al. 1980), was labeled by digoxigenin using PCR with M13 primers and DNA clone pSc119.2 as a template.
FISH on flow-sorted chromosomes was performed according to Kubaláková et al. (2003). The sites of digoxigenin-labeled probe hybridization were detected using anti-digoxigenin-FITC raised in sheep (Roche Molecular Biochemicas, Mannheim, Germany) and the signal was amplified using anti-sheep-FITC (Vector Laboratories, Burlingame, CA). Biotin-labeled probes were detected using Cy3-labeled streptavidin and amplified using biotinylated streptavidin and another layer of Cy3 streptavidin. Chromosome preparations were counterstained with 0.2 μg/ml DAPI and mounted in Vectashield antifade solution (Vector Laboratories). The preparations were evaluated using an Olympus BX60 microscope equipped with optical filter sets appropriate for DAPI, fluorescein, and Cy3 fluorescence. The images of DAPI, fluorescein, and Cy3 fluorescence were acquired separately with a b/w CCD camera, which was interfaced to a PC running the ISIS software (Metasystems, Altussheim, Germany). The images were superimposed after contrast and background optimization.
RESULTS
Preliminary experiments indicated that the protocol for cell cycle synchronization in hexaploid wheat (Vrána et al. 2000) was not suitable for durum wheat. The root-tip cells arrested with 2 mm HU recovered much later than expected. Subsequently, we found that at 1.25 mm HU, the recovery pattern approached that observed in hexaploid wheat. Maximum mitotic activity (53% cells in mitosis) was observed 5 hr after the release from the HU block. The timing of the APM block to accumulate synchronized cells in metaphase was established after analyzing chromosome suspensions obtained from root tips treated with APM at various intervals during the recovery. Flow karyotypes with the least amount of debris background and highest resolution of chromosome peaks were obtained 5 hr after the recovery from HU and 2 hr APM treatment.
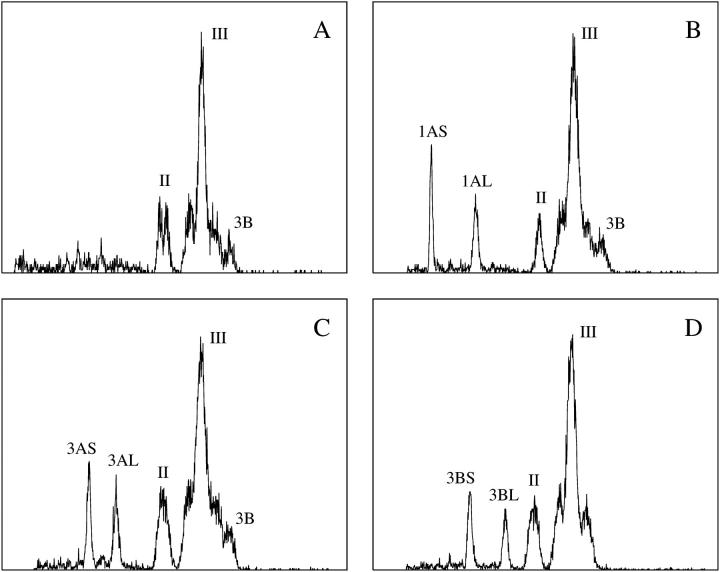
Unlike the cell cycle synchronization, no modification of the chromosome isolation protocol (Vrána et al. 2000) was needed. Suspensions of intact chromosomes were obtained after fixation in 2% formaldehyde (5°, 20 min). On average, 5 × 105 chromosomes could be isolated from 30 root tips. The analysis of DAPI-stained chromosome suspensions prepared from durum wheat cv. Langdon resulted in flow karyotypes with two composite peaks labeled II and III, and a peak corresponding to chromosome 3B (Figure 1A). Similar flow karyotypes were also obtained for cv. Creso and others cultivars (not shown). To label the peaks on durum wheat flow karyotype, we followed the procedure for labeling of hexaploid wheat as proposed by Vrána et al. (2000). The composite peak I of the hexaploid wheat flow karyotype represented three chromosomes of the D genome and hence is absent in durum wheat.
Figure 1.—
Flow karyotypes (histograms of relative fluorescence intensities) obtained after the analysis of DAPI-stained chromosome suspensions prepared from durum wheat “Langdon” with a standard karyotype and from three dDt lines (B–D). (A) The karyotype of Langdon consists of two composite peaks (II and III) representing specific groups of chromosomes and a peak representing chromosome 3B. Flow karyotypes of dDt lines show extra peaks representing short and long chromosome arms. (B) Langdon dDt1A (2n = 26 + 2t1AL + 2t1AS). Note that peak II represents only chromosome 6A in this line. (C) LD222 dDt3A (2n = 26 + 2t3AL + 2t3AS). (D) LD222 dDt3B (2n = 26 + 2t3BL + 2t3BS). x-axis, relative DAPI fluorescence intensity; y-axis, number of events.
With the aim of identifying the chromosome content of the three peaks in the flow karyotype of durum wheat, chromosomes were sorted onto microscope slides and subjected to FISH with probes for the Afa family repeat, GAA microsatellites, 5S rDNA, and pSc119.2 (Figure 2A). The analysis showed that peak II represented chromosomes 1A and 6A, while peak III represented chromosomes 2A, 3A, 4A, 5A, 7A, and all B-genome chromosomes with the exception of chromosome 3B. Our observations imply that only chromosome 3B, which was discriminated as a separated peak, could be sorted with a high purity (>90%) in durum wheat.
Figure 2.—
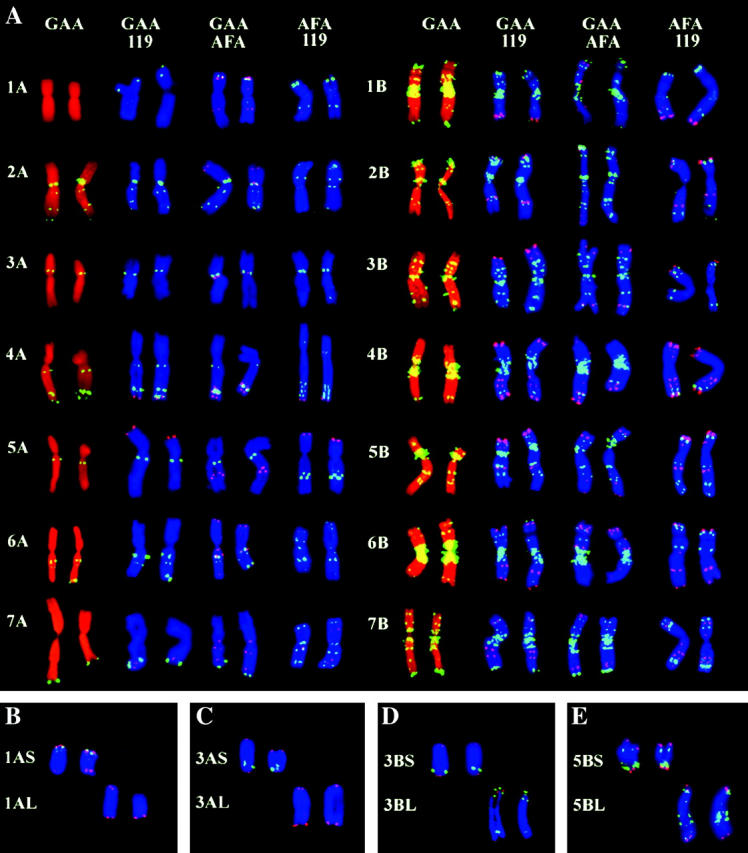
Genomic distribution of repetitive DNA sequences obtained after FISH on flow-sorted chromosomes. Labeled DNA probes were detected with either fluorescein (yellow-green) or Cy3 (red). Chromosomes were counterstained with either propidium iodide (red) or DAPI (blue). For each chromosome, two representative examples are given. (A) Distribution of three repetitive DNA sequences, pSc119.2 (119), GAA microsatellite (GAA), and Afa family repeat (AFA), on A- and B-genome chromosomes of the durum wheat Langdon. (B–E) Chromosome arms sorted from ditelosomic lines of durum wheat after FISH with probes for GAA microsatellite (yellow-green) and telomeric repeat (red).
We took advantage of performing FISH on large numbers of sorted chromosomes by analyzing the genomic distribution of a set of repetitive DNA sequences. Representative examples of fluorescent-labeling patterns that were obtained are given in Figure 2A. In general, the fluorescent-labeling patterns obtained with the probes were similar to the pattern observed for group A and B chromosomes of hexaploid wheat for the GAA satellite and Afa family repeat (Pedersen and Langridge 1997) and for the pSc119.2 and Afa family repeats (Mukai et al. 1993). However, some minor additional bands were observed in durum wheat for the three DNA repeats. For example, small bands of Afa repeats were localized on the durum chromosomes 2A–7A, 2B, 4B, and 5B (Figure 2A). These bands are missing in the hexaploid wheat chromosomes (Pedersen and Langridge 1997). The idiogram representing the genomic distribution of the four repeat sequences in durum wheat is shown in Figure 3. The differences in fluorescent-labeling patterns facilitated identification of each of the 28 chromosome arms of durum wheat.
Figure 3.—
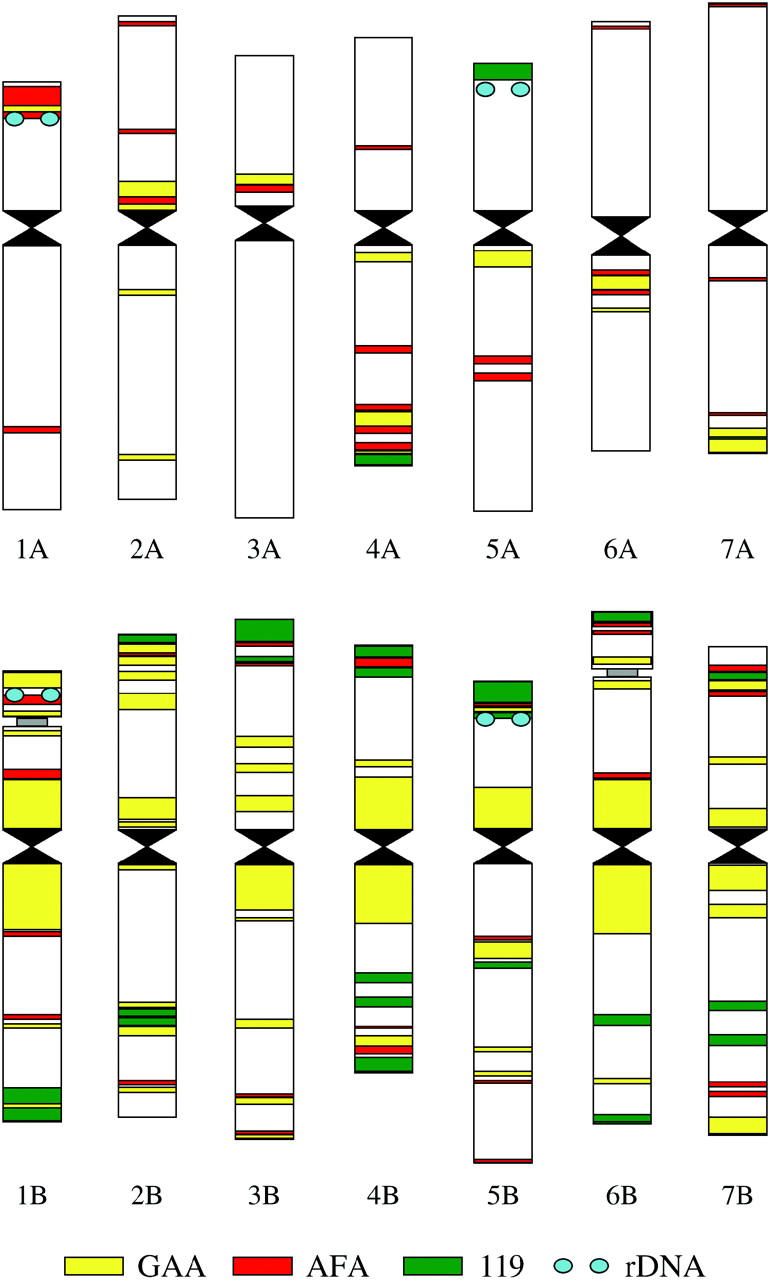
Idiogram of the durum wheat Langdon showing genomic distribution of four repetitive DNA sequences: GAA microsatellite (GAA), Afa family repeat (AFA), pSc119.2 (119), and 5S rDNA (rDNA). Differences in labeling pattern facilitate identification of each of the 28 chromosome arms.
The successful preparation of suspensions of intact chromosomes suitable for flow cytometry stimulated experiments focused on sorting single chromosome arms from dDt lines. The analysis of a representative set of seven dDt lines, which included chromosome 1AS with the lowest relative DAPI fluorescence and chromosome 3BL with the second-highest DAPI fluorescence (after 5BL), indicated that every chromosome arm of the wheat A- and B-genome chromosomes could be discriminated and sorted (Figure 1, B–D). Microscopic analysis of sorted chromosome fractions subjected to FISH with probes for GAA microsatellite and telomeric repeats (Figure 2, B–D) showed that the chromosome arms could be sorted at very high purities ranging from 90 to 98%. Replacement of a wild-type chromosome with a pair of telosomes resulted not only in the appearance of two additional peaks on the flow karyotype representing the two telosomes, but also in the change of the chromosome content of the composite and/or the presence of the original chromosome peaks. As a consequence, peak II in dDt1A represented only the wild-type chromosome 6A (Figure 1B), and the peak representing chromosome 3B was absent in dDt line 3B (Figure 1D).
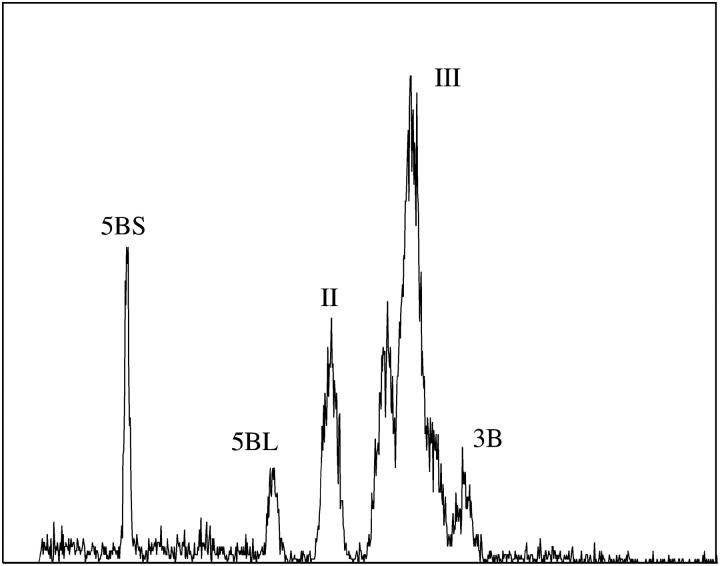
The ability to analyze chromosomes of T. turgidum var. durum prompted us to analyze chromosome suspensions prepared from a strain of T. turgidum var. melanopus with 15 chromosome pairs. Resulting flow karyotypes resembled those of double-ditelosomic lines of T. turgidum var. durum. In addition to peaks II, III, and 3B, two peaks appeared to the left of peak II (Figure 4). Chromosomes from these peaks were sorted onto microscope slides and subjected to FISH with probes for GAA satellite and telomeric repeat. The analysis of fluorescence-labeling patterns identified these chromosomes as 5BS and 5BL (Figure 2E).
Figure 4.—
Flow karyotype (histogram of relative fluorescence intensity) obtained after the analysis of DAPI-stained chromosome suspension from a strain of T. turgidum var. melanopus with 2n = 30. The karyotype consists of two composite peaks (II and III) representing specific groups of chromosomes and a peak representing chromosome 3B. In addition, two peaks that represent chromosomes 5BS and 5BL are well discriminated. x-axis, relative DAPI fluorescence intensity; y-axis, number of events.
DISCUSSION
The availability of chromosome-specific and chromosome-arm-specific BAC libraries prepared from DNA of flow-sorted chromosomes would greatly facilitate the assembly of a global physical map and the preparation for sequencing of the gene-containing regions in wheat (Gill et al. 2004). In line with this, the primary focus of this study was to assess the feasibility of chromosome sorting in durum wheat. To identify flow-sorted chromosomes and to determine the purity in sorted fractions, we used FISH with probes for repetitive DNA sequences that facilitated identification of sorted chromosomes in our previous study (Kubaláková et al. 2003). Until this study, chromosome identification in durum wheat had been done after C-banding (Venora et al. 2002) rather than using FISH. Thus, as the first step in this work, we determined genomic distribution of the GAA microsatellite, pSc119.2, Afa family, and 5S rDNA repeat sequences. Fluorescent patterns thus obtained facilitated the unambiguous identification of every chromosome arm of durum wheat. To our knowledge, the molecular karyotype presented here is the first of its kind in this species.
FISH analysis of repeated DNA sequences revealed minor differences in their genomic distribution between the hexaploid wheat (Mukai et al. 1993; Pedersen and Langridge 1997) and the durum wheat (this study). Recent molecular data indicate differences in molecular chromosome organization among the genomes of diploid, tetraploid, and hexaploid wheat (Wicker et al. 2003; Gu et al. 2004). It is thus tempting to speculate on the species specificity of the distributions. On the other hand, a number of observations indicate variations in C-banding and the GAA-banding among hexaploid wheat lines (Friebe and Gill 1994; Kubaláková et al. 2002). Thus, the final conclusion regarding the species specificity of genomic distribution of the DNA repeats observed in this study can be made only after extensive analysis of a range of hexaploid and tetraploid wheat cultivars.
The utility of flow karyotyping and chromosome sorting in durum wheat was demonstrated by analyzing the chromosome suspensions prepared from a strain of T. turgidum var. melanopus with 2n = 30. We have demonstrated that the two telocentric chromosomes in this strain correspond to the short and the long arms of chromosome 5B. This observation confirms previous assignment of these two telosomes to the arms of chromosome 5B on the basis of chromosome-pairing studies (Nishikawa et al. 1986).
Kubaláková et al. (2002) demonstrated that problems with the discrimination and sorting of single chromosomes in hexaploid wheat could be avoided by employing telosomic lines. Of a total of 42 chromosome arms, 40 could be discriminated and sorted. Peaks of chromosome arms 3BL and 5BL overlapped with peak I and the arms could be sorted only as isochromosomes (Kubaláková et al. 2002). Our results indicate that all arms of the A- and B-genome chromosomes, including chromosomes 3BL and 5BL, can be sorted from durum wheat using telosomic lines. Similar to hexaploid wheat, only one wild-type chromosome 3B can be sorted from durum wheat. Interestingly, another wild-type wheat chromosome 6A can also be sorted in durum wheat from peak II in the double-ditelosomic line for 1A.
Although not tested in this work, our results indicate the suitability of durum wheat stocks for sorting D-genome chromosomes, which cannot be sorted individually in hexaploid wheat. Chromosomes 1D, 4D, and 6D, whose peaks are expected to appear to the left of peak II, should be sortable from the respective substitution lines in durum wheat. Predicted peak positions of the remaining D chromosomes overlap with peak II. Thus, their sorting would require the availability of D-chromosome substitution lines in which chromosomes 1A and 6A were in the ditelosomic forms.
As shown by FISH analysis of sorted particles, the chromosome arms could be sorted with a purity approaching 100%. The analysis was done rigorously using a probe for the GAA microsatellite to identify individual chromosomes and a probe for telomeric repeat to discriminate between telosomes and arms that could result from chromosome breakage during the preparation of chromosome suspensions and sorting. Thus, it is now possible to sort every wheat chromosome arm to a high purity. Furthermore, we have demonstrated that the use of double-ditelosomic lines allows sorting of short and long arms simultaneously. This strategy could be useful in shortening the total time for sample preparation and sorting when large numbers (>106) of chromosome arms are needed for use in constructing specific BAC libraries (Janda et al. 2004; Šafář et al. 2004).
This study marks a further step in the development of flow cytogenetics for wheat. Any chromosome arm may be sorted in high purity and large quantity. Flow-sorted plant chromosomes were shown to be useful in a number of studies and their use could be advantageous over other approaches (Doležel et al. 2004). For example, large numbers of chromosomes sorted onto a small area of a microscope slide make localization of DNA sequences by FISH relatively simple. This facilitates discovery of rare structural changes (Kubaláková et al. 2003) among many other possible applications. Furthermore, sorted chromosomes can be linearly stretched and used for FISH with unparalleled spatial resolution (Valárik et al. 2004). Perhaps the most attractive application of sorted chromosome arms is the construction of specific BAC libraries that represent only a few percent of the complex wheat genome. Successful creation of a subgenomic BAC library specific for chromosomes 1D, 4D, and 6D (Janda et al. 2004), a chromosome 3B-specific BAC library (Šafář et al. 2004), and BAC library specific for the short arm of chromosome 1B (J. Janda, J. Šafář, M. Kubaláková, J. Bartoš, P. Kovářová, P. Suchánková, S. Pateyron, J. Číhalíková, P. Sourdille, H. Šimková, M. Bernard, A. Lukaszewski, B. Chalhoub and J. Doležel, unpublished results) confirm that such libraries can be produced. Finally, one can envisage using flow-sorted wheat chromosome arms for targeted isolation of low-copy sequences after DNA renaturation (Cot) analysis (Peterson et al. 2002), as probes for screening DNA microarrays (Gribble et al. 2004), and for HAPPY mapping (Thangavelu et al. 2003).
Acknowledgments
We thank our colleagues Michaela Libosvárová and Radka Tušková for excellent technical assistance. A gift sample of amiprophos-methyl from the Agriculture Division, Bayer (Kansas City, MO), is acknowledged. This work was supported by research grants from the Czech Science Foundation (522/03/0354 and 521/04/0607) and by a research contract from Ente per le Nuove tecnologie, l'Energia e l'Ambiente (ENEA) as part of the Analisi del Genoma del Frumento Duro per l'Identificazione di Geni Utili al Miglioramento della Tolleranza a Carenze Idriche ed alla Salinità (FRUMISIS) project supported by the Italian Ministry of Agriculture and Forest Policy.
References
- Bedbrook, J. R., J. Jones, M. O'Dell, R. D. Thompson and R. B. Flavell, 1980. A molecular description of telomeric heterochromatin in Secale species. Cell 19: 545–560. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., and I. J. Leitch, 1995. Nuclear-DNA amounts in angiosperms. Ann. Bot. 76: 113–176. [Google Scholar]
- Cenci, A., N. Chantret, X. Kong, Y. Gu, O. D. Anderson et al., 2003. Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp. durum). Theor. Appl. Genet. 107: 931–939. [DOI] [PubMed] [Google Scholar]
- Doležel, J., P. Binarová and S. Lucretti, 1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol. Plant. 31: 113–120. [Google Scholar]
- Doležel, J., S. Lucretti and I. Schubert, 1994. Plant chromosome analysis and sorting by flow cytometry. Crit. Rev. Plant Sci. 13: 275–309. [Google Scholar]
- Doležel, J., M. Kubaláková, J. Bartoš and J. Macas, 2004. Flow cytogenetics and plant genome mapping. Chromosome Res. 12: 77–91. [DOI] [PubMed] [Google Scholar]
- Erayman, M., D. Sandhu, D. Sidhu, M. Dilbirligi, P. S. Baenziger et al., 2004. Demarcating the gene-rich regions of wheat genome. Nucleic Acids Res. 32: 3546–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris, J. D., J. P. Fellers, S. A. Brooks and B. S. Gill, 2003. A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 164: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe, B., and B. S. Gill, 1994. C-band polymorphism and structural rearrangements detected in common wheat (Triticum aestivum). Euphytica 78: 1–5. [Google Scholar]
- Fukui, K., N. Ohmido and G. S. Khush, 1994. Variability in rDNA loci in the genus Oryza detected through fluorescence in situ hybridization. Theor. Appl. Genet. 87: 893–899. [DOI] [PubMed] [Google Scholar]
- Gamborg, O. L., and L. R. Wetter, 1975 Plant Tissue Culture Methods. National Research Council of Canada, Saskatoon, SK, Canada.
- Gill, B. S., R. Appels, A.-M. Botha-Oberholster, C. R. Buell, J. L. Bennetzen et al., 2004. A workshop report on wheat genome sequencing: international genome research on wheat consortium. Genetics 168: 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble, S. M., H. Fiegler, D. C. Burford, E. Prigmore, F. Yang et al., 2004. Applications of combined DNA microarray and chromosome sorting technologies. Chromosome Res. 12: 35–43. [DOI] [PubMed] [Google Scholar]
- Gu, Y. Q., D. Coleman-Derr, X. Y. Kong and O. D. Anderson, 2004. Rapid genome evolution revealed by comparative sequence analysis of orthologous regions from four triticeae genomes. Plant Physiol. 135: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., S. A. Brooks, W. Li, J. P. Fellers, H. N. Trick et al., 2003. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploidy genome of bread wheat. Genetics 164: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., A. Sirikhachornkit, X. J. Su, J. Faris, B. Gill et al., 2002. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99: 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, J., J. Bartoš, J. Šafář, M. Kubaláková, M. Valárik et al., 2004. Construction of a subgenomic BAC library specific for chromosomes 1D, 4D and 6D of hexaploid wheat. Theor. Appl. Genet. 109: 1337–1345. [DOI] [PubMed] [Google Scholar]
- Joppa, L. R., 1993. Chromosome engineering in tetraploid wheat. Crop Sci. 33: 908–913. [Google Scholar]
- Joppa, L. R., and N. D. Williams, 1988. Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30: 222–228. [Google Scholar]
- Kejnovský, E., J. Vrána, S. Matsunaga, P. Souček, J. Široký et al., 2001. Localization of male-specifically expressed MROS genes of Silene latifolia by PCR on flow-sorted sex chromosomes and autosomes. Genetics 158: 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, H., 1944. Discovery of the DD-analyzer, one of the ancestors of vulgare wheats. Agric. Hortic. 19: 889–890. [Google Scholar]
- Kubaláková, M., J. Macas and J. Doležel, 1997. Mapping of repeated DNA sequences in plant chromosomes by PRINS and C-PRINS. Theor. Appl. Genet. 94: 758–763. [Google Scholar]
- Kubaláková, M., J. Vrána, J. Číhalíková, H. Šimková and J. Doležel, 2002. Flow karyotyping and chromosome sorting in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 104: 1362–1372. [DOI] [PubMed] [Google Scholar]
- Kubaláková, M., M. Valárik, J. Bartoš, J. Vrána, J. Číhalíková et al., 2003. Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 46: 893–905. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Luo, M. C., C. Thomas, F. M. You, J. Hsiao, O. Y. Shu et al., 2003. High-throughput fingerprinting of bacterial artificial chromosomes using the SNaPshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82: 378–389. [DOI] [PubMed] [Google Scholar]
- Lysák, M. A., J. Číhalíková, M. Kubaláková, H. Šimková, G. Künzel et al., 1999. Flow karyotyping and sorting of mitotic chromosomes of barley (Hordeum vulgare L.). Chromosome Res. 7: 431–444. [DOI] [PubMed] [Google Scholar]
- McFadden, E. S., and E. R. Sears, 1946. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37: 81–89. [DOI] [PubMed] [Google Scholar]
- Mukai, Y., Y. Nakahara and M. Yamamoto, 1993. Simultaneous discrimination of the 3 genomes in hexaploid wheat by multicolour fluorescence in situ hybridisation using total genomic and highly repeated DNA probes. Genome 36: 489–494. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., H. Tsujimoto, K. Isono and T. Sasakuma, 1995. Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome 38: 479–486. [DOI] [PubMed] [Google Scholar]
- Neumann, P., D. Požárková, J. Vrána, J. Doležel and J. Macas, 2002. Chromosome sorting and PCR-based physical mapping in pea (Pisum sativum L.). Chromosome Res. 10: 63–71. [DOI] [PubMed] [Google Scholar]
- Nishikawa, K., A. Takagi, T. Ban, H. Ohtsuka and Y. Furuta, 1986. Spontaneous reciprocal translocation in cultivated form of emmer wheat. Jpn. J. Genet. 61: 361–370. [Google Scholar]
- Pedersen, C., and P. Langridge, 1997. Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40: 589–593. [DOI] [PubMed] [Google Scholar]
- Peterson, D. G., S. R. Schulze, E. B. Sciara, S. A. Lee, J. E. Bowers et al., 2002. Integration of Cot analysis, DNA cloning, and high-throughput sequencing facilitates genome characterization and gene discovery. Genome Res. 12: 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Požárková, D., A. Koblížková, B. Román, A. M. Torres, S. Lucretti et al., 2002. Development and characterization of microsatellite markers from chromosome 1-specific DNA libraries of Vicia faba. Biol. Plant. 45: 337–345. [Google Scholar]
- Román, B., Z. Satovic, D. Požárková, J. Macas, J. Doležel et al., 2004. Development of a composite map in Vicia faba, breeding applications and future prospects. Theor. Appl. Genet. 108: 1079–1088. [DOI] [PubMed] [Google Scholar]
- Šafář, J., J. Bartoš, J. Janda, A. Bellec, M. Kubaláková et al., 2004. Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J. 39: 960–968. [DOI] [PubMed] [Google Scholar]
- Sears, E. R., 1966 Nullisomic-tetrasomic combination in hexaploid wheat, pp. 29–45 in Chromosome Manipulation and Plant Genetics, edited by R. Riley and K. R. Lewis. Oliver & Boyd, Edinburgh.
- Sears, E. R., 1969. Wheat cytogenetics. Annu. Rev. Genet. 3: 451–468. [Google Scholar]
- Thangavelu, M., A. B. James, A. Bankier, G. J. Bryan, P. H. Dear et al., 2003. HAPPY mapping in a plant genome: reconstruction and analysis of a high-resolution physical map of a 1.9 Mbp region of Arabidopsis thaliana chromosome 4. Plant Biotech. J. 1: 23–31. [DOI] [PubMed] [Google Scholar]
- Tsunewaki, K., 1963. An emmer wheat with 15 chromosome pairs. Can. J. Genet. Cytol. 5: 462–466. [Google Scholar]
- Valárik, M., H. Šimková, E. Hřibová, J. Šafář, M. Doleželová et al., 2002. Isolation, characterization and chromosome localization of repetitive DNA sequences in bananas (Musa spp.). Chromosome Res. 10: 89–100. [DOI] [PubMed] [Google Scholar]
- Valárik, M., J. Bartoš, P. Kovářová, M. Kubaláková, J. H. de Jong et al., 2004. High-resolution FISH on super-stretched flow-sorted plant chromosomes. Plant J. 37: 940–950. [DOI] [PubMed] [Google Scholar]
- Venora, G., S. Blangiforti, M. R. Castiglione, D. Pignone, F. Losavio et al., 2002. Chromatin organisation and computer aided karyotyping of Triticum durum Desf. cv. Timilia. Caryologia 55: 91–98. [Google Scholar]
- Vláčilová, K., D. Ohri, J. Vrána, J. Číhalíková, M. Kubaláková et al., 2002. Development of flow cytogenetics and physical genome mapping in chickpea (Cicer arietinum L.). Chromosome Res. 10: 695–706. [DOI] [PubMed] [Google Scholar]
- Vrána, J., M. Kubaláková, H. Šimková, J. Číhalíková, M. A. Lysák et al., 2000. Flow-sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 156: 2033–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., N. Yahiaoui, R. Guyot, E. Schlagenhauf, Z. D. Liu et al., 2003. Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and A(m) genomes of wheat. Plant Cell 15: 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, G. Tranquilli, M. Helguera, T. Fahima et al., 2003. Positional cloning of the wheat vernalization gene Vrn1. Proc. Natl. Acad. Sci. USA 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]