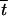Abstract
In this article, we study the effect of self-fertilization on the evolution of a modifier allele that alters the recombination rate between two selected loci. We consider two different life cycles: under gametophytic selfing, a given proportion of fertilizations involves gametes produced by the same haploid individual, while under sporophytic selfing, a proportion of fertilizations involves gametes produced by the same diploid individual. Under both life cycles, we derive approximations for the change in frequency of the recombination modifier when selection is weak relative to recombination, so that the population reaches a state of quasi-linkage equilibrium. We find that gametophytic selfing increases the range of epistasis under which increased recombination is favored; however, this effect is substantial only for high selfing rates. Moreover, gametophytic selfing affects the relative influence of different components of epistasis (additive × additive, additive × dominance, dominance × dominance) on the evolution of the modifier. Sporophytic selfing has much stronger effects: even a small selfing rate greatly increases the parameter range under which recombination is favored, when there is negative dominance × dominance epistasis. This effect is due to the fact that selfing generates a correlation in homozygosity at linked loci, which is reduced by recombination.
SEX is the combination of two complementary events—meiosis and syngamy—resulting in the alternation of a diploid and a haploid phase during the life cycle; by contrast, an asexual life cycle can be viewed simply as a succession of mitoses. Both life cycles can be compared to understand the relative advantages of sexual vs. asexual reproduction, and this approach has been useful to identify the costs (Maynard Smith 1971; Williams and Mitton 1973) and benefits of meiosis owing to the processes of segregation and recombination (see below). However, a whole variety of sexual life cycles does exist, depending in particular on how syngamy occurs. Although both the evolution of mating systems and the evolution of sex and recombination have received considerable attention, the interplay between mating systems and the evolution of recombination has received less attention.
The famous twofold cost of meiosis notoriously depends on the mating system (Charlesworth 1980); it can, for example, vanish in selfing hermaphrodites. Similarly, the advantage of sexual reproduction due to segregation greatly varies depending on the mating system (Uyenoyama and Bengtsson 1989; Agrawal and Chasnov 2001; Otto 2003). However, despite considerable work and progress in understanding when recombination may evolve and provide an advantage to sexual reproduction, we still largely ignore how the mating system influences the evolution of recombination. Some simulations have been performed in this direction (Charlesworth et al. 1977, 1979; Holsinger and Feldman 1983), but no clear conclusion has emerged as to exactly how nonrandom mating might affect the general conditions under which recombination is favored.
Different theories have been proposed to explain the evolution and maintenance of high rates of recombination in higher organisms. Apart from theories based on the idea of a mechanistic advantage of recombination (e.g., through DNA repair), recombination is thought to be beneficial because it reduces linkage disequilibria (LD) between loci. This last hypothesis (termed a generative hypothesis) consists of a variety of possible underlying processes that can be classified according to the mechanisms generating LD (Kondrashov 1993; Otto and Lenormand 2002). First, LD can be produced by epistatic selection, in which case recombination is favored when epistasis is weak and negative (Feldman et al. 1980; Kondrashov 1982, 1988; Barton 1995) and not too variable among pairs of loci (Otto and Feldman 1997) or when epistasis fluctuates over short periods of time (Charlesworth 1976; Barton 1995). Second, LD can be produced by migration and directional selection, in which case recombination is favored when directional selection at different loci covaries negatively between habitats (Lenormand and Otto 2000). Third, LD can be produced by drift and directional selection, a process referred to as the Hill-Robertson effect (Felsenstein 1974), in which case recombination is favored when populations are not too large (Fisher 1930; Muller 1932; Otto and Barton 1997, 2001). These different sources of LD can also occur simultaneously, which affects the conditions for the evolution of recombination (Lenormand and Otto 2000; Otto and Barton 2001).
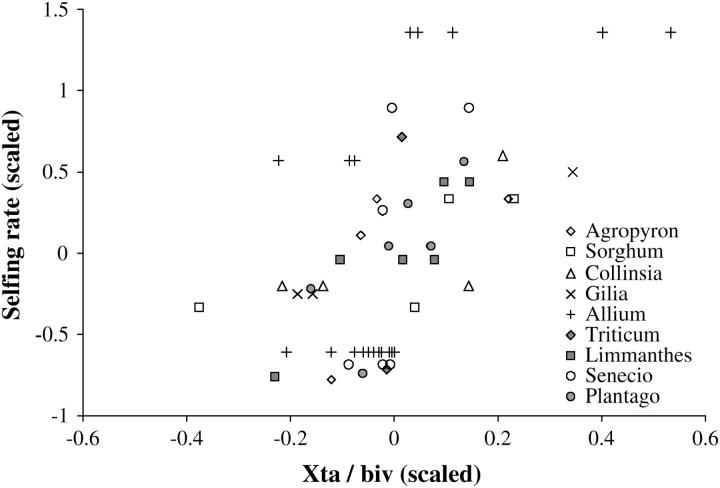
To date, most analytical models on the evolution of recombination have considered randomly mating populations; however, biological data and simulation models suggest that nonrandom mating may be an important factor. These data and models have focused on the case of partial self-fertilization. For example, cytological observations indicate that self-pollinating species of flowering plants tend to have higher chiasma frequencies than closely related outcrossing species, suggesting that recombination rates may correlate with the amount of selffertilization (Grant 1958; Stebbins 1958; Ved Brat 1965; Zarchi et al. 1972; Arroyo 1973; Gibbs et al. 1975; Sharma et al. 1992). Figure 1 displays data collected from these studies. In general, quantitative estimates of rates of self-fertilization are difficult to obtain, but the different species can be classified into different broad classes, from “complete outcrossers” (for example, in the case of auto-incompatible species) to “complete selfers” (for example, in the case of cleistogamous species). From this classification, we attributed a number to each species, representing its degree of selfing. The y-axis of Figure 1 represents the difference between the degree of selfing of each species and the average over all measured species of the same genus, divided by the genus average. The x-axis represents the number of chiasmata per bivalent (measured in general at metaphase I of meiosis); again, each value corresponds to the deviation of each species relative to the genus average, divided by the genus average. Variables on both axes are strongly correlated in every genus and overall (Spearman Rho is 0.535 overall on the 57 points, P < 10−4). However, it is important to note here that all these data have been obtained from male meioses; data from female meioses are thus needed to determine if the same correlation exists in both cases, given that the ratio of the number of chiasmata in male/female meioses also covaries with the mating system (Lenormand and Duteil 2005).
Figure 1.—
Cytological data on selfing rates and numbers of chiasmata per bivalent at meiosis. Each symbol corresponds to a different species or subspecies. x-axis, deviation of the number of chiasmata per bivalent relative to the genus average, divided by the genus average; y-axis, deviation of the degree of selfing relative to the genus average, divided by the genus average. Data are from Grant (1958) (Agropyron, Sorghum, Collinsia, and Gilia), Ved Brat (1965) (Allium), Zarchi et al. (1972) (Triticum), Arroyo (1973) (Limmanthes), Gibbs et al. (1975) (Senecio), and Sharma et al. (1992) (Plantago).
Furthermore, simulation models have shown that self-fertilization increases the strength of indirect selection acting on recombination modifiers and that under some conditions modifiers increasing recombination decrease in frequency under random mating, but increase in frequency under partial selfing, even when the selfing rate is small (Charlesworth et al. 1977, 1979; Holsinger and Feldman 1983). However, the mechanisms by which self-fertilization affects the evolution of recombination remain difficult to understand from these simulations.
In this article, we present an analytical study of the effects of partial selfing on the evolution of recombination, using multilocus techniques that have been applied previously to the study of the evolution of recombination (Barton 1995; Otto and Feldman 1997; Lenormand 2003). Our model is deterministic (infinite population size), and therefore, the evolution of recombination will be driven by epistatic interactions between loci. We consider two different life cycles. Under gametophytic selfing, we assume that a given proportion of fertilizations involves gametes produced by the same haploid individual (generating zygotes, which are homozygous at all loci). Gametophytic selfing corresponds to the most extreme form of inbreeding, occurring, for example, in homosporous ferns (McCauley et al. 1985; Soltis and Soltis 1986). It has also been used by Otto (2003) to consider the effects of inbreeding on segregation and the evolution of sex. Under gametophytic selfing, we assume that selection can occur during both the haploid and the diploid stages of the life cycle. The second life cycle corresponds to sporophytic selfing: in this case, a given proportion of fertilizations involves gametes produced by the same diploid individual (as in most selfing plants and animals). Under sporophytic selfing, we assume that selection occurs only during the diploid phase of the life cycle. We will see that gametophytic selfing tends to increase the range of epistasis values where recombination is favored; it also increases strongly selection pressures on recombination and affects the relative importance of the different components of epistasis (additive × additive, additive × dominance, dominance × dominance). Under sporophytic selfing, dominance × dominance epistasis generates a selective pressure on the evolution of recombination that is absent under random mating. If dominance × dominance epistasis is negative, even a small rate of sporophytic selfing greatly increases the range of parameters under which recombination can increase.
METHODS
Throughout the article, we consider three linked loci, i, j, and k (present in that order along the chromosome), where j, k are selected loci, while locus i affects the recombination rate between j and k. Two alleles, noted 0 and 1, segregate at each locus. We use the multilocus formalism of Barton and Turelli (1991) (extended by Kirkpatrick et al. 2002) to write recurrence equations for allele frequencies and for genetic associations, measuring the statistical associations among alleles at different loci and/or on different chromosomes. Definitions of the different parameters and variables are given in Table 1. Following previous usage, we denote pl the frequency of allele 1 at locus l in the population, and ql = 1 − pl. We then define two indicator variables Xl and X*l, which equal 1 if allele 1 is present at locus l on the maternally inherited (Xl) or paternally inherited X*l chromosome of a given individual and 0 otherwise. In the haploid stage, we have only one Xl variable per haploid individual. ζ variables are defined as
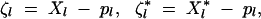 |
1 |
and products of ζ-variables are given by
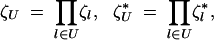 |
2 |
where U is a set of loci, the possible sets being ⊘, {i}, {j}, {k}, {i, j}, {i, k}, {j, k}, and {i, j, k}. By convention, 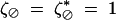 . Genetic associations CU measured in the haploid phase (before haploid selection) are defined as
. Genetic associations CU measured in the haploid phase (before haploid selection) are defined as
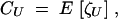 |
3 |
where U is a set of loci, and where E means the average over all individuals in the population. Genetic associations in diploid organisms are given by
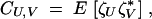 |
4 |
where U and V are sets of loci. For example, Cij measures the linkage disequilibrium between loci i and j in the haploid phase, while Cij,⊘ measures the linkage disequilibrium between i and j on maternal chromosomes, during the diploid phase. Throughout, we assume no sex-specific selection nor any effect of the sex-of-origin of chromosomes, and therefore we will always have CU,V = CV,U.
TABLE 1.
Notation
| i, j, k | Indices for loci; j and k are selected loci, and i modifies the recombination rate between j and k | |
|---|---|---|
| pl, ql = 1 − pl | Frequency of allele 1 (pl) and of allele 0 (ql) at locus l | |
| Xl, Xl* | Indicator variables that equal 1 if allele 1 is present at locus l on the maternally inherited chromosome (Xl) or paternally inherited chromosome Xl* of a given individual and 0 otherwise |
|
| ζl = Xl − pl | Scaled indicator variable | |
| ζU | Product of ζ-variables over the set of loci U | |
| CU, CUH | Genetic association between loci in the set U during the haploid phase (in the gametophytic selfing model), before and after haploid selection |
|
| CU,VS, CU,VD | Genetic association between loci in the sets U and V on homologous chromosomes of diploid individuals, before and after diploid selection (in the sporophytic selfing model, CU,VS is noted CU,V) |
|
| aUH, eUH | Effect of haploid selection acting on the set of loci U (in the gametophytic selfing model) and epistasis between these loci on a multiplicative scale |
|
| aU,VD, eU,VD | Effect of diploid selection acting on the sets of loci U and V present on homologous chromosomes and epistasis between these loci on a multiplicative scale; D superscripts are dropped in the sporophytic selfing model |
|
| s | Direct selection acting on loci j and k, under the fitness matrix given by Table 2 | |
| h | Dominance of alleles 1 at loci j and k, under the fitness matrix given by Table 2 | |
| ea×a, ea×d, ed×d | Additive × additive, additive × dominance, and dominance × dominance epistasis between loci j and k, under the fitness matrix given by Table 2 |
|
| α | Rate of self-fertilization (either gametophytic or sporophytic) | |
| rij, rjk | Recombination rate between loci i and j and between loci j and k (without modifier) | |
| dr | Effect of the modifier on rjk | |
| hM | Dominance of allele 1 at the modifier locus (locus i) | |
| tS,T | Probability that a gamete produced by an individual contains the set S of loci derived from one of its parents and the set T from the other |
|
|
Population average of tS,T | |
| δtS,T|i, δtS,T|i,i | Additive effect of the modifier on tS,T and effect of dominance at the modifier locus | |
| ε | Scaling factor |
Gametophytic selfing:
The life cycle under gametophytic selfing is represented in Figure 2A. Each generation starts after meiosis, at the beginning of the haploid phase. In this phase, the population can be described by seven haploid genotype frequencies (there are eight genotypes, and their frequencies sum to 1) or by the three allele frequencies pi, pj, and pk and the four gametic linkage disequilibria Cij, Cik, Cjk, and Cijk. We assume that selection can occur during the haploid phase, followed by syngamy. At syngamy, a proportion α of zygotes are formed by two gametes produced by the same haploid individual (and are thus homozygous at all loci), while a proportion 1 − α are formed by gametes sampled at random. Finally, selection occurs between diploid organisms, followed by meiosis. Changes in allele frequencies and genetic associations caused by selection have been derived in Kirkpatrick et al. (2002), while recursions for meiosis in the presence of a recombination modifier are given in Barton (1995). Therefore, the only new part here corresponds to the recursions for syngamy. In the following, we use superscripts H, S, and D to denote variables measured after haploid selection, syngamy, and diploid selection, respectively.
Figure 2.—
Model life cycles. (A) Gametophytic selfing; (B) sporophytic selfing. In each case, the arrow marks the start of the life cycle, at which point we measure allele frequencies and genetic associations.
Haploid selection:
To represent haploid selection, Barton and Turelli (1991) and Kirkpatrick et al. (2002) define selection coefficients aHU such that the fitness of a haploid individual, relative to the average fitness in the population, can be written as
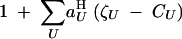 |
5 |
aHU represents the effect of selection acting on the set of loci U; since we assume that selection acts only at loci j and k, we have only three coefficients ajH, akH, and ajkH, where ajH and akH represent direct selection at loci j and k, while ajkH represents the interaction between these loci.
Recursions for allele frequencies and genetic associations, using these aUH coefficients are given by Equations 9, 10, and 15 in Kirkpatrick et al. (2002). In particular, the change in frequency of the recombination modifier during haploid selection is given by
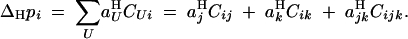 |
6 |
Syngamy:
Allele frequencies do not change during syngamy. Associations after syngamy are given by
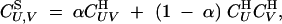 |
7 |
where α is the rate of gametophytic selfing, and U and V are sets of loci. For instance, 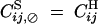 ,
, 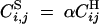 , while
, while 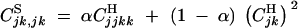 . Repeated indices in associations can be eliminated using the relation
. Repeated indices in associations can be eliminated using the relation
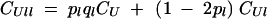 |
8 |
(Kirkpatrick et al. 2002, Equation 5), so that, for example, Cjjkk = pjqjpkqk + (1 − 2pj)(1 − 2pk)Cjk.
Diploid selection:
Diploid selection is represented by defining aU,VD selection coefficients, such that the fitness of a diploid organism, relative to the average fitness of the population, can be written as
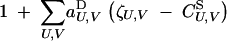 |
9 |
The sum in Equation 9 is over all sets of loci U and V, including the empty set. Again, aU,VD represents the effect of selection acting on the set of loci (U, V); for example aj,⊘D and ak,⊘D represent direct selection acting on loci j and k, while aj,jD and ak,kD represent the effects of dominance at these loci (in the additive case, 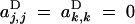 ). In principle, the notation allows for different selective effects depending upon the sex-of-origin of chromosomes (for example, it is possible to have aj,⊘D ≠ a⊘,jD), but because we assume no sex-of-origin effect,
). In principle, the notation allows for different selective effects depending upon the sex-of-origin of chromosomes (for example, it is possible to have aj,⊘D ≠ a⊘,jD), but because we assume no sex-of-origin effect, 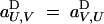 for all sets U and V. Interactions between pairs of genes at loci j and k are represented by coefficients ajk,⊘D and aj,kD; without cis-trans effects,
for all sets U and V. Interactions between pairs of genes at loci j and k are represented by coefficients ajk,⊘D and aj,kD; without cis-trans effects, 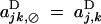 . Finally, interactions among three and four alleles at loci j and k are represented by coefficients ajk,jD, ajk,kD, and ajk,jkD.
. Finally, interactions among three and four alleles at loci j and k are represented by coefficients ajk,jD, ajk,kD, and ajk,jkD.
The exact expressions of aU,VD coefficients are complicated functions of the fitnesses and frequencies of the different genotypes, but they take simpler forms when calculated to first order in the selective differences. Throughout the article, we consider the case of directional selection acting on loci j and k and use the symmetric fitness matrix given in Table 2, where s is the selection coefficient of alleles 1 at loci j and k, h is their dominance coefficient, and ea×a, ea×d, and ed×d measure additive × additive, additive × dominance, and dominance × dominance epistasis between loci j and k. Without epistasis (ea×a = ea×d = ed×d = 0) and under random mating, this selection regime does not generate any gametic linkage disequilibrium between loci j and k (Cjk = 0). aU,VD coefficients can be obtained by equating the fitnesses of the different genotypes with expressions derived from Equation 9; appendix A gives expressions for the aU,VD coefficients under the fitness matrix given in Table 2 to different orders in s, ea×a, ea×d, and ed×d.
TABLE 2.
Example of fitness matrix for diploid selection
| 00 | 01 | 11 | |
|---|---|---|---|
| 00 | 1 | 1 + hs | 1 + s |
| 01 | 1 + hs | (1 + hs)2 + ea×a | (1 + hs)(1 + s) + 2ea×a + ea×d |
| 11 | 1 + s | (1 + hs)(1 + s) + 2ea×a + ea×d | (1 + s)2 + 4ea×a + 4ea×d + ed×d |
The entries correspond to the different genotypes at loci j and k; ea×a, ea×d, and ed×d correspond to additive × additive, additive × dominance, and dominance × dominance epistatic effects, respectively.
Recursions for allele frequencies and genetic associations after diploid selection, in terms of the aU,VD coefficients, are given by Equations 9, 10, and 15 in Kirkpatrick et al. (2002). In particular, the change in frequency of the recombination modifier is given by
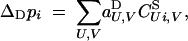 |
10 |
where the sum is over all sets U and V, including the empty set.
Meiosis:
We denote the recombination rate between loci i and j as rij. The recombination rate between j and k depends on the individual's genotype at locus i: individuals having genotype 00, 01, and 11 at this locus have recombination rate rjk, rjk + hMdr, and rjk + dr, respectively, between loci j and k. Therefore, dr measures the effect of the recombination modifier, while hM is the dominance of allele 1 at locus i. We assume no interference among crossing-over events.
Meiosis does not change allele frequencies. A general recursion for genetic associations in the presence of a recombination modifier is given in Barton (1995). The method consists in defining coefficients tS,T, measuring the probability that a gamete produced by an individual contains the set S of loci derived from one of its parents and the set T from the other. For example, ti,jk is the proportion of gametes containing locus i from one parent (either mother or father) and loci j and k from the other. In the presence of a recombination modifier, some tS,T will depend on the individual genotype at locus i and can be written under the form
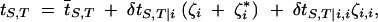 |
11 |
where 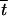 S,T is the population average, δtS,T|i represents the additive effect of the modifier on tS,T, while δtS,T|i,i represents the effect of dominance at the modifier locus. For all S and T,
S,T is the population average, δtS,T|i represents the additive effect of the modifier on tS,T, while δtS,T|i,i represents the effect of dominance at the modifier locus. For all S and T, 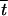 S,T, δtS,T|i, and δtS,T|i,i can be expressed as functions of rij, rjk, dr, hM, and pi (Barton 1995). Genetic associations after meiosis are then given by
S,T, δtS,T|i, and δtS,T|i,i can be expressed as functions of rij, rjk, dr, hM, and pi (Barton 1995). Genetic associations after meiosis are then given by
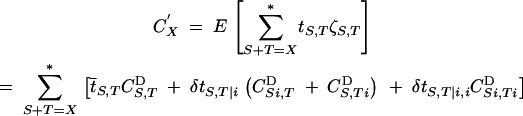 |
12 |
(where E means the average over all individuals, just before meiosis). To go from the first to the second line, one uses Equation 11 and replaces expectations of products of ζ-variables by associations before meiosis. As in Barton (1995), the asterisk in Equation 12 means that the sum is over all distinct partitions of the set X. For example, for X = ijk, the partitions {S = i, T = jk} and {S = jk, T = i} contribute to a single term in the sum. This stands in contrast to sums like in Equation 10, where, for example, {U = jk, V = j} and {U = j, V = jk} represent two different terms.
Sporophytic selfing:
The life cycle under sporophytic selfing is represented in Figure 2B. Each generation starts after fertilization; selection then occurs (in the diploid phase), followed by meiosis and syngamy. A proportion α of fertilizations involves gametes produced by the same diploid individual, while a proportion 1 − α involves gametes sampled at random from the whole population. Under sporophytic selfing, the population cannot be described only in terms of haplotype frequencies: one has to keep track of 36 diploid genotype frequencies or of 3 allele frequencies and of 32 genetic associations (measured at the beginning of the life cycle, in juvenile diploids): 9 associations between pairs of genes (such as Ci,i, Cij,⊘, Ci,k), 10 associations between three genes (such as Cijk,⊘, Cij,i, Cik,j), 9 associations between four genes (such as Cijk,i, Cij,ij, Cik,jk), 3 associations between five genes (Cijk,ij, Cijk,ik, Cijk,jk), and the association between the six genes, Cijk,ijk. Because of this higher number of variables, the analysis of the model becomes more tedious, but nevertheless leads to simple results, at least to leading order in the dominance × dominance epistasis.
As in the previous section, recursions for diploid selection are obtained by defining aU,VD selection coefficients and using Equations 9, 10, and 15 in Kirkpatrick et al. (2002). Recursions for meiosis and syngamy are obtained as follows. Recursions for associations between genes present on the same chromosome are not affected by selfing and are given by
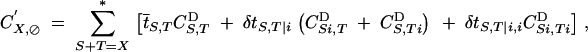 |
13 |
which is equivalent to Equation 12. Recursions for associations between genes present on homologous chromosomes are then given by
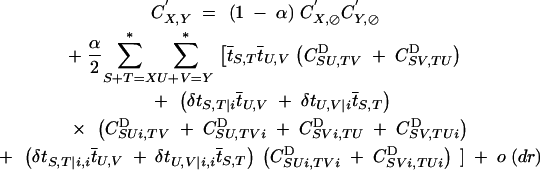 |
14 |
This expression is obtained using the same method as in Barton (1995): one considers all the possible recombination events that can have produced each of the two chromosomes of a newly formed zygote; if this zygote has been produced by selfing, one thus has to sum over all products tS,T and tU,V (such that S + T = X and U + V = Y).
Approximations:
To obtain recursions on allele frequencies and genetic associations, we implemented the general expressions described above in a Mathematica 5.0 notebook (available upon request); these full recursions are complicated expressions, which are not given here for space reasons. From these expressions, useful approximations can be obtained when selection is weak relative to recombination: in this case, genetic associations equilibrate fast relative to the change in allele frequencies, and the population quickly reaches a state described as quasi-linkage equilibrium (QLE), where associations change slowly over time (Barton and Turelli 1991; Nagylaki 1993). In principle, self-fertilization must not be too high for the QLE approximation to hold, because selfing reduces the effectiveness of recombination. Expressions of genetic associations at QLE, for given allele frequencies, are obtained by solving recursions for associations at equilibrium (that is, solving equations of the form 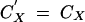 under gametophytic selfing, and
under gametophytic selfing, and 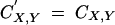 under sporophytic selfing). This way, associations can be expressed in terms of allele frequencies and of the different parameters of the model.
under sporophytic selfing). This way, associations can be expressed in terms of allele frequencies and of the different parameters of the model.
We consider different assumptions concerning the strength of selection and the frequency of self-fertilization; to do this, we introduce a small scaling term ε and assume that the modifier effect dr is of order ε, while selection coefficients aU,V and the selfing rate α can be of different orders in ε. At QLE, genetic associations will be of different orders, depending on the assumptions made. To obtain approximations to leading orders of associations at QLE, we perform a perturbation analysis: we first do not make any assumption on the order of magnitude of genetic associations and look for terms that are of order 1 at QLE (they are obtained by solving recursions expressed to the order 1). We then assume that all associations are of order ε, except those that were found to be of order 1 (which are replaced by their expression to the order 1, plus a term in ε), and solve recursions expressed to the order ε, to find associations that are effectively of order ε at QLE. We repeat the same procedure for the orders ε2, ε3, … , until all associations needed to express the change in frequency of the modifier have been obtained.
RESULTS
Gametophytic selfing:
Under gametophytic selfing we obtained that, as under random mating, recombination only decreases when selection and epistasis are of the same order of magnitude (as is apparent from our results). To find cases where recombination increases, one has to assume that epistasis is weak relative to selection. Therefore, throughout this section, we assume that selection coefficients ajH, akH, aj,⊘D, ak,⊘D, aj,jD, and ak,kD are of order ε, while ajkH, ajk,⊘D, aj,kD, ajk,jD, ajk,kD, and ajk,jkD are of order ε2. We also assume that the modifier has a small effect: dr is of order ε. We do not make any assumption on α. In this case, after solving recursions to express genetic associations at QLE, one obtains that Cij and Cik are of order ε4, Cjk is of order ε2, and Cijk is of order ε3. Recursions for these associations to leading orders in ε are given in appendix B.
The coefficients ajk,⊘D, aj,kD, ajk,jD, ajk,kD, and ajk,jkD measure additive epistatic effects (as relative fitness in Equation 9 is written as a sum over these coefficients); to the order ε2, they depend both on multiplicative epistatic parameters (Table 2, ea×a, ea×d, and ed×d) and on direct selection. However, these coefficients can be written in the form
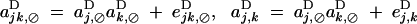 |
15 |
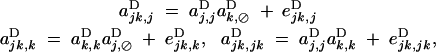 |
16 |
where eU,VD coefficients represent multiplicative epistatic effects (for example, 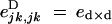 ). This can be seen from the expressions given in appendix A. Similarly, in the case of haploid selection, ajkH can be written in the form ajHakH + ejkH, where ejkH represents multiplicative epistasis (Barton and Turelli 1991).
). This can be seen from the expressions given in appendix A. Similarly, in the case of haploid selection, ajkH can be written in the form ajHakH + ejkH, where ejkH represents multiplicative epistasis (Barton and Turelli 1991).
General solution at QLE:
The change in frequency at the modifier locus is obtained from Equations B1, B2, B3, B5, and B7 in appendix B:
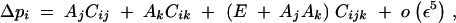 |
17 |
with
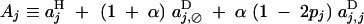 |
18 |
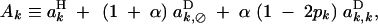 |
19 |
and
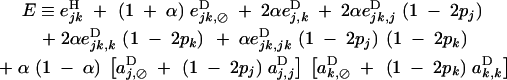 |
20 |
QLE values of Cij, Cik, and Cijk (denoted with a hat) are obtained from the recursions given in appendix B. Assuming that the modifier has an additive effect (hM = 1/2), we obtain
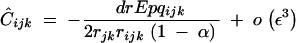 |
21 |
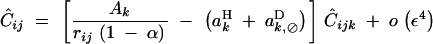 |
22 |
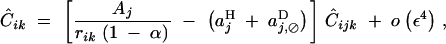 |
23 |
where pqijk = piqipjqjpkqk. In Equation 21, rijk is the probability that at least one crossing over occurs between loci i, j, and k, in the absence of the modifier. Since we assume no interference between recombination events between i and j and between j and k, we have rijk = 1 − (1 − rij)(1 − rjk). Equations 17–23 give the frequency change at the modifier locus at QLE,
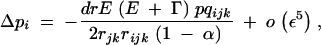 |
24 |
where
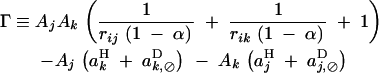 |
25 |
Equation 24 shows that, as under random mating, recombination always decreases when direct selection and epistasis are of the same order of magnitude. Indeed in this case Γ becomes negligible relative to E, and Δpi has the same sign as −dr. In the absence of selfing (α = 0), Equation 24 corresponds to the result obtained by Barton (1995) for the change in frequency of a recombination modifier in a panmictic population (a modifier increasing recombination increases in frequency when Γ < E < 0). Finally, Δpi under arbitrary dominance at the modifier locus is given by Equation 24, multiplied by a factor 2[hM + pi(1 − 2hM)].
QLE under haploid selection:
When selection occurs only during the haploid phase of the life cycle, Equation 24 becomes
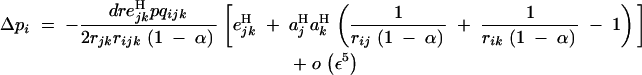 |
26 |
Equation 26 takes the same form as under random mating (Equation 12 in Barton 1995), all recombination rates (dr, rij, rik, rjk, and rijk) being multiplied by a factor 1 − α; this reflects the fact that selfing reduces the effectiveness of recombination. As a result, the minimum value of epistasis for a modifier increasing recombination to be selected, given by the term between brackets in Equation 26, decreases as α increases. Therefore, under haploid selection, gametophytic selfing increases the range of negative epistasis values over which increased recombination is selected.
We tested the validity of Equation 26, using a model of directional selection, where haploid individuals carrying zero, one, and two alleles 1 at loci j and k have fitnesses 1, 1 + s, and (1 + s)2 + e, respectively. In this case, when s is of order ε and e of order ε2, one obtains that 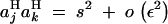 , and
, and 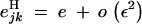 ; thus, Equation 26 predicts that a modifier increasing recombination should increase in frequency when
; thus, Equation 26 predicts that a modifier increasing recombination should increase in frequency when
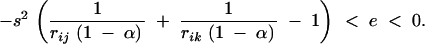 |
27 |
Figure 3 compares this prediction with deterministic simulations. In the simulations we iterate exact recursions for genotype frequencies; we first let the system equilibrate in the absence of the modifier and then introduce the modifier in small frequency and see if it increases (Figure 3, solid circles) or decreases (Figure 3, open circles) in frequency. Simulations confirm that gametophytic selfing increases the range of (negative) epistasis values over which increased recombination is favored; they also show that Equation 27 gives good predictions when selection is weak (Figure 3A, s = −0.01), except when the selfing rate is close to 1, while it does not give such good predictions for stronger selection (Figure 3B, s = −0.05), when the selfing rate is moderate to high. This is due to the fact that selfing reduces the effectiveness of recombination and therefore reduces the range of selection coefficients and recombination rates under which the QLE approximation is valid.
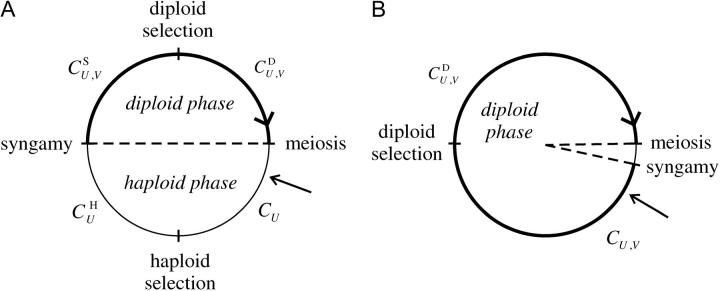
Figure 3.—
Range of epistasis under which increased recombination is favored, under directional haploid selection, as a function of the rate of gametophytic selfing, α. Equation 27 predicts that recombination should increase in the shaded area. Circles correspond to simulation results: solid circle, a modifier increasing recombination increases in frequency when rare; open circle, it decreases in frequency. Parameter values are: (A) s = −0.01 and (B) s = −0.05, rij = 0.2, rjk = 0.2. In the simulations dr = 0.01, hM = 1/2, and u = 10−5.
QLE under diploid selection:
In the case of diploid selection, it can be seen from Equations 20 and 24 that self-fertilization generates indirect selection on the recombination modifier even in the absence of epistasis (when all eU,VD coefficients equal zero). For example, without epistasis or dominance, 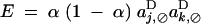 , and Equation 24 shows that a modifier increasing recombination will always be selected against (when aj,⊘D and ak,⊘D have the same sign). Indeed, self-fertilization generates positive linkage disequilibrium between loci j and k, even in the absence of epistasis; at QLE, one obtains (without epistasis or dominance)
, and Equation 24 shows that a modifier increasing recombination will always be selected against (when aj,⊘D and ak,⊘D have the same sign). Indeed, self-fertilization generates positive linkage disequilibrium between loci j and k, even in the absence of epistasis; at QLE, one obtains (without epistasis or dominance)
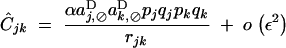 |
28 |
Linkage disequilibrium Cjk is due to the fact that, even under a completely multiplicative fitness matrix, the marginal fitnesses of the different types of chromosomes are not multiplicative, because under partial selfing genotype frequencies are not given by simple products of gamete frequencies.
To consider the effects of dominance and epistasis under diploid selection, we used the fitness matrix given in Table 2. To obtain simpler expressions, we considered the case where alleles 1 at loci j and k are deleterious (s < 0) and produced by recurrent mutations occurring at a small rate u (this small mutation rate should not affect significantly our expressions of associations at QLE, calculated in the absence of mutation); in this case, pj and pk remain small. By neglecting terms in pj2 and pk2 in Equation 24, one obtains that a modifier increasing recombination increases in frequency if ea×a lies between
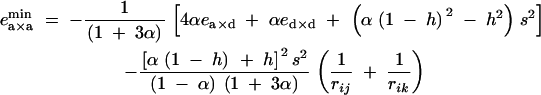 |
29 |
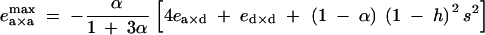 |
30 |
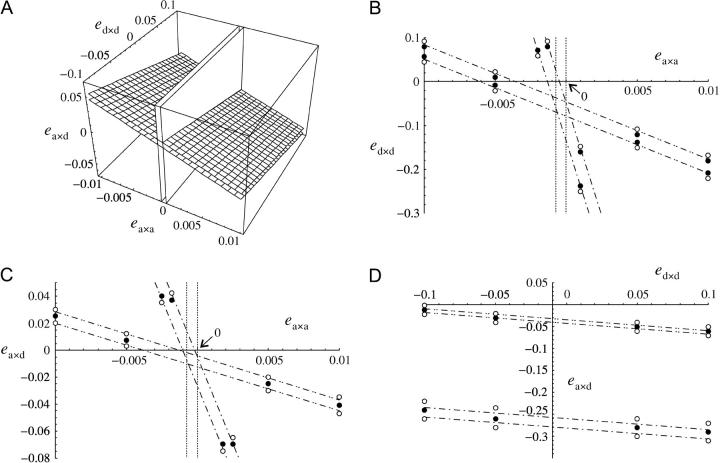
Figure 4 represents the range of epistasis values under which a modifier increasing recombination increases in frequency, for different values of α, at mutation-selection equilibrium. When α is small (Figure 4, 10−5), recombination increases only when ea×a is weak and negative, almost independently of ea×d and ed×d. As α increases, mutant homozygotes become more frequent, and ea×d and ed×d play a more important role; however, the parameter range under which recombination is selected remains narrow. In some sense, there is still a requirement for negative epistasis, as at least one of the parameters ea×a, ea×d, and ed×d must be negative for increased recombination to evolve. At high selfing rates, Equations 29 and 30 and simulations show that increased recombination is favored over a wider range of epistasis values: the space between the planes in Figure 4A increases (not shown). When α = 1, the modifier becomes neutral again, however, as recombination has no effect in this case.
Figure 4.—
Range of epistasis values under which a modifier increasing recombination increases in frequency, under gametophytic selfing, and under the fitness matrix given in Table 2. (A) Increased recombination is selected between the two vertical planes when α = 10−5 and between the two other planes when α = 10−1. (B–D) Regions of ea×a, ea×d, and ed×d, where increased recombination is selected, when α = 10−5 (dotted lines), α = 10−2 (dashed-dotted lines), and α = 10−1 (dashed-double-dotted lines). Each time, a modifier increasing recombination is predicted to increase in frequency between the two lines (obtained from Equations 29 and 30). Circles correspond to simulation results for α = 10−2 and 10−1: solid circle, the modifier increases in frequency when rare; open circle, it decreases in frequency. Parameter values are: (B) s = −0.1, h = 0.1, rij = 0.2, rik = 0.2, ea×d = 0.01; (C) ed×d = 0.01; and (D) ea×a = 0.01. In the simulations, dr = 0.01, hM = 1/2, and u = 10−5.
Sporophytic selfing:
Results under sporophytic selfing are qualitatively different from those under gametophytic selfing. In particular, modifiers increasing recombination can increase in frequency even when direct selection and epistasis are of the same order of magnitude (which is not the case under random mating and under gametophytic selfing). Since selection occurs only during the diploid phase, we drop D superscripts from aUVD coefficients. In the following, we again assume that epistasis is weak relative to direct selection: ajk, aj,k, ajk,j, ajk,k, and ajk,jk are of order ε2, while aj,⊘, ak,⊘, aj,j, and ak,k are of order ε. We also give approximations for the case where all selection coefficients are of the same order. Concerning the rate of self-fertilization, we consider the cases where α is of order 1, of order ε, and of order ε2. Finally, we still assume that the modifier effect dr is of order ε. Recursions for associations that influence the change in frequency of the modifier, for the different cases that we consider, are given in appendix C. Table 3 gives the leading-order terms of Δpi at QLE, under weak selection and weak epistasis, and for the different assumptions on the order of α. In the following, we give only results for hM = 1/2 (additive modifier); results are qualitatively similar under arbitrary hM, and, in many cases, the change in frequency at the modifier locus can be obtained simply by multiplying the expressions obtained by a factor 2[hM + pi(1 − 2hM)]; we indicate when this is the case. Expressions for arbitrary dominance of the modifier effect are given in appendix C.
TABLE 3.
Expressions for the change in frequency at the recombination modifier locus at QLE, under sporophytic selfing
| Weak selection | Weak epistasis | |
|---|---|---|
| α = O(1) | Δpi = ajk,jkCijk,jk + o(ε2) | Δpi = aj,jCij,j + ak,kCik,k + ajk,jkCijk,jk + o(ε3) |
| α = O(ε) | Δpi = ajk,⊘Cijk,⊘ + ajk,jkCijk,jk + o(ε3) | Δpi = ajk,jkCijk,jk + o(ε4) |
| α = O(ε2) | Δpi = ajk,⊘Cijk,⊘ + o(ε3) | Δpi = aj,⊘Cij,⊘ + ak,⊘Cik,⊘ + ajk,⊘Cijk,⊘ + ajk,jkCijk,jk + o(ε5) |
The modifier effect dr is assumed to be of order ε; under weak selection, all aU,V coefficients are of order ε, while under weak epistasis aj,⊘, ak,⊘, aj,j, and ak,k are of order ε and ajk,⊘, aj,k, ajk,j, ajk,k, and ajk,jk are of order ε2. The different rows correspond to different orders of magnitude of the selfing rate α.
α of order 1:
Under weak epistasis and strong selfing, the change in frequency of the modifier takes the form
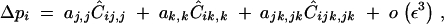 |
31 |
where again the hat stands for genetic associations at QLE. Indeed, under sporophytic selfing, the association Cijk,jk is generated only by selfing and by the effect of the modifier (even when selection is absent) and is therefore of order ε. When hM = 1/2, Ĉijk,jk is given by
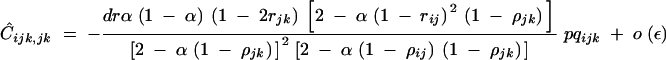 |
32 |
(appendix C), where ρij = 2rij(1 − rij), ρjk = 2rjk(1 − rjk), and again pqijk = piqipjqjpkqk. Equation 32 shows that Ĉijk,jk has the same sign as −dr; this reflects the fact that, under partial selfing, the correlation in homozygosity between loci j and k is lower among individuals carrying an allele that increases recombination between j and k. The associations Cij,j and Cik,k are generated by Cijk,jk and by selection at loci j and k and are of order ε2; at QLE, they are given by
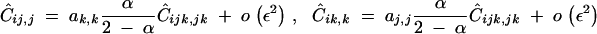 |
33 |
(appendix C). Other associations of the form CUi,V produce terms of higher order in the expression of Δpi. Equations 31 and 33 lead to
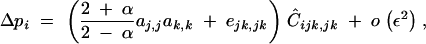 |
34 |
where ejk,jk equals ajk,jk − aj,jak,k and measures multiplicative dominance × dominance epistasis. Because Ĉijk,jk has the sign of −dr, we obtain that a modifier increasing recombination increases in frequency if
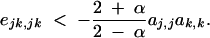 |
35 |
Applying the coefficients from appendix A, this condition becomes, under the fitness matrix given in Table 2,
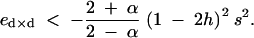 |
36 |
When epistasis and direct selection are of the same order of magnitude, this condition becomes ∼ed×d < 0: recombination is favored when double homozygotes are less fit than expected on the basis of single homozygotes, because recombination during sporophytic selfing then produces fewer double homozygotes. This mechanism thus provides an advantage to alleles that increase recombination, in the presence of negative dominance × dominance epistasis. Condition (36) indicates that the maximum value of ed×d for recombination to be selected is in fact slightly negative; this is due to the fact that, by reducing the proportion of double homozygotes, recombination also decreases the variance in fitness, which reduces the efficiency of natural selection and therefore gives a cost to recombination. In the absence of dominance × dominance epistasis (ed×d = 0), the advantage of recombination mentioned above is absent, and this cost selects against recombination. These mechanisms by which recombination evolves under sporophytic selfing are very different from those under random mating, as is discussed later.
α of order ε:
We now assume that the selfing rate is small: α is of order ε. In this case, Ĉijk,jk becomes of order ε2 and is given by Equation 32, expressed to the first order in α:
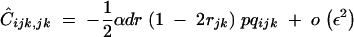 |
37 |
Solving recursions for genetic associations indicates that, at QLE, Ĉijk,⊘ is of order ε3, while other associations involving a single i index are of higher order. As a consequence, the change in frequency at the modifier locus is simply given by
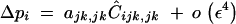 |
38 |
However, if epistasis terms are stronger (i.e., if all aU,V coefficients are of order ε), Ĉijk,⊘ becomes of the same order as Ĉijk,jk, while other associations involving a single i index are of higher order. In that case, we obtain
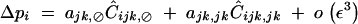 |
39 |
Solving the recursion for Cijk,⊘ gives
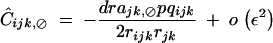 |
40 |
(appendix C). Indeed, selfing affects Ĉijk,⊘ only through terms of order ε3, which are neglected here. Equations 10, 37, and 40 lead to
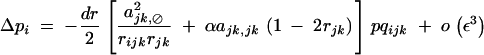 |
41 |
Therefore, under weak selection and weak selfing, we find that a modifier increasing recombination increases in frequency when dominance × dominance epistasis is sufficiently negative,
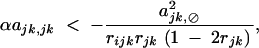 |
42 |
which, using Equations 54 and 55, gives
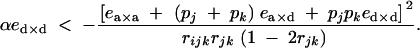 |
43 |
Under arbitrary dominance at the modifier locus, Equation 41 is multiplied by a factor 2[hM + pi(1 − 2hM)], which does not affect condition (42). When epistasis is weak (ajk,jk and ajk,⊘ of order ε2), this condition becomes ∼ajk,jk < 0, in agreement with Equation 38.
α of order ε2:
Finally, when the selfing rate is of order ε2, Ĉijk,jk and Ĉijk,⊘ are of order ε3, Ĉij,⊘ and Ĉik,⊘ are of order ε4, while other associations involving a single i index are of higher order, giving
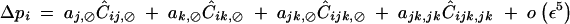 |
44 |
At QLE, Ĉijk,jk is still given by Equation 37, while other associations in (44) are given by
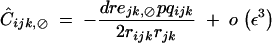 |
45 |
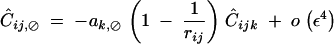 |
46 |
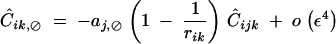 |
47 |
(see appendix C), giving for the change in frequency of the modifier,
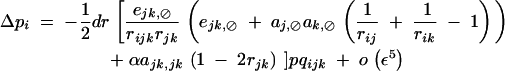 |
48 |
We checked this expression against numerical simulations, using the fitness matrix given by Table 2, again assuming that alleles 1 at loci j and k are deleterious (s < 0) and maintained at mutation-selection equilibrium, so that pj and pk remain small. In this case, the expressions given in appendix A give: aj,⊘ak,⊘ ≈ s2h2 + o(ε2), ejk,⊘ ≈ ea×a + o(ε2), and ajk,jk ≈ (1 − 2h)2s2 + ed×d + o(ε2). From this and from Equation 48, we expect that a modifier increasing recombination should increase in frequency if
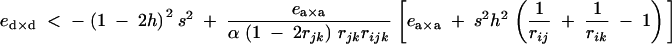 |
49 |
Figure 5 compares this prediction with simulation results obtained by iterating exact recursions for genotype frequencies. When α is very small (Figure 5, 10−5), recombination increases only when ea×a is weakly negative, almost independently of ed×d, as under random mating; as α increases, the range of ea×a under which recombination is selected greatly increases, as long as ed×d < 0. As α becomes strong (Figure 5, 10−1), approximation (49) still gives correct results and shows that recombination increases as long as ed×d is lower than a limit value (without lower bound).
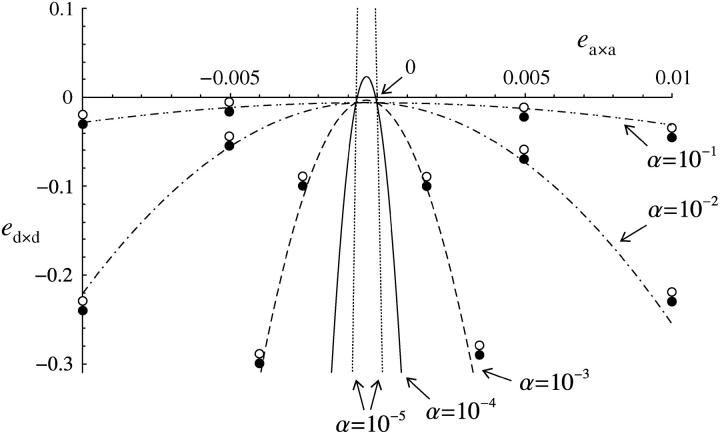
Figure 5.—
Maximum value of dominance × dominance epistasis (ed×d) for a modifier increasing recombination to be selected, under sporophytic selfing and under the fitness matrix given in Table 2, as a function of additive × additive epistasis (ea×a). The curves correspond to our analytical result for weak epistasis and weak selfing (Equation 49), for α = 10−5 (dotted curves), 10−4 (solid curve), 10−3 (dashed curve), 10−2 (dashed-dotted curve), and 10−1 (dashed-double-dotted curve). Circles correspond to simulation results for α = 10−3, 10−2, and 10−1: solid circle, the modifier increases in frequency when rare; open circle, it decreases in frequency. Parameter values are: rij = rjk = 0.2, s = −0.1, h = 0.1; in the simulations dr = 0.01, hM = 1/2, ea×d = 0.01, u = 10−5.
The axes of Figure 5 are not drawn to the same scale, the scale for ed×d being much larger than that for ea×a; we used these different scales to be able to represent the narrow parameter range under which recombination is selected for under random mating or when selfing is very small (α = 10−5), while at the same time presenting simulation results for strong absolute values of ed×d. However, drawing the figure in this way may give a false impression of the parameter range where recombination is favored. Figure 6 presents similar graphs, but where axes are drawn to the same scale. Decreasing the strength of selection (from s = −0.1 in Figure 6A to s = −0.05 in Figure 6B) reduces the area in which recombination is selected under random mating (this area is too small to be represented in Figure 6B), while it increases the area in which recombination is selected for under selfing (the threshold value for ed×d is higher in Figure 6B than in Figure 6A).
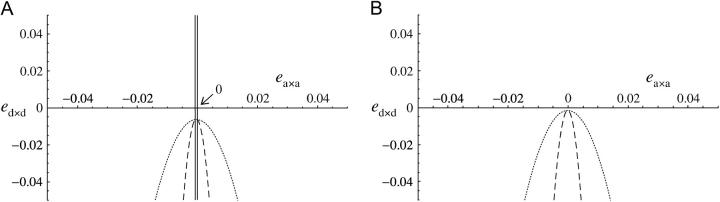
Figure 6.—
Values of ea×a and ed×d for which a modifier increasing recombination increases in frequency, under sporophytic selfing and under the fitness matrix given in Table 2. Solid lines, α = 0; dashed lines, α = 0.01; dotted lines, α = 0.1. The modifier increases in frequency between the solid lines in A and below the dashed and dotted lines in A and B. Parameter values are: (A) s = −0.1 and (B) s = −0.05, h = 0.1, rij = 0.2, rjk = 0.2.
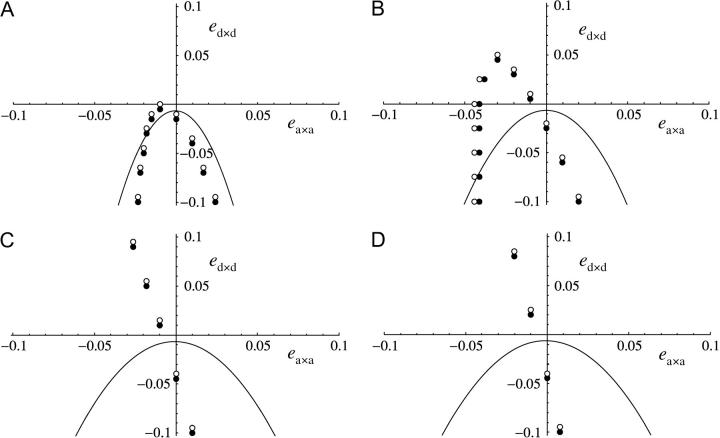
The QLE approximation does not hold under strong selfing. Comparisons between the prediction of Equation 49 and simulations are shown in Figure 7, for s = −0.1 and α = 0.3, 0.6, 0.9, and 0.99. As α gets large, the QLE approximation becomes clearly wrong: Equation 49 predicts that a modifier increasing recombination increases in frequency below the curves in Figure 7 (that is, for sufficiently negative ed×d), while simulations show that, when selfing is strong, recombination can increase even when ed×d is positive. Under strong selfing, selection on the recombination modifier becomes less dependent on ed×d and more dependent on ea×a, which in most cases has to be negative (recombination increases in Figure 7, C and D, left sides).
Figure 7.—
Values of ea×a and ed×d for which a modifier increasing recombination increases in frequency, under stronger sporophytic selfing and under the fitness matrix given in Table 2. (A) α = 0.3, (B) α = 0.6, (C) α = 0.9, and (D) α = 0.99. Other parameters are: s = −0.1, h = 0.1, ea×d = 0.01, rij = 0.2, rjk = 0.2, dr = 0.01, hM = 0.5, u = 10−5. The curves correspond to Equation 49, and the circles show simulation results; solid circle, the modifier increases in frequency when rare; open circle, it decreases in frequency.
DISCUSSION
Our model shows that self-fertilization has important effects on the evolution of recombination. Both sporophytic and gametophytic selfing generate correlations in homozygosity at different loci (associations such as Cjk,jk). While such correlations do not depend on recombination under gametophytic selfing, they are decreasing functions of recombination under sporophytic selfing, as individuals who recombine more produce fewer double homozygotes when they self. This mechanism generates an association Cijk,jk between the recombination modifier (locus i) and loci j and k under sporophytic selfing, even in the absence of selection at these loci. By contrast, under random mating, all associations between the recombination modifier and other loci are built by selection acting at these loci and by the modifier effect. This association Cijk,jk has important qualitative and quantitative effects. For example, in the case where direct selection and epistasis are of the same order, the evolution of modifiers that increase recombination becomes possible (a necessary condition being that dominance × dominance epistasis is negative) while it is not the case under random mating (Barton 1995). For example, in the case of recurrent deleterious mutations, increased recombination can evolve only when additive × additive epistasis is weakly negative when mating is random. This condition changes very fast as the rate of sporophytic selfing increases (Figure 5), selfing rates as low as 10−2 having substantial effects. As selfing increases, the condition for the evolution of recombination depends more and more on ed×d (which has to be negative, without a lower bound) and less and less on ea×a (in particular, recombination can increase when ea×a is positive, as long as ed×d is sufficiently negative, as can be seen in Figure 5). Importantly, self-fertilization not only increases the parameter range under which recombination is favored, but also strongly increases the strength of selection on recombination, because when α is strong, Cijk,jk is of higher order than the other associations between the modifier and the selected loci.
These results are in agreement with simulation results obtained by Charlesworth et al. (1979) and confirmed by Holsinger and Feldman (1983). Charlesworth et al. considered a fitness matrix where dominance × dominance epistatic effects are measured by four coefficients k1, k2, k3, and k4 (Table 2 in Charlesworth et al. 1979). It is possible to show that, under their fitness matrix, the coefficient ajk,jk equals −k1 − k2 − k3 − k4, to the first order in selective differences. Charlesworth et al. found that, when all ki are equal (and equal to k), increased recombination is selected when k is positive, provided that self-fertilization occurs (even at a small rate); under random mating, recombination does not increase. This result agrees with our Equations 36 and 42.
The mechanisms by which recombination evolves under sporophytic selfing (when ed×d < 0) and under random mating are also qualitatively different. Under random mating, and when epistasis is negative, recombination decreases the average fitness of offspring, while it increases their variance in fitness. This increase in variance drives the evolution of the modifier, provided that the initial decrease in fitness is not too high (which can be the case only when epistasis is weak). Recombination has the opposite effect under sporophytic selfing when ed×d < 0. Indeed, because sporophytic selfing generates an excess of double homozygotes (Cjk,jk > 0), it increases the variance in fitness. Recombination, however, reduces this excess of double homozygotes (Cjk,jk decreases as rjk increases, as can be seen from Equation C3) and thereby reduces the variance in fitness. However, when ed×d < 0, individuals who recombine more produce offspring that are more fit on average (because they produce fewer double homozygotes through selfing). As a result, conditions for higher rates of recombination to evolve are reversed: while recombination increases when epistasis is weakly negative under random mating, it increases under selfing when ed×d is more negative than a limit value (Equation 35).
Under gametophytic selfing, correlations in homozygosity at different loci do not depend on recombination, because selfing occurs among identical gametes (produced by a haploid gametophyte). Under this mating system, the association Cijk,jk is generated by selection at loci j and k and by the modifier effect, and results are more similar to those obtained under random mating (this can be seen from Equation 17, which gives the change in frequency of the modifier under gametophytic selfing, and which takes the same form as under random mating). Gametophytic selfing has, however, two important effects. First, it decreases the effectiveness of recombination, thereby increasing hitchhiking effects. As a consequence, the range of epistasis under which increased recombination can evolve is increased—this effect, however, is important mostly at high selfing rates (Figure 3). Second, by increasing homozygosity, gametophytic selfing increases the effects of additive × dominance and of dominance × dominance epistasis under diploid selection (Figure 4). In the case of deleterious mutations, the combination of ea×a, ea×d, and ed×d determines when modifiers that increase recombination can evolve (rather than just ea×a), and, in some cases, such modifiers evolve when ea×a is positive.
We expect that results for other sources of inbreeding, such as sib mating or population structure, should be qualitatively similar to those obtained under sporophytic selfing; indeed, inbreeding (and population structure) generates correlations in homozygosity, which in general decrease as recombination rates increase (gametophytic selfing being here an exception). Therefore, the result that negative dominance × dominance epistasis selects for increased recombination would probably be obtained from other models of inbreeding. These different cases remain to be investigated using specific models.
Our results stress the fact that the different components of epistasis play different roles in the evolution of recombination under inbreeding: for example, under sporophytic selfing, increased recombination can evolve when ea×a is positive, as long as ed×d is sufficiently negative. Therefore, it appears important to measure these different components separately (not only ea×a) to assess the deterministic theories for the evolution of recombination. At present, most experimental studies measuring epistasis between mutations affecting fitness have been performed on haploid organisms (de Visser et al. 1996, 1997a,b; Elena and Lenski 1997; Elena 1999; de la Peña et al. 2000; Wloch et al. 2001) or on diploids made homozygous at all loci (de Visser and Hoekstra 1998; Peters and Keightley 2000; Whitlock and Bourguet 2000), in which case the different components of epistasis cannot be separated. However, some studies on diploid plants have tested for dominance × dominance epistasis (Willis 1993; Falconer and Mackay 1996 and references therein); in these studies, different components of fitness are measured over successive generations of inbreeding. In the absence of dominance × dominance epistasis, the decline in fitness with inbreeding is expected to be linear, while negative ed×d leads to a greater than linear decrease (Crow and Kimura 1970, p. 80). Some of these studies found linear decreases, while others obtained accelerating decreases. However, this method is biased against finding negative dominance × dominance epistasis, because the worst genotypes can be lost by selection during the course of the experiment; therefore, linear decreases in fitness with inbreeding do not prove that dominance × dominance epistasis is absent.
In a recent article, Otto (2003) studied the hypothesis that sex might have evolved to promote segregation. She found that segregation evolves only under a restricted range of parameters with random mating, but under a much wider range in the presence of inbreeding (represented in her model by gametophytic selfing). With inbreeding, selection on a modifier of segregation is also much stronger, and she concluded that sexual reproduction is more likely to have been driven by the benefits of segregation than by the benefits of recombination. However, our model demonstrates that under sporophytic selfing, selection on a recombination modifier is also much stronger than under random mating (when there is dominance × dominance epistasis); therefore, which of the two effects of sex (segregation or recombination) brings more benefits is not so clear; models including both effects are needed to answer this question, in parallel with measures of dominance × dominance epistasis.
Acknowledgments
We thank Sally Otto, Sergey Gavrilets, François Rousset, Mike Whitlock, and an anonymous reviewer for helpful comments and discussions on previous versions of this manuscript. This work was supported by grant Action Concertée Incitative Jeunes Chercheurs no. 0693 from the French Ministry of Research.
APPENDIX A
Under the fitness matrix given by Table 2, to the first order in s, ea×a, ea×d, and ed×d (weak selection), aU,VD coefficients are given by
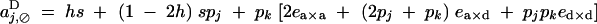 |
A1 |
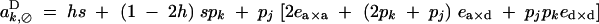 |
A2 |
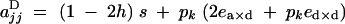 |
A3 |
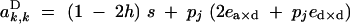 |
A4 |
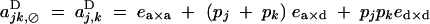 |
A5 |
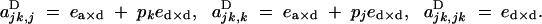 |
A6 |
To the second order in s, and to the first order in ea×a, ea×d, and ed×d (weak epistasis), aU,VD coefficients are given by
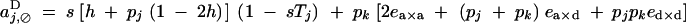 |
A7 |
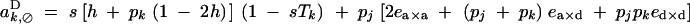 |
A8 |
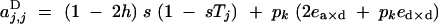 |
A9 |
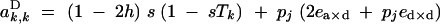 |
A10 |
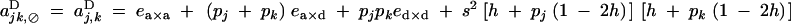 |
A11 |
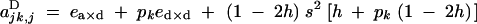 |
A12 |
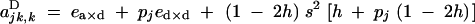 |
A13 |
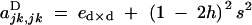 |
A14 |
with
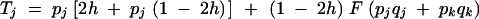 |
A15 |
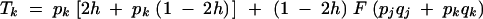 |
A16 |
and F is the neutral inbreeding coefficient, equal to α/(2 − α) under sporophytic selfing and α under gametophytic selfing.
APPENDIX B
Here we give recursions for gametophytic selfing under weak epistasis: ajH, akH, aj,⊘D, ak,⊘D, aj,jD, and ak,kD are of order ε, while ajkH, ajk,⊘D, aj,kD, ajk,jD, ajk,kD, and ajk,jkD are of order ε2.
Haploid selection:
The change in allele frequency at the modifier locus (locus i) due to selection during the haploid phase is given by
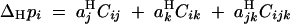 |
B1 |
(from Kirkpatrick et al. 2002, Equation 10). The recursions for linkage disequilibria to leading orders in ε are given by
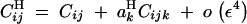 |
B2 |
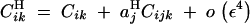 |
B3 |
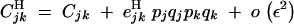 |
B4 |
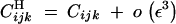 |
B5 |
(from Kirkpatrick et al. 2002, Equations 9 and 15).
Diploid selection:
The change in allele frequency at the modifier locus during diploid selection is given by
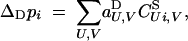 |
B6 |
which, using Equations 7 and 8, gives
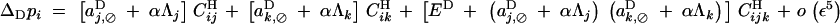 |
B7 |
with
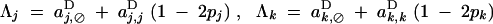 |
B8 |
and
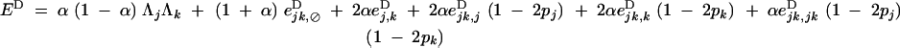 |
B9 |
Genetic associations after diploid selection are given by
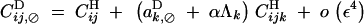 |
B10 |
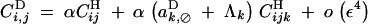 |
B11 |
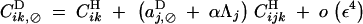 |
B12 |
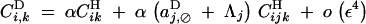 |
B13 |
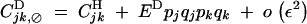 |
B14 |
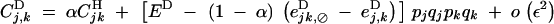 |
B15 |
Finally, we have
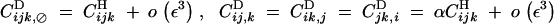 |
B16 |
Recombination:
Recombination does not affect allele frequencies. Recursions for genetic associations in the presence of the recombination modifier are the same as in Barton (1995),
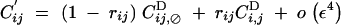 |
B17 |
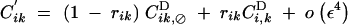 |
B18 |
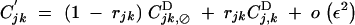 |
B19 |
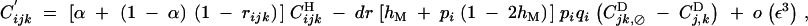 |
B20 |
where, rijk is the probability that at least one crossing over occurs between loci i, j, and k, in the absence of the modifier.
APPENDIX C
In the absence of selection, selfing increases homozygosity and generates correlations in homozygosity at different loci, leading to positive associations between identical sets of loci present on homologous chromosomes. Equilibrium values for these associations under sporophytic selfing are obtained by solving recursions derived from Equations 13 and 14, with 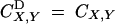 , and all δtS,T (which represent the modifier effect) equal to zero. One obtains for the association between homologous genes at a single locus,
, and all δtS,T (which represent the modifier effect) equal to zero. One obtains for the association between homologous genes at a single locus,
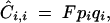 |
C1 |
where F = α/(2 − α) measures the deviation from Hardy-Weinberg equilibrium at a single locus, due to selfing. Associations between pairs of homologous genes at two loci are given by
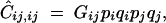 |
C2 |
where
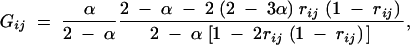 |
C3 |
measuring the correlation between homozygosity at two linked loci (again due to selfing). Finally, one has
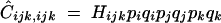 |
C4 |
with
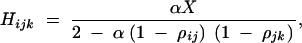 |
C5 |
where
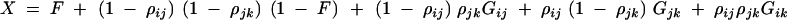 |
C6 |
and ρij = 2rij(1 − rij) and ρjk = 2rjk(1 − rjk).
α of order 1:
When α is of order 1, Ĉijk,jk is of order ε; indeed, Cijk,jk is generated only by selfing and by the modifier effect. To linear order in dr, one obtains from Equations 13 and 14
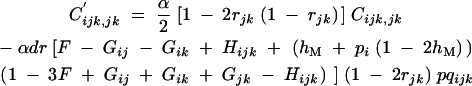 |
C7 |
with pqijk = piqipjqjpkqk. Solving Equation C7 for 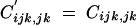 gives
gives
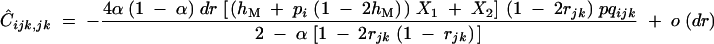 |
C8 |
with
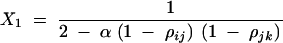 |
C9 |
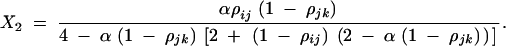 |
C10 |
Except dr, all terms in the fraction of Equation C8 are positive when rij and rjk are in the range [0, 1/2], and α, hM, and pi are in [0, 1]; therefore, Ĉijk,jk has the same sign as −dr.
Ĉij,j and Ĉik,k are of order ε2 and are obtained by solving the recursions
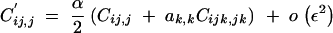 |
C11 |
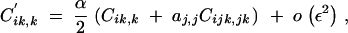 |
C12 |
giving at QLE,
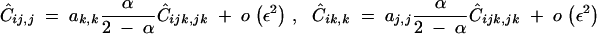 |
C13 |
Under weak epistasis, Ĉijk,j, Ĉij,jk, Ĉijk,k, and Ĉij,ik are also of order ε2, while other associations involving a single i index are of higher order.
α of order ε:
When α is of order ε, Cijk,jk becomes of order ε2 and is given by
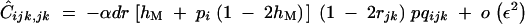 |
C14 |
(from Equation C8). Under weak epistasis, Cijk,⊘ is of order 3 at QLE, while other associations involving a single i index are of higher order. Under weak selection (when all aU,V coefficients are of order ε), Cijk,⊘ is of order ε2 and is obtained by solving the recursions
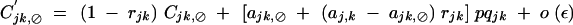 |
C15 |
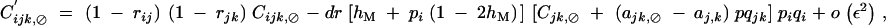 |
C16 |
giving at QLE,
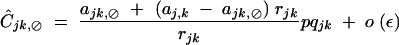 |
C17 |
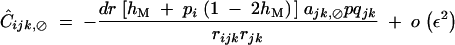 |
C18 |
with rijk = 1 − (1 − rij)(1 − rjk).
α of order ε2:
When α is of order ε2, Ĉijk,jk is of order ε3 and is given by Equation C14. Under weak epistasis, Ĉijk,⊘ is of order ε3 and is obtained by solving
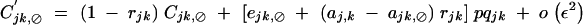 |
C19 |
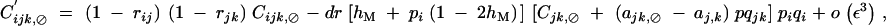 |
C20 |
giving at QLE,
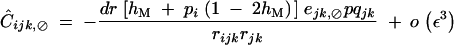 |
C21 |
Finally, Ĉij,⊘ and Ĉik,⊘ are of order ε4 and are obtained by solving
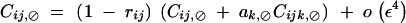 |
C22 |
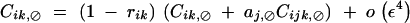 |
C23 |
Other associations involving a single i index are of higher order in ε.
References
- Agrawal, A., and J. Chasnov, 2001. Recessive mutations and the maintenance of sex in structured populations. Genetics 158: 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo, M. T. K., 1973. Chiasma frequency evidence on the evolution of autogamy in Limnanthes floccosa (Limnanthaceae). Evolution 27: 679–688. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., 1995. A general model for the evolution of recombination. Genet. Res. 65: 123–144. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and M. Turelli, 1991. Natural and sexual selection on many loci. Genetics 127: 229–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1976. Recombination modification in a fluctuating environment. Genetics 83: 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1980. The cost of sex in relation to the mating system. J. Theor. Biol. 84: 655–671. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., B. Charlesworth and C. Strobeck, 1977. Effects of selfing on selection for recombination. Genetics 86: 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., B. Charlesworth and C. Strobeck, 1979. Selection for recombination in partially self-fertilizing populations. Genetics 93: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970 An Introduction to Population Genetics Theory. Harper & Row, New York.
- de la Peña, M., S. F. Elena and A. Moya, 2000. Effect of deleterious mutation-accumulation on the fitness of RNA bacteriophage MS2. Evolution 54: 686–691. [DOI] [PubMed] [Google Scholar]
- de Visser, J. A. G. M., and R. F. Hoekstra, 1998. Synergistic epistasis between loci affecting fitness: evidence in plants and fungi. Genet. Res. 71: 39–49. [Google Scholar]
- de Visser, J. A. G. M., R. F. Hoekstra and H. van den Ende, 1996. The effect of sex and deleterious mutations on fitness in Chlamydomonas. Proc. R. Soc. Lond. Ser. B 263: 193–200. [Google Scholar]
- de Visser, J. A. G. M., R. F. Hoekstra and H. van den Ende, 1997. a An experimental test for synergistic epistasis and its application in Chlamydomonas. Genetics 145: 815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser, J. A. G. M., R. F. Hoekstra and H. van den Ende, 1997. b Test of interaction between genetic markers that affect fitness in Aspergillus niger. Evolution 51: 1499–1505. [DOI] [PubMed] [Google Scholar]
- Elena, S. F., 1999. Little evidence for synergism among deleterious mutations in a nonsegmented RNA virus. J. Mol. Evol. 49: 703–707. [DOI] [PubMed] [Google Scholar]
- Elena, S. F., and R. E. Lenski, 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390: 395–397. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996 Introduction to Quantitative Genetics. Addison Wesley Longman, Harlow, UK.
- Feldman, M. W., F. B. Christiansen and L. D. Brooks, 1980. Evolution of recombination in a constant environment. Proc. Natl. Acad. Sci. USA 77: 4838–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1930 The Genetical Theory of Natural Selection. Oxford University Press, Oxford.
- Gibbs, P. E., C. Milne and M. Vargas Carillo, 1975. Correlation between the breeding system and recombination index in five species of Senecio. New Phytol. 75: 619–626. [Google Scholar]
- Grant, V., 1958. The regulation of recombination in plants. Cold Spring Harbor Symp. Quant. Biol. 23: 337–363. [DOI] [PubMed] [Google Scholar]
- Holsinger, K. E., and M. W. Feldman, 1983. Linkage modification with mixed random mating and selfing: a numerical study. Genetics 103: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, M., T. Johnson and N. H. Barton, 2002. General models of multilocus evolution. Genetics 161: 1727–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1982. Selection against harmful mutations in large sexual and asexual populations. Genet. Res. 40: 325–332. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1993. Classification of hypotheses on the advantage of amphimixis. J. Hered. 84: 372–387. [DOI] [PubMed] [Google Scholar]
- Lenormand, T., 2003. The evolution of sex dimorphism in recombination. Genetics 163: 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand, T., and J. Duteil, 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3 (3): e63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand, T., and S. P. Otto, 2000. The evolution of recombination in a heterogeneous environment. Genetics 156: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., 1971. What use is sex? J. Theor. Biol. 30: 319–335. [DOI] [PubMed] [Google Scholar]
- McCauley, D. E., D. P. Whittier and L. M. Reilly, 1985. Inbreeding and the rate of self-fertilization in a grape fern, Botrychium dissectum. Am. J. Bot. 72: 1978–1981. [Google Scholar]
- Muller, H. J., 1932. Some genetic aspects of sex. Am. Nat. 66: 118–138. [Google Scholar]
- Nagylaki, T., 1993. The evolution of multilocus systems under weak selection. Genetics 134: 627–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., 2003. The advantages of segregation and the evolution of sex. Genetics 164: 1099–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., and N. H. Barton, 1997. The evolution of recombination: removing the limits to natural selection. Genetics 147: 879–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., and N. H. Barton, 2001. Selection for recombination in small populations. Evolution 55: 1921–1931. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and M. W. Feldman, 1997. Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Popul. Biol. 51: 134–147. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and T. Lenormand, 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3: 252–261. [DOI] [PubMed] [Google Scholar]
- Peters, A. D., and P. D. Keightley, 2000. A test for epistasis among induced mutations in Caenorhabditis elegans. Genetics 156: 1635–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, N., P. Koul and A. K. Koul, 1992. Genetic systems of six species of Plantago (Plantaginaceae). Plant Syst. Evol. 181: 1–9. [Google Scholar]
- Soltis, D. E., and P. S. Soltis, 1986. Electrophoretic evidence for inbreeding in the fern Botrychium virginianum (Ophioglossaceae). Am. J. Bot. 73: 588–592. [Google Scholar]
- Stebbins, G. L., 1958. Longevity, habitat and release of genetic variability in the higher plants. Cold Spring Harbor Symp. Quant. Biol. 23: 365–377. [DOI] [PubMed] [Google Scholar]
- Uyenoyama, M. K., and B. Bengtsson, 1989. On the origin of meiotic reproduction: a genetic modifier model. Genetics 123: 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ved Brat, S., 1965. Genetic systems in Allium. III. Meiosis and breeding systems. Heredity 20: 325–339. [Google Scholar]
- Whitlock, M. C., and D. Bourguet, 2000. Factors affecting the genetic load in Drosophila: synergistic epistasis and correlations among fitness components. Evolution 54: 1654–1660. [DOI] [PubMed] [Google Scholar]
- Williams, G. C., and J. B. Mitton, 1973. Why reproduce sexually? J. Theor. Biol. 39: 545–554. [DOI] [PubMed] [Google Scholar]
- Willis, J. H., 1993. Effects of different levels of inbreeding on fitness components in Mimulus guttatus. Evolution 47: 864–876. [DOI] [PubMed] [Google Scholar]
- Wloch, D. M., R. H. Borts and R. Korona, 2001. Epistatic interactions of spontaneous mutations in haploid strains of the yeast Saccharomyces cerevisiae. J. Evol. Biol. 14: 310–316. [Google Scholar]
- Zarchi, Y., G. Simchen, J. Hillel and T. Schaap, 1972. Chiasmata and the breeding system in wild populations of diploid wheats. Chromosome 38: 77–94. [Google Scholar]