Abstract
Highly repetitive satellite DNA sequences are main components of heterochromatin in higher eukaryotic genomes. It is well known that satellite repeats can expand and contract dramatically, which may result in significant genome size variation among genetically related species. The origin of satellite repeats, however, is elusive. Here we report a satellite repeat, Sobo, from a diploid potato species, Solanum bulbocastanum. The Sobo repeat is mapped to a single location in the pericentromeric region of chromosome 7. This single Sobo locus spans ∼360 kb of a 4.7-kb monomer. Sequence analysis revealed that the major part of the Sobo monomer shares significant sequence similarity with the long terminal repeats (LTRs) of a retrotransposon. The Sobo repeat was not detected in other Solanum species and is absent in some S. bulbocastanum accessions. Sobo monomers are highly homogenized and share >99% sequence identity. These results suggest that the Sobo repeat is a recently emerged satellite and possibly originated by a sudden amplification of a genomic region including the LTR of a retrotransposon and its flanking genomic sequences.
TANDEMLY repetitive DNA elements are frequently referred to as satellite DNAs because they were first isolated from satellite bands upon centrifugation in density gradients. Satellite DNAs are major constituents of large complex eukaryotic genomes. As a classical example, the HS satellite of the kangaroo rat Dipodomys ordii contains TTAGGG as its principal constituent and comprises 30% of the genomic DNA (Hatch and Mazrimas 1974; Fry and Salser 1977). Satellite sequences can diverge rapidly and their evolutionary patterns often do not follow any obvious phylogenetic order. The HS satellite of D. ordii in a close relative, D. deserti, is barely detectable (Mazrimas and Hatch 1972; Hatch and Mazrimas 1974). Over 160 families of satellite repeats have been described in higher plants (Macas et al. 2002). Satellite sequences reported in plants often show erratic distributions and great differences in their abundance among closely related species (Schmidt and Heslop-Harrison 1993; Kubis et al. 1997; Macas et al. 2000; Frello et al. 2004). Some satellite elements, however, are nearly universal within taxa. For example, the 2D8 satellite repeat discovered in potato is present in almost all species throughout the genus Solanum (Stupar et al. 2002).
It has been well established that satellite repeats are often associated with heterochromatic features and located mainly in the centromeric and telomeric regions. There is increasing evidence that many satellite repeats are structural or functional components of eukaryotic chromosomes. Satellite DNAs are often the major DNA components of the functional centromeres of complex eukaryotic species (Henikoff et al. 2001; Jiang et al. 2003). Nevertheless, the origin of satellite repeats has been elusive. Several hypotheses were proposed to explain the birth of satellite DNAs. A tandem repeat may be derived from nonrepetitious sequences by repeated and random unequal crossing overs (Smith 1976) or may be generated by both replication slippage and unequal crossing over and subsequently expanded by amplification mechanisms, which may create a new satellite (Walsh 1987). Salser et al. (1976) proposed a “library” hypothesis to explain the evolution of satellite repeats among genetically related species. According to this hypothesis, related species share a common library of satellite sequences. Specific members within the library may be amplified in some species and are present at low levels in other species (Salser et al. 1976; Fry and Salser 1977). Therefore, the appearance of most “new” satellites may not occur de novo, but due to amplification of one of the satellites present at a low level in the “library” (Salser et al. 1976).
Potato provides an excellent model to study the evolution of repetitive DNA elements. The asexual propagation of the potato species may minimize meiotic mechanisms that would remove newly emerged repeats (Stupar et al. 2002). Solanum bulbocastanum (2n = 2x = 24) is a diploid tuber-forming wild species distributed throughout central and southern Mexico to Guatemala (Spooner et al. 2004). This species has been regarded as self-incompatible on the basis of extensive pollination studies of different S. bulbocastanum accessions (Graham et al. 1959). Because of the paucity of seed balls on plants under natural conditions, asexual production through tubers is probably the main mode of propagation of this species.
Here we report the isolation and characterization of a tandem repeat specific to S. bulbocastanum. This repeat, named Sobo, was investigated with both molecular and cytogenetic approaches. Our results suggest that the Sobo repeat has emerged recently and is possibly derived from a DNA amplification event. The implications of the Sobo repeat for the origin of satellite DNAs are discussed.
MATERIALS AND METHODS
Plant materials:
S. bulbocastanum clone PT29 (PI 243510) was used in cytological and DNA analyses. Additional accessions or clones of the three subspecies of S. bulbocastanum (ssp. bulbocastanum, ssp. dolichophyllum, and ssp. partitum) were provided by James Bradeen (University of Minnesota) or obtained from the Potato Introduction Station (Sturgeon Bay, WI) (Table 1). The haploid clone USW1 (2n = 2x = 24), derived from S. tuberosum cv. Katahdin, was used in Southern blot analysis. DNA of other Solanum species used in this study, including S. etuberosum, S. ochranthum, S. morelliforme, S. bulbocastanum, S. cardiophyllum, S. lesteri, S. capsicibaccatum, S. chacoense, S. boliviense, S. infundibuliforme, S. agrimonifoluim, S. albornozii, S. verrucosum, S. multidissectum, S. oplocense, S. curtilobum, S. fendleri, S. iopetalum, and tomato (Lycopersicon esculentum), were kindly provided by David Spooner, U.S. Department of Agriculture-Agricultural Research Service and Department of Horticulture, University of Wisconsin (Madison, WI).
TABLE 1 .
Distribution of the Sobo repeat amongS. bulbocastanum accessions
| Subspecies | Accession | Origin | Southern blot |
Materials for FISH |
Interphase FISHa | |
|---|---|---|---|---|---|---|
| 1 | bulbocastanum | PI 243345C | Federal District, Mexico | ND | Tuber | No signal |
| 2 | bulbocastanum | PI 243505C | Federal District, Mexico | ND | Tuber | No signal |
| 3 | bulbocastanum | PI 243513B | Morelos, Mexico | ND | Tuber | No signal |
| 4 | dolichophyllum | PI 253210F | Morelos, Mexico | ND | Tuber | One signal |
| 5 | bulbocastanum | PI 275187A | Michoacan, Mexico | ND | Tuber | No signal |
| 6 | bulbocastanum | PI 275188E | Mexico | ND | Tuber | Two signals |
| 7 | dolichophyllum | PI 255516 | Jalisco, Mexico | ND | Tuber | Two signals |
| 8 | bulbocastanum | PI 275185 | Federal District, Mexico | Present | Seeds | Zero, one, two signals |
| 9 | bulbocastanum | PI 275192 | Tlaxcala, Mexico | Present | Seeds | Zero, one, two signals |
| 10 | bulbocastanum | PI 275196 | Oaxaca, Mexico | Present | Seeds | Two signals |
| 11 | bulbocastanum | PI 347757 | Michoacan, Mexico | Absent | Seeds | No signal |
| 12 | bulbocastanum | PI 498223 | Oaxaca, Mexico | Present | Seeds | Two signals |
| 13 | partitum | PI 275200 | Huehuetenango, Guatemala | Absent | Seeds | No signal |
| 14 | bulbocastanum | PI 243510 (clone PT29) | Mexico Federal District | Present | Tuber | One signal |
ND, no data.
For lines that were obtained as seeds, we germinated 10 seeds from each line. Root tips were collected and mixed for cytological preparation. If nuclei with zero, one, or two FISH signals were all observed, it indicates that the original plant contains a hemizygous Sobo locus. If nuclei with either zero or two FISH signals were observed, it indicates that the original plant contains either no or a homozygous Sobo locus.
Cloning, sequencing, and sequence analysis:
A bacterial artificial chromosome (BAC) clone 28G12 was isolated from a S. bulbocastanum library (Song et al. 2000). BAC DNA was isolated using a standard alkaline lysis method and was digested with HindIII. The digested DNA fragments were resolved on an agarose gel. The two HindIII fragments, 3.8 kb and 866 bp, respectively, were cloned into pUC18 vector. One 3.8-kb clone and one 866-bp clone were completely sequenced using the GPS-1 Genome Priming System (New England Biolabs, Beverly, MA) following the manufacturer's instructions. Ends of additional clones from both fragments were also sequenced. Sequence analyses were performed using the Lasergene 99 software package (DNAStar, Madison, WI), dotter (Sonnhammer and Durbin 1995), and Staden package (Staden 1996). Sequence similarity searches were performed using BLAST and FASTA (Pearson and Lipman 1988; Altschul et al. 1997), and detection of conserved protein domains was done using RPS-Blast (Marchler-Bauer et al. 2003).
Fluorescence in situ hybridization:
The fluorescence in situ hybridization (FISH) procedures were described previously (Jiang et al. 1995; Jackson et al. 1998). Probe DNA was labeled with biotin-dUTP or digoxigenin-dUTP in standard nick translation reactions and was detected with avidin-FITC and antidigoxigenin-rhodamine. Images were digitally captured with a CCD camera attached to an Olympus microscope and a Macintosh computer. The length of the fiber-FISH signals were digitally measured using IPLab software and converted to kilobases using a conversion ratio of 3.21 kb/μm (Cheng et al. 2002). Plasmid pTa71, which contains the coding sequences for the 18S·26S ribosomal RNA genes of wheat (Gerlach and Bedbrook 1979), was used as a rDNA probe.
Southern blot hybridization:
Genomic DNAs were isolated from young leaves of greenhouse-grown plants. Restriction enzyme digestions of the DNA samples were performed and fractionated by agarose gel electrophoresis. DNAs were transferred to Hybond-H+ membrane (Amersham Biosciences, Piscataway, NJ). Prehybridization and hybridization experiments were performed at 65°. The membranes were hybridized with 32P-labeled DNA probe. After an overnight hybridization, membranes were sequentially washed with 2× SSC plus 0.1% SDS, then with 1× SSC plus 0.1% SDS, and finally with 0.5× SSC plus 0.1% SDS for 20 min each. The membranes were exposed to X-ray films and developed.
RESULTS
Isolation and cytological characterization of the Sobo repeat from S. bulbocastanum:
In an effort to characterize the major repetitive DNA families in the potato genome, we screened a BAC library of S. bulbocastanum using the genomic DNA of S. bulbocastanum as a probe. A BAC clone, 28G12, showed strong hybridization. BAC 28G12 contains a 150-kb insert based on pulsed-field gel electrophoresis (PFGE) (data not shown). HindIII digestion of 28G12 DNA produced only two bands, 866 bp and 3.8 kb, respectively, in addition to the vector band (7.4 kb) (Figure 1). This result suggests that BAC 28G12 contains a tandemly repeated element, which we named the “Sobo” (Solanum bulbocastanum) repeat.
Figure 1.—
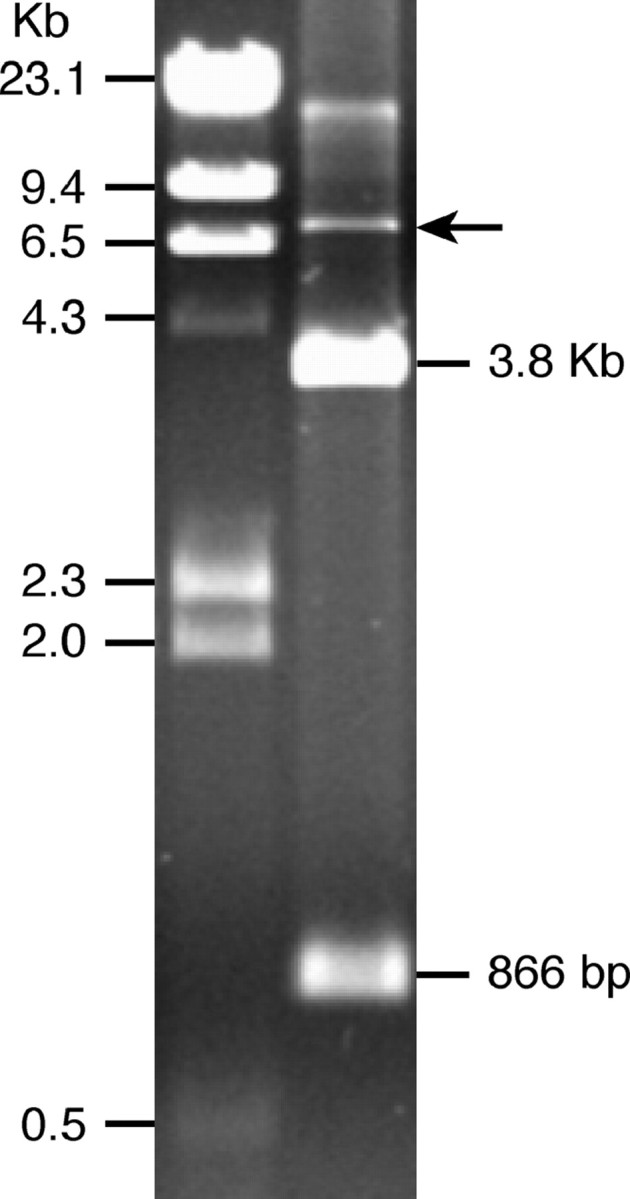
Agarose gel electrophoresis of BAC 28G12 following HindIII digestion (right lane). BAC 28G12 produces two bands with sizes of 3.8 kb and 866 bp, respectively. Arrow points to the BAC vector.
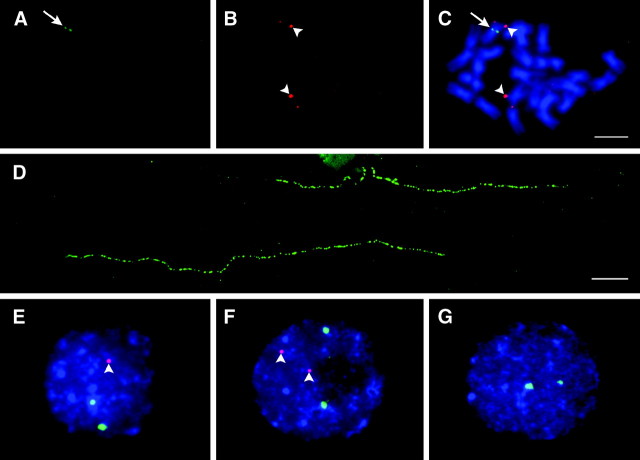
FISH analysis using BAC 28G12 as a probe revealed that this BAC hybridizes to only 1 of the 24 S. bulbocastanum (PI 243510; clone PT29) chromosomes (Figure 2, A–C). This hemizygous Sobo locus was mapped to the pericentromeric region of chromosome 7, which was identified by co-FISH mapping using the chromosome 7-specific BAC clone 38O2 (Dong et al. 2000) (Figure 2, A–C). Fiber-FISH was used to estimate the DNA size of the Sobo locus (Figure 2D). To better estimate the variation caused by the variable stretching degree of the genomic DNA fibers, we included the known BAC clone 220C03 in the same cytological preparations as an internal control. BAC 220C03 contains ∼100 kb of single-copy sequences based on PFGE analysis (Bradeen et al. 2003). The average length of 16 fiber-FISH signals derived from BAC 220C03 was 34.6 μm (data not shown), which converts into 111 kb of DNA using the conversion rate of 3.21 kb/μm (Cheng et al. 2002). This estimation is close to 100 kb estimated by PFGE, indicating that the 3.21 kb/μm conversion ratio is relatively accurate. The average length of 28 fiber-FISH signals derived from the Sobo repeat was 112 ± 38 μm. Using the 3.21 kb/μm conversion rate, we estimate that the Sobo repeat spans 360 ± 122 kb.
Figure 2.—
Cytogenetic mapping of the Sobo locus in S. bulbocastanum. (A) A single FISH signal derived from BAC 28G12 (arrow) was observed in the S. bulbocastanum accession PI 243510 (clone PT29). (B) FISH signals (arrowheads) derived from the chromosome 7-specific BAC clone 38O2. (C) FISH signals are merged with the somatic metaphase chromosomes of PT29. The Sobo locus (arrow) is located in the centromere-proximal region of chromosome 7. Bar, 5 μm. (D) A microscopic field containing two fiber-FISH signals derived from the Sobo repeat in S. bulbocastanum accession PI 243510 (PT29). (E–G) Representative interphase nuclei containing 1, 2, or 0 FISH sites (red signals, arrowheads) from S. bulbocastanum accessions PI 275185, PI 498223, and PI 347757, respectively. The 18S·26S ribosomal RNA genes (green signals) were used as a reference probe. Bar, 10 μm.
The Sobo repeat shares sequence similarity with the long terminal repeat of the Sore1 retrotransposon:
The two HindIII fragments, 866 and 3829 bp, respectively, were subcloned into the pUC18 vector. One plasmid clone derived from each of these two fragments was fully sequenced. Restriction mapping revealed that these two fragments are from a single monomeric unit and that the 866-bp fragment represents the first part of the monomer (data not shown). Thus, these two fragments are combined as a single 4689-bp Sobo element (AY849929 in GenBank) by adding the 866- and 3829-bp elements and subtracting a 6-bp HindIII cloning site in the junction.
Dot-plot analysis and sequence similarity searches of the Sobo monomer revealed a complex structure, including inverted and direct subrepeats within the monomer and regions of homology to several different genomic sequences (Figure 3). The region between nucleotides 368–837 is homologous to a segment of a BAC clone from S. bulbocastanum chromosome 8 (AY303171 in GenBank, nucleotides 145173–145650, FASTA expectation value 4.4e−30); this sequence is of unknown origin and does not show similarity to any other entry in the GenBank. The region between nucleotides 3390 and 4100 containing tandem subrepeats is highly similar (expectation value 1.3e−34) to the satellite repeat Sb4A of S. brevidens (Preiszner et al. 1994).
Figure 3.—
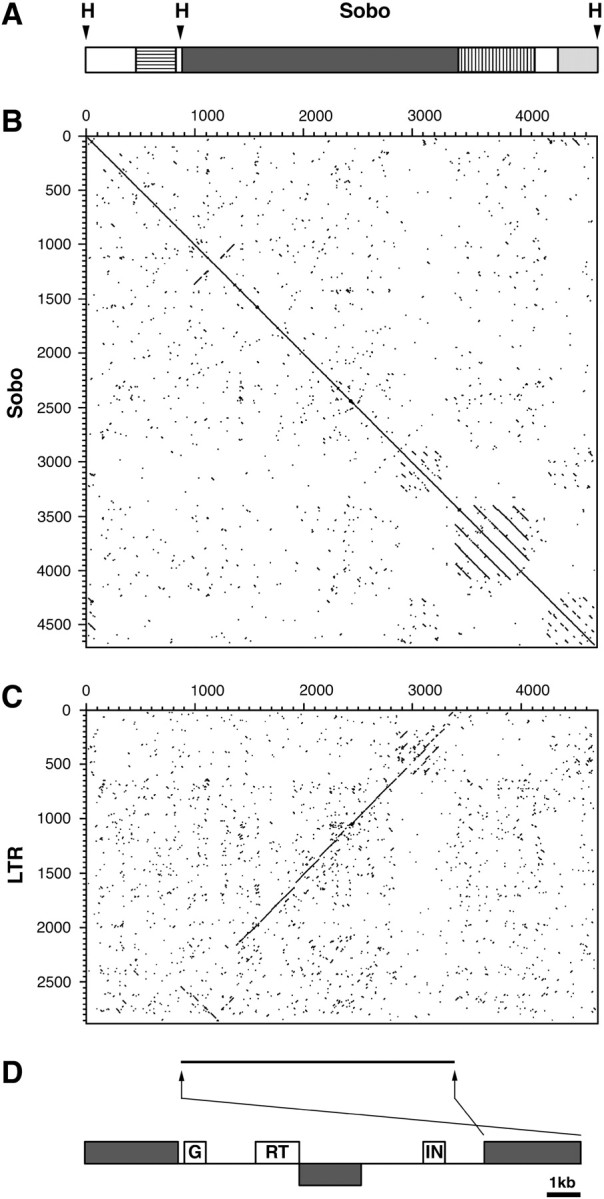
Structure of the Sobo monomer sequence. (A) Schematic of the Sobo monomer showing positions of two HindIII sites (H) and regions of homology to the S. bulbocastanum sequence AY303171 (horizontal lines), the satellite repeat Sb4A of S. brevidens (vertical lines), and the LTR of retrotransposon Sore1 of S. demissum (dark shading). The 3′-end of the Sobo monomer includes imperfect tandem repeats with scattered homology to the structurally similar region within the LTR of Sore1 (light shading). (B) The dot-plot comparison of the Sobo sequence with itself and (C) with the right LTR of the Sore1 element. Sequence similarity >67% (at least 40 identical nucleotides within a window of 60) is represented with a dot or diagonal line. (D) A scheme of the Sore1 element in AC149290 showing positions of the LTR sequences (shading; the insertion of a partial LTR sequence within the element is in reverse orientation) and putative gag (G), reverse transcriptase (RT), and integrase (IN) coding domains. Horizontal line and arrows under the dot-plot show position and orientation of the LTR-like sequence within the Sobo element.
The majority of the Sobo monomer (nucleotides 860–3380 and 4300–4688; Figure 3, A and C) displays similarity to a long terminal repeat (LTR) of a retrotransposon identified in a genomic sequence of S. demissum (AC149290, position 123425–138040). This element, designated as Sore1 (Solanum retrotransposon 1), is 14,616 bp long, contains LTRs of 2699 and 2857 bp, respectively, and is flanked by a 5-bp target site duplication (TSD). On the basis of the sequence divergence of its LTRs (85.6% sequence similarity) and the presence of a mutation within the TSD and numerous mutations that interrupt open reading frames coding for retroelement proteins (data not shown), this Sore1 element may be an ancient copy. In addition, there is an insertion of a part of an inverted LTR sequence within the coding region of the element (Figure 3D). However, detection of conserved protein domains using RPS-BLAST with intact segments of the element-coding regions allowed us to identify putative gag, reverse transcriptase, and integrase domains (expectation values of 4e−10, 6e−10, and 1e−11, respectively). The order of these domains indicated that this element belongs to the Ty3/gypsy group of retrotransposons.
We sequenced the ends of four additional plasmid clones derived from the 866-bp fragment and detected no sequence polymorphism. Similarly, we sequenced the ends of seven additional clones derived from the 3829-bp fragment. No polymorphism was detected among the ∼300 bp that spans the 3′ sequence ends. The 5′ sequence ends of the seven clones, however, can be divided into two subgroups. Sequences within each subgroup are identical. The sequences between two subgroups contain eight single nucleotide polymorphism sites scattered within a 580-bp region. Collectively, the sequencing data show that the monomers of the Sobo repeats share >99% sequence identity.
The Sobo element is present only in S. bulbocastanum:
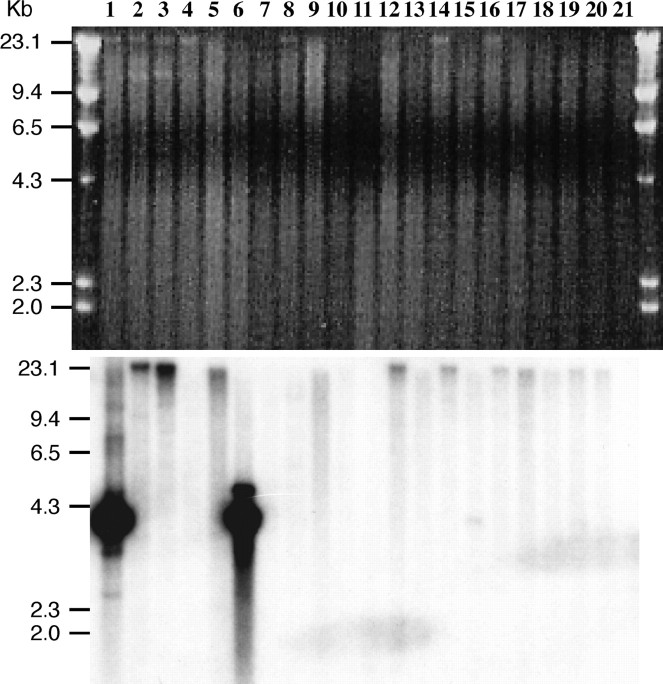
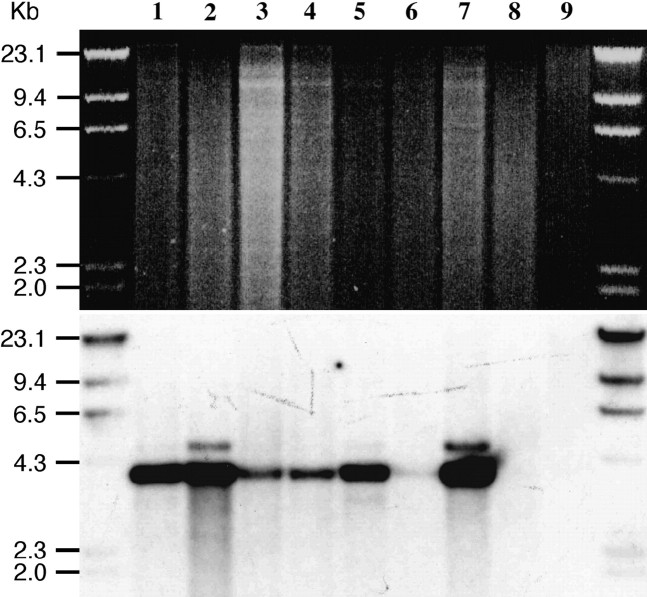
To survey the distribution of the Sobo repeat, we conducted Southern blot analysis of DNA from tomato and 19 Solanum species using the entire Sobo monomer sequence as a probe. None of these species, except S. bulbocastanum, hybridized to the Sobo repeat even under low hybridization stringency (Figure 4). We also conducted a separate Southern blot experiment including only S. tuberosum, S. bulbocastanum, and S. demissum and the Sobo element was not detected in S. demissum (data not shown). We further analyzed nine different accessions of three subspecies of S. bulbocastanum (S. bulbocastanum ssp. bulbocastanum, ssp. dolichophyllum, and ssp. partitum). The Sobo repeat was detected in six of the seven S. bulbocastanum ssp. bulbocastanum accessions (Figure 5).
Figure 4.—
Distribution of the Sobo repeat in Solanum and related species. HindIII-digested DNAs from S. bulbocastanum PI 243510 (PT29) (lane 1), S. tuberosum (USW1) (lane 2), S. etuberosum PI 558303 (lane 3), S. ochranthum PI 230519 (lane 4), S. morelliforme PI 275223 (lane 5), S. bulbocastanum PI 275198 (lane 6), S. cardiophyllum PI 347759 (lane 7), S. lesteri PI 442694 (lane 8), S. capsicibaccatum PI 205560 (lane 9), S. chacoense PI 414153 (lane 10), S. boliviense PI 545963 (lane 11), S. infundibuliforme PI 498246 (lane 12), S. agrimonifoluim PI 243352 (lane 13), S. albornozii PI 498206 (lane 14), S. verrucosum PI 275260 (lane 15), S. multidissectum PI 210043 (lane 16), S. oplocense PI 435079 (lane 17), S. curtilobum PI 225649 (lane 18), S. fendleri PI 498004 (lane19), S. iopetalum PI 275182 (lane 20), and tomato (L. esculentum) PI 1334 (lane 21) were hybridized with the Sobo repeat. Only two S. bulbocastanum accessions (lanes 1 and 6) show hybridizations.
Figure 5.—
Southern analysis of a set of nine S. bulbocastanum accessions. HindIII-digested DNAs from S. bulbocastanum ssp. bulbocastanum PI 243510 (PT29) (lane 1), PI 275198 (lane 2), PI 275185 (lane 3), PI 275192 (lane 4), PI 275196 (lane 5), PI 347757 (lane 6), PI 498223 (lane 7), S. bulbocastanum ssp. dolichophyllum PI 595473 (lane 8), and S. bulbocastanum ssp. partitum PI 275200 (lane 9) were probed with the Sobo repeat.
We hypothesized that the absence of the Sobo repeat in certain S. bulbocastanum accessions may be due to segregation of the hemizygous Sobo locus in natural populations. We conducted interphase FISH analysis of seven additional clonally maintained accessions (Table 1, lines 1–7). No signals were observed in four accessions (Table 1, Figure 2G). The other three accessions contain one or two hybridization sites (Table 1, Figure 2, E and F). We also obtained seeds of six accessions (Table 1, lines 8–13), which were also included in Southern blot analysis (Figure 5). FISH analysis using a population of 10 seeds from each accession was conducted. FISH results confirmed that two of the six accessions (PI 347757 and PI 275200) do not contain the Sobo repeat (Table 1). All of the seeds from two accessions (PI 275169 and PI 498223) contain two signals, presumably representing a homozygous Sobo locus (Table 1). Furthermore, two of the six accessions (PI 275185 and PI 275192) displayed variation in the number of Sobo FISH signals from 0 to 2. This indicates that the parents of these seeds likely had a hemizygous Sobo constitution and the number of Sobo foci were therefore segregating in these populations. Collectively, the FISH results in these S. bulbocastanum accessions confirmed that the Sobo repeat is present as a single locus and is segregating in some populations.
The Sobo sequences are heavily methylated:
Most satellite repeats in higher eukaryotic genomes are heavily methylated. We used restriction combinations of EcoRII/BstNI and HpaII/MspI to test if the Sobo sequences are methylated. Methylation-sensitive EcoRII and methylation-insensitive BstNI cleave the CC(A/T)GG site, depending on the methylation status of the interior cytosine. Similarly, HpaII and MspI recognize the CCGG sequence, but neither can cut when the 5′ C is methylated. Only MspI, not HpaII, can cleave if the internal C is methylated. The sequenced Sobo element contains a single CCGG site but does not contain a CC(A/T)GG site. Southern hybridization detected signals only in the high molecular weight band in EcoRII- and HpaII-digested DNA. However, several smaller bands were detected in BstNI- and MspI-digested DNA (Figure 6). These results indicate that the internal C of the CC(A/T)GG and CCGG sites are methylated.
Figure 6.—
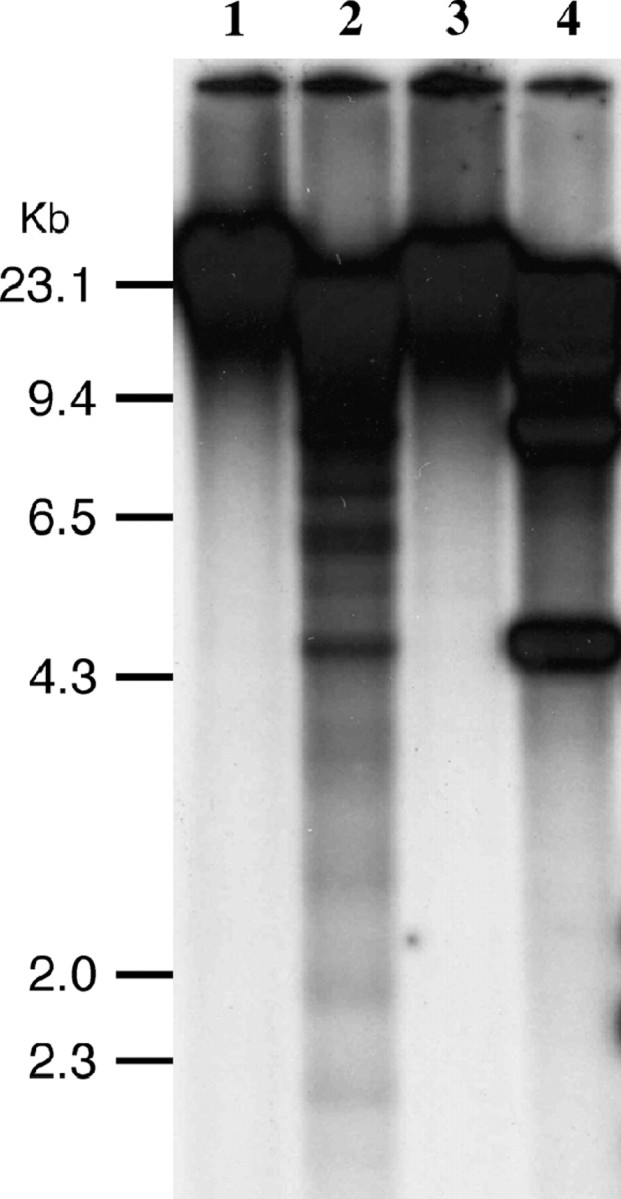
Methylation analysis of the Sobo element. Genomic DNA isolated from S. bulbocastanum (PT29) was digested by EcoRII (lane 1), BstNI (lane 2), HpaII (lane 3), and MspI (lane 4) and probed with the Sobo repeat.
DISCUSSION
Several lines of evidence suggest that Sobo is a recently amplified satellite repeat:
A survey of various Solanum species revealed that the Sobo element is present only in S. bulbocastanum. Several of these species, such as S. cardiophyllum, are closely related to S. bulbocastanum (Spooner et al. 2004). In addition, the Sobo repeat is either absent or in a hemizygous condition in several S. bulbocastanum accessions (Table 1). These findings indicate that the Sobo repeat likely emerged sometime after the speciation of S. bulbocastanum.
Sequencing analysis of multiple subclones showed that the Sobo monomers are highly homogenized. We found only two subgroups of Sobo monomers that are distinguished by few single nucleotide polymorphisms. HindIII digestion of BAC 28G12, which contains an ∼150-kb insert, resulted in only two bands (Figure 1), providing additional evidence of the homogeneity of the Sobo repeats. DNA sequences with no function are expected to accumulate mutations in the absence of selection (Charlesworth et al. 1994). Therefore, lack of sequence divergence among different Sobo monomers may be explained by its recent amplification.
The Sobo monomer includes regions of similarity to several unrelated repetitive sequences (Figure 3). Thus, considering its sequence homogeneity, the Sobo monomer seems to originate by amplification of a single genomic locus containing these sequences.
Several mechanisms have been proposed to explain the origin and evolution of tandemly repeated DNA sequences. Unequal exchange between tandem arrays on sister chromatids or between arrays on homologous chromosomes has generally been assumed to be the most common mechanism of tandem repeat expansion or contraction (Smith 1976). Unequal exchange, however, does not appear to explain the origin of the Sobo repeat. In general, recombination is rare in pericentromeric regions in Solanaceae species (Sherman and Stack 1995). Additionally, the Sobo repeat has only one FISH focus in several S. bulbocastanum accessions. Our FISH technique should allow us to detect 5- to 10-kb (one to two Sobo monomer copies) DNA targets; thus there should be no technical limitations in identifying Sobo foci with the FISH approach. Therefore, our data indicate that in these accessions Sobo is in a hemizygous condition with no paralogous loci; there are no homologous or paralogous loci with which to recombine the Sobo repeat.
Extrachromosomal circular DNAs (eccDNA) have been discovered in a number of species (Gaubatz 1990; Cohen et al. 2003). Novel satellite repeats might originate from amplification of eccDNA through rolling circle replication, followed by reinsertion into the genome (Walsh 1987; Charlesworth et al. 1994). The origin of the Sobo repeat may be explained by this mechanism. The Sobo locus spans ∼360 kb of highly homogenized DNA. In addition, the monomer size, 4.7 kb, is very large compared to most previously reported satellite repeats (Macas et al. 2002). Emergence of such a locus cannot be explained by other mechanisms, such as replication slippage, which would involve a short stretch of DNA in each cycle. The species-specific and hemizygous status of the Sobo repeat in several S. bulbocastanum accessions suggests that it is likely derived from a single amplification event, rather than from a series of events. If a new repeat is derived from mechanisms related to unequal exchanges or concerted evolution (Smith 1976; Dover 1986), it may require multiple events to eventually become a long and highly homogenized array.
We demonstrated that the major part of the Sobo monomer is related to the LTR of a Sore1 element (Figure 3A). The structure of the Sobo repeat suggests that the original monomer may have originated by recombination-based excision of a genomic region between two LTRs separated with other repetitive sequences, giving rise to the eccDNA that could be amplified and reintegrated into the genome in a form of the long array of homogenous monomers. Surprisingly, the Sobo repeat does not hybridize to the rest of the S. bulbocastanum genome in both FISH and Southern hybridization. It suggests that the original Sobo monomer was derived from the LTR of an ancient and significantly diverged Sore1 element. This hypothesis is supported by the fact that the S. demissum Sore1 element, which shows sequence similarity with Sobo, is also an ancient element. Alternatively, potato genomes may contain very few copies of the Sore1 retrotransposon.
Several previous reports demonstrate sequence similarities between satellite repeats and transposons (Pasero et al. 1993; Rossi et al. 1993; Langdon et al. 2000; Cheng and Murata 2003). These results suggest that transposons can be the original sources of satellite DNA. The supernumerary B chromosome of rye contains the E3900 satellite family (Blunden et al. 1993). The 3984-bp monomer of this satellite repeat contains an ∼530-bp fragment that shares extensive similarity to the coding region for the gag protein of the crwydryn retrotransposon (Langdon et al. 2000). The remainder of the E3900 repeat appears to be unrelated to retrotransposon sequences. The monomer of another satellite repeat associated with the rye B chromosome, the D1100 family (Sandery et al. 1990; Houben et al. 1996), contains sequences related to Tnr1, a miniature inverted-repeat transposoable element reported in rice (Langdon et al. 2000). The sequence complexity of genomic clones containing the E3900 and D1100 repeats indicated that these two satellite families have undergone extensive amplification and rearrangements (Langdon et al. 2000). Therefore, it is not known if the transposon-related sequences were involved in the original formation of the satellite repeats or were later recruited in the E3900 and D1100 families. Cheng and Murata (2003) reported a centromere-specific tandem repeat in wheat that is likely derived from the centromere-specific retrotransposons. On the basis of the colocalization of the tandem repeat and the retrotransposons, most of the members of this tandem repeat are possibly still associated with the retrotransposons. It is not clear if this centromeric tandem repeat is organized as long arrays and whether these arrays have evolved independently from the centromeric retrotransposons.
In summary, although several previous reports showed sequence similarities between satellite repeats and transposons, it is not known if the transposon sequences contributed to the formation of the original monomers of the related satellite repeats. The Sobo repeat provides the first evidence that a satellite repeat can be amplified from a part of a retrotransposon in a sudden and dramatic fashion.
Acknowledgments
We thank David Spooner for providing DNA samples of the Solanum species and Weiwei Jin for helping with Southern blot hybridization experiments. This research is supported by Hatch funds to J.J. and by the Grant Agency of the Czech Republic (grant no. GA204/04/1207) to J.M.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunden, R., T. J. Wilkes, J. W. Forster, M. M. Jimenez, M. J. Sandery et al., 1993 Identification of the E3900 family, a second family of rye B chromosome specific repeated sequences. 36: 706–711. [DOI] [PubMed]
- Bradeen, J. M., S. K. Naess, J. Song, G. T. Haberlach, S. M. Wielgus et al., 2003. Concomitant reiterative BAC walking and fine genetic mapping enable physical map development for the broad-spectrum late blight resistance region, RB. Mol. Genet. Genomics 269 603–611. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., P. Sniegowski and W. Stephan, 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371 215–220. [DOI] [PubMed] [Google Scholar]
- Cheng, Z. J., and M. Murata, 2003. A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. Genetics 164 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z. K., C. R. Buell, R. A. Wing and J. Jiang, 2002. Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res. 10 379–387. [DOI] [PubMed] [Google Scholar]
- Cohen, S., K. Yacobi and D. Segal, 2003. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 13 1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., J. Song, S. K. Naess, J. P. Helgeson, C. Gebhardt et al., 2000. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor. Appl. Genet. 101 1001–1007. [Google Scholar]
- Dover, G. A., 1986. Molecular drive in multigene families: how biological novelties arise, spread and are assimilated. Trends Genet. 2 159–165. [Google Scholar]
- Frello, S., M. Orgaard, N. Jacobsen and J. S. Heslop-Harrison, 2004. The genomic organization and evolutionary distribution of a tandemly repeated DNA sequence family in the genus Crocus (Iridiaceae). Hereditas 141 81–88. [DOI] [PubMed] [Google Scholar]
- Fry, K., and W. Salser, 1977. Nucelotide sequence of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 12 1069–1084. [DOI] [PubMed] [Google Scholar]
- Gaubatz, J. W., 1990. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat. Res. 237 271–292. [DOI] [PubMed] [Google Scholar]
- Gerlach, W. L., and J. R. Bedbrook, 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 7 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, K. M., J. S. Niederhauser and L. Servin, 1959. Studies on fertility and late blight resistance in Solanum bulbocastanum Dun. in Mexico. Can. J. Bot. 37 41–48. [Google Scholar]
- Hatch, F. T., and J. A. Mazrimas, 1974. Fractionation and characterization of satellite DNAs of the kangroo rat (Dipodomys ordii). Nucleic Acids Res. 1 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Houben, A., R. G. Kynast, U. Heim, H. Hermann, R. N. Jones et al., 1996. Molecular cytogenetic characterisation of the terminal heterochromatic segment of the B-chromosome of rye (Secale cereale). Chromosoma 105 97–103. [DOI] [PubMed] [Google Scholar]
- Jackson, S. A., M. L. Wang, H. M. Goodman and J. Jiang, 1998. Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41 566–572. [PubMed] [Google Scholar]
- Jiang, J., B. S. Gill, G. L. Wang, P. C. Ronald and D. C. Ward, 1995. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., J. B. Birchler, W. A. Parrott and R. K. Dawe, 2003. A molecular view of plant centromeres. Trends Plant Sci. 8 570–575. [DOI] [PubMed] [Google Scholar]
- Kubis, S., J. S. Heslop-Harrison and T. Schmidt, 1997. A family of differentially amplified repetitive sequences in the genus Beta reveals genetic variation in Beta vulgaris subspecies and cultivars. J. Mol. Evol. 44 310–320. [DOI] [PubMed] [Google Scholar]
- Langdon, T., C. Seago, R. N. Jones, H. Ougham, H. Thomas et al., 2000. De novo evolution of satellite DNA on the rye B chromosome. Genetics 154 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas, J., D. Pozarkova, A. Navratilova, M. Nouzova and P. Neumann, 2000. Two new families of tandem repeats isolated from genus Vicia using genomic self-priming PCR. Mol. Gen. Genet. 263 741–751. [DOI] [PubMed] [Google Scholar]
- Macas, J., T. Meszaros and M. Nouzova, 2002. PlantSat: a specialized database for plant satellite repeats. Bioinformatics 18 28–35. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer et al., 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazrimas, J. A., and F. T. Hatch, 1972. A possible relationship between satellite DNA and the evolution of kangaroo rat species (genus Dipodomys). Nat. New Biol. 240 102–105. [DOI] [PubMed] [Google Scholar]
- Pasero, P., N. Sjakste, C. Blettry, C. Got and M. Marilley, 1993. Long-range organization and sequence-directed curvature of Xenopus laevis satellite 1 DNA. Nucleic Acids Res. 21 4703–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, W. R., and D. J. Lipman, 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiszner, J., I. Takacs, M. Bilgin, J. Gyorgyey, D. Dudits et al., 1994. Organization of a Solanum brevidens repetitive sequence related to the TGRI subtelomeric repeats of Lycopersicon esculentum. Theor. Appl. Genet. 89 1–8. [DOI] [PubMed] [Google Scholar]
- Rossi, M. S., C. G. Pesce, O. A. Reig, A. R. Kornblihtt and J. Zorzopulos, 1993. Retroviral-like features in the monomer of the major satellite DNA from the South American rodents of the genus Ctenomys. DNA Seq. 3 379–381. [DOI] [PubMed] [Google Scholar]
- Salser, W., S. Bowen, D. Browne, F. El Adli, N. Fedoroff et al., 1976. Investigation of the organization of mammalian chromosomes at the DNA sequence level. Fed. Proc. 35 23–35. [PubMed] [Google Scholar]
- Sandery, M. J., J. W. Forster, R. Blunden and R. N. Jones, 1990. Identification of a family of repeated sequences on the rye B-chromosome. Genome 33 908–913. [DOI] [PubMed] [Google Scholar]
- Schmidt, T., and J. S. Heslop-Harrison, 1993. Variability and evolution of highly repeated DNA sequences in the genus Beta. Genome 36 1074–1079. [DOI] [PubMed] [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1995. Two-dimensional spreads of synaptonemal complexes from Solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. P., 1976. Evolution of repeated DNA sequences by unequal crossover. Science 191 528–535. [DOI] [PubMed] [Google Scholar]
- Song, J., F. Dong and J. Jiang, 2000. Construction of a bacterial artificial chromosome (BAC) library for potato molecular cytogenetics research. Genome 43 199–204. [PubMed] [Google Scholar]
- Sonnhammer, E. L. L., and R. Durbin, 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167 GC1–GC10. [DOI] [PubMed] [Google Scholar]
- Spooner, D. M., R. G. van den Berg, A. Rodríguez, J. Bamberg, R. J. Hijmans et al., 2004. Wild potatoes (Solanum section Petota) of North and Central America. Syst. Bot. Monogr. 68 1–209. [Google Scholar]
- Staden, R., 1996. The Staden sequence analysis package. Mol. Biotechnol. 5 233–241. [DOI] [PubMed] [Google Scholar]
- Stupar, R. M., J. Song, A. L. Tek, Z. Cheng, F. Dong et al., 2002. Highly condensed potato pericentromeric heterochromatin contains rDNA-related tandem repeats. Genetics 162 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, J. B., 1987. Persistence of tandem arrays: implications for satellite and simple-sequence DNAs. Genetics 115 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]