Abstract
Little is known about the physical makeup of heterochromatin in the soybean (Glycine max L. Merr.) genome. Using DNA sequencing and molecular cytogenetics, an initial analysis of the repetitive fraction of the soybean genome is presented. BAC 076J21, derived from linkage group L, has sequences conserved in the pericentromeric heterochromatin of all 20 chromosomes. FISH analysis of this BAC and three subclones on pachytene chromosomes revealed relatively strict partitioning of the heterochromatic and euchromatic regions. Sequence analysis showed that this BAC consists primarily of repetitive sequences such as a 102-bp tandem repeat with sequence identity to a previously characterized ∼120-bp repeat (STR120). Fragments of Calypso-like retroelements, a recently inserted SIRE1 element, and a SIRE1 solo LTR were present within this BAC. Some of these sequences are methylated and are not conserved outside of G. max and G. soja, a close relative of soybean, except for STR102, which hybridized to a restriction fragment from G. latifolia. These data present a picture of the repetitive fraction of the soybean genome that is highly concentrated in the pericentromeric regions, consisting of rapidly evolving tandem repeats with interspersed retroelements.
OUR knowledge of the structural makeup of the soybean (Glycine max L. Merr.) genome is superficial. The genome size of soybean is 1100 Mb (Arumuganathan and Earle 1991), the chromosome number 2n = 40, and the repetitive fraction, based on Cot analyses, ranges between 40 and 60% (Goldberg 1978; Gurley et al. 1979). Despite more than a decade of genomics, we still know little about the DNA composition of the repetitive fraction, the distribution of genes relative to repeats, the molecular structure of the heterochromatic/euchromatic regions, and how duplicated regions of the genome have evolved structurally.
It has long been suspected that the soybean genome has undergone multiple rounds of duplication as evidenced by the number of RFLP fragments in mapping experiments (Shoemaker et al. 1996), sequence analysis of expressed sequence tags (ESTs) (Schlueter et al. 2004), and the construction of a bacterial artificial chromosome (BAC)-based physical map (Wu et al. 2004). Analysis of ESTs has shown that there have been at least two rounds of duplication, ∼15 and 44 MYA (Schlueter et al. 2004). Despite the relatively large genome size, little has been published about the repetitive fraction of the soybean genome. A few tandem repeats (SB92, Vahedian et al. 1995; STR120, Morgante et al. 1997) and retroelements (Laten and Morris 1993; Graham et al. 2002) have been described, but the sequence composition and chromosomal distribution for much of the repetitive DNA that accounts for 40–60% of the soybean genome remains unknown.
In another legume, Medicago truncatula, cytological evidence has shown that chromosome arms are almost exclusively euchromatic and that the majority of the heterochromatin (repetitive sequences) is found in pericentromeric regions (Kulikova et al. 2001). This same study indicated that genes in M. truncatula are overwhelmingly localized to the euchromatic arms. In many cereals such as maize, wheat, and barley, repetitive sequences are dispersed throughout the chromosomes and there is little evidence of demarcated euchromatic and heterochromatic regions (Mroczek and Dawe 2003). Previous cytogenetic analysis of soybean pachytene chromosomes has shown that ∼36% of the physical length is heterochromatic and that most of this is pericentromeric or localized to a few highly heterochromatic short arms (Singh and Hymowitz 1988).
We are attempting to determine the molecular organization of the soybean genome by defining the types of sequences that are repetitive within the soybean genome and concurrently determining the chromosomal location of these sequences. We began by using fluorescence in situ hybridization (FISH) to map genetically anchored BACs to molecular linkage groups (Pagel et al. 2004). With a series of anchored BACs spanning MLG L we further sought to determine the molecular organization of this chromosome by integrating BACs and regions of the linkage map to the chromosomal map. We hypothesized that the centromeric regions would be composed of a series of highly repetitive DNA sequences, including tandem repeats, interspersed with retroelements, as observed in other plant species (reviewed in Jiang et al. 2003), and that the heterochromatic regions would be delimited from euchromatin, reflecting the organization of chromatin in M. truncatula (Kulikova et al. 2001).
MATERIALS AND METHODS
FISH and fiber-FISH:
Mitotic chromosomes were prepared by acetocarmine squashes using the meristematic portion of hydroxyquinoline-treated root tips. Pachytene chromosomes were prepared using squashes of anthers from flowers fixed in 3:1 ethanol to galacial acetic acid. Slides were screened using a phase-contrast microscope and kept at −80° until used for FISH. Nuclei were isolated for fiber-FISH following established protocols (Zhong et al. 1996) except that a 22-μm filtration was added.
For FISH, ∼1 μg of plasmid (or BAC) DNA was labeled with either biotin-UTP or digoxigenin-UTP (Hoffman-La Roche) using nick translation. Chromosomes were hybridized following previously published protocols (Jiang et al. 1996b) except that detection was with AlexaFluor 488-streptavidin (Molecular Probes, Eugene, OR) for biotin and mouse antidigoxigenin (Hoffman-La Roche) followed by Alexafluor 568 anti-mouse (Molecular Probes) for digoxigenin-labeled probes. Digital mapping followed previously published protocols (Jackson et al. 1999). Grayscale digital images were captured using either an Olympus BX60 with an Hamimatsu Orca ER CCD camera controlled with MetaMorph (Universal Imaging, West Chester, PA) or a Nikon E400 with an Optronics MagnaFire CCD camera controlled by ImagePro (Media Cybernetics). Images were further analyzed using MetaMorph and final publication images were prepared using Adobe Photoshop v7.0 for Macintosh.
DNA isolation and Southern analysis:
G. max cv. Resnik 2000 seeds were provided by Niels Nielsen [U. S. Department of Agriculture (USDA)-Agricultural Research Service (ARS), Purdue University). Seeds for all other Glycine species were provided by the USDA Soybean Germplasm Collection, USDA-ARS, University of Illinois, Champaign-Urbana (Table 1).
TABLE 1 .
Species used in Glycine species blots including accession numbers and lane numbers on the blots
|
Species |
PI no. or cultivar |
Lane |
|---|---|---|
| G. tabacina | 509494 | 1 |
| G. falcata | 612231 | 2 |
| G. argyrea | 595792 | 3 |
| G. pindanica | 595818 | 4 |
| G. clandestina | 440961 | 5 |
| G. canescens | 440933 | 6 |
| G. latrobeana | 505184 | 7 |
| G. stenophita | 546981 | 8 |
| G. curvata | 505164 | 9 |
| G. tomentella | 441006 | 10 |
| G. cyrtaloba | 373993 | 11 |
| G. rubiginosa | 591588 | 12 |
| G. pescadrensis | 505195 | 13 |
| G. latifolia | 321393 | 14 |
| G. arenaria | 505204 | 15 |
| G. soja | 597457 | 16 |
| Vigna radiata | cv. nm92 | 17 |
|
G. max |
cv. Resnik 2000 |
18 |
Plant genomic DNA was extracted from young, frozen leaf tissue using a standard CTAB extraction protocol. BAC DNA was extracted using a QIAGEN (Chatsworth, CA) large-construct kit, with the following modification to the kit protocol: during the final precipitation step (QIAGEN protocol step 17), 4 μl of d-glycogen were added to the isopropanol to aid in DNA precipitation.
Two micrograms of DNA from BACs 009M21 and 076J21 (Iowa State University, Marek and Shoemaker 1997) and 123O07 (University of Minnesota, Danesh et al. 1998) were digested in a 37° water bath overnight with 30 units HindIII restriction enzyme (New England Biolabs, Beverly, MA) and separated on an 1% agarose gel. For plant species, 1 μg of plant genomic DNA from each species was restriction digested with 6 units HindIII, 5 units HpaII, or 6 units MspI (New England Biolabs) in a 37° water bath overnight and separated on a 0.8% agarose gel. DNA from the gels was blotted onto Zeta-Probe GT genomic tested blotting membrane (Bio-Rad, Hercules, CA).
The membrane was prehybridized for at least 30 min in Church hybridization buffer (1% BSA/1 mm, EDTA/7% SDS/0.5 m sodium phosphate) at 58°. Probes were prepared using the Rediprime II random prime labeling system (Amersham Biosciences). Before the probes were used for hybridization, they were purified using the QIAquick nucleotide removal kit (QIAGEN). The probe was hybridized to the membrane at 58° overnight. After hybridization, the membrane was washed in 1.5× SSC/0.1% SDS for 30 min at 58°, then in 1× SSC/0.1% SDS for 30 min. The membrane was exposed to autoradiography film overnight at −80°. Alternatively, the membrane was exposed overnight to a Fujifilm BAS-MS imaging plate and digitally scanned using a Fuji FLA-5000 Bio Imaging Analyzer.
BAC DNA sequencing and analysis:
BAC 076J21 was sheared and shotgun cloned as previously described (SanMiguel et al. 2002) (bankit659374, GenBank no. AY748457). Sequence coverage of 11× was generated by sequencing 1152 of these clones with both T3 and T7 primers. These sequence reads were assembled using the PhredPhrap script of the phred/phrap/consed package (Ewing et al. 1998; Gordon et al. 1998). Further analysis was done using the Genetics Computer Group (GCG) package (Accelrys), the Artemis viewer (Rutherford et al. 2000), Dotter (Sonnhammer and Durbin 1995), and the Tandem Repeat Finder (Benson 1999). Sequence comparisons were made to the February 15, 2004 v. 140.0 release of GenBank. For EST comparison, the NCBI Blast server was used with default settings and the top 100 are reported. Soybean genome shotgun sequences (GSS) were generated from 4-kb randomly sheared genomic fragments cloned into ToPo (Invitrogen, San Diego) and sequenced using T3/T7 primers (GenBank CL867099, CL868434).
RESULTS
Identification of two BAC clones derived from centromeric heterochromatin:
Two soybean BAC libraries were screened with SSR and RFLP markers to derive a set of genetically anchored BAC clones (Marek et al. 2001). These anchored BAC clones were subsequently used for FISH to integrate the genetic and chromosome-based maps (Pagel et al. 2004). Two BAC clones from MLG L, 076J21 (position 34.6 cM) and 09M21 (position 32.4 cM), both selected with SSR markers, were found to map to pericentromeric regions of all 40 soybean chromosomes (Figure 1a). It was not clear at the resolution of mitotic chromosomes if these BACs were derived from either centromeric or pericentromeric regions or if they were found at interstitial heterochromatic locations (Figure 1a, inset); therefore, further molecular characterization was undertaken.
Figure 1.—
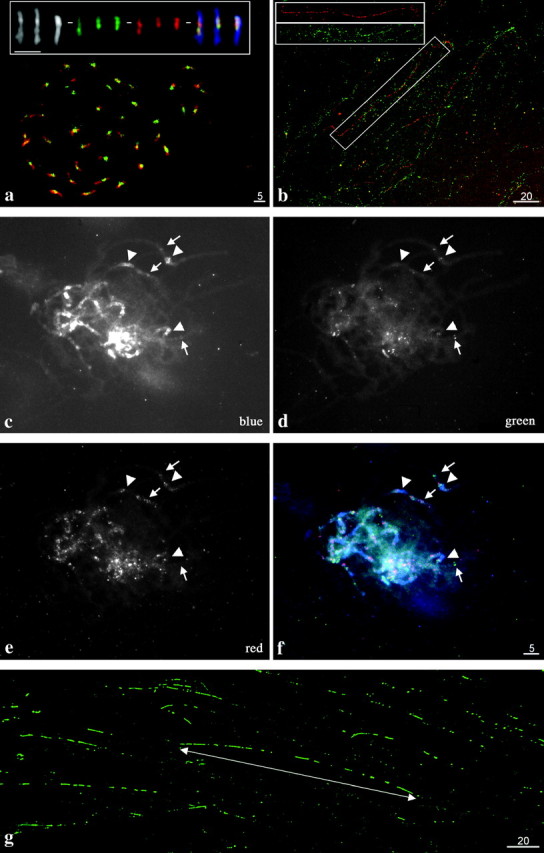
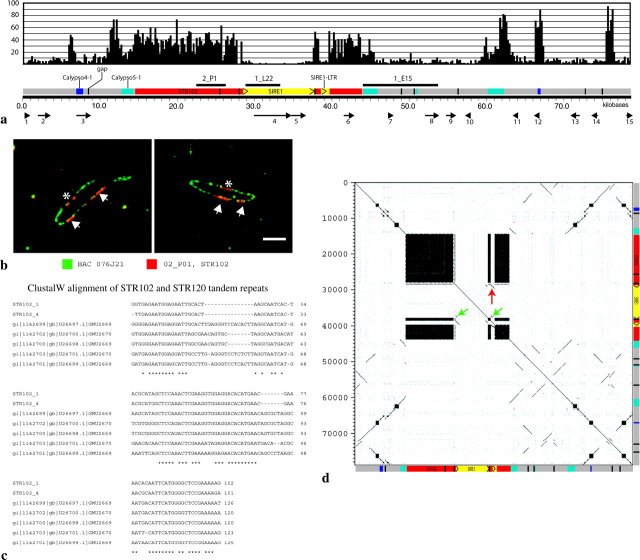
Fluorescence in situ hybridization analysis of BACs and subclones to chromosomes and extended DNA fibers of soybean. (a) FISH of BAC 076J21 (green) to mitotic chromosomes (red). (Inset) Three chromosomes from another preparation showing, from left to right: DAPI stained (black and white), 076J21 (green), 09M21 (red), and merged. (b) Fiber-FISH of BACs 076J21 (red) and 09M21 (green) to DNA fibers of soybean showing little overlap in FISH signal. (Inset) An image of a single fiber with the two color channels shown separately. (c–f) FISH to pachytene chromosomes of soybean with subclones of BAC 076J21. Arrows indicate heterochromatic regions and arrowheads indicate centromeric heterochromatin. (c) DAPI-stained chromosomes. (d) STR102 [2_P01]. (e) SIRE 1 [1_L22] and Calypso 5-1 [1_E15] pooled. (f) Merged image. (g) Fiber-FISH analysis of STR102 [2_P01] on extended DNA fibers of soybean showing long interrupted arrays of STR102 [2_P01]. Line with arrows indicates a 435.6-kb cluster of repeats.
BACs 09M21 and 076J21 have sequences in common but are not entirely redundant:
It was not clear if these two BACs differed in DNA content as they both colocalized to entire centromeric regions. Three approaches were undertaken to test whether these two BACs had DNA sequences in common. First, FISH of BACs 076J21 (red) and 09M21 (green) on extended genomic fibers (fiber-FISH) of soybean showed that the signals from the BACs were not entirely coincident (Figure 1b). BAC 076J21 had long stretches of hybridization signals that did not overlap with any hybridization signal from BAC 09M21 (Figure 1b, inset). Second, both BACs were digested with HindIII, gel blotted, and reciprocally hybridized with the other BAC. These reciprocal Southern analyses showed that although some restriction fragments did hybridize to the other BAC, others did not (Figure 2). Third, we performed ∼1.2× draft coverage sequencing of BAC 09M21, which has a ∼40-kb insert (data not shown), and compared it to an 11× coverage sequence of 076J21. This revealed that ∼29% of the sequences from 09M21 had strong matches (E < e−04) to 076J21 (sequence results below) and that most of the matches were in the SIRE1 element and calypso-like retroelements; none of the sequences from 09M21 had significant matches to either STR102 or STR120 (described below). Even though these BACs hybridized to the same chromosomal region, DNA sequencing, fiber-FISH, and Southern analyses show that they are not entirely redundant in sequence composition.
Figure 2.—
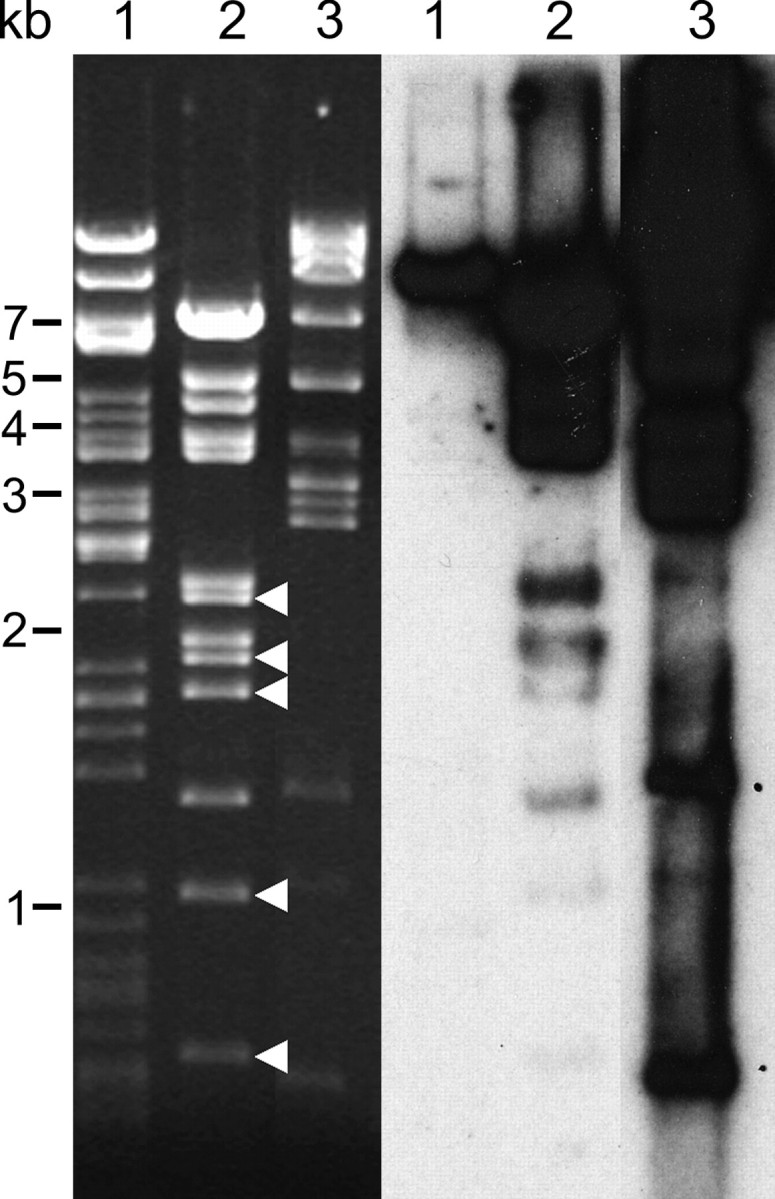
Southern analysis of shared sequences between BACs 09M21 and 076J21. (Left) Ethidium-bromide-stained gel of restriction-digested BACs before blotting. Lane 1, BAC 123E07; lane 2, BAC 09M21; lane 3, BAC 076J21. (Right) Hybridization of 076J21 to gel blot. BAC 123E07 is derived from a euchromatic region of soybean and only the BAC vector cross-hybridizes (∼7.4 kb). BAC 09M21 shares some sequences with 076J21 but some restriction bands (arrowheads) show little cross-hybridization.
Organization of DNA sequences within BAC 076J21:
To more fully understand the DNA sequence composition, we sequenced BAC 076J21 to 11× coverage using a shotgun approach. Due to the highly repetitive nature of this BAC, assembly of the DNA sequence reads was difficult. However, it was possible to collapse 10 sequence contigs into one 79,623-bp scaffold (Figure 3a), and the order and orientation of the contigs with respect to one another was inferred by forward/reverse sequence reads from clones putatively spanning gaps between the contigs. The full set of contigs comprised 104,573 bases with phred/phrap-generated quality scores >20, ∼32 kb smaller than the PFGE size estimate of 136 kb of the BAC clone (data not shown). A caveat to this assembly is that sequence scaffolds of BACs comprised primarily of repetitive DNA elements are often difficult to assemble and may contain errors.
Figure 3.—
Sequence analysis of BAC 076J21. (a) Schematic of BAC 076J21 (79,623 bp). Black bars above sequence diagram are the subclones used for Southern analysis and FISH. Across the top, a 200-bp sliding window (x-axis) was used to map the number of BLASTN hits (y-axis) from a search against the soybean GSS sequences along the length of 076J21. FGENESH-predicted genes are shown with arrows along the bottom of the diagram. (b) Digital mapping of 2_P01 [STR102] onto BAC 076J21 using FISH. Three clusters are seen, two of which (arrows) may border the SIRE1 element. The other (star) did not assemble into the sequence scaffold. (c) ClustalW was used to align two representatives of the STR102 repeat (STR102_1 and STR102_4) with five STR120 members from GenBank. Asterisks denote complete identity among all seven sequences at a nucleotide position. (d) A dot plot was made using Dotter (Sonnhammer and Durbin 1995) with a word size of 20 nt of 076J21 against itself. A schematic of the BAC with internal structures is shown on both axes. The LTRs of the SIRE1 element are indicated with green arrows and the solo LTR with a red arrow.
FISH mapping directly on the BAC plasmid (digital mapping, Jackson et al. 1999) was employed to assess the overall accuracy of the assembly. Three STR102 clusters (102-bp tandem repeat) are apparent in 076J21 (Figure 3b), but only two are present in the final sequence scaffold. Quantitative analysis of the digital mapping data revealed that the STR102 repeat accounts for 25.5% (SD 4.1). This would correspond to 35 kb of sequence but only 19 kb are present in sequence contigs. Further, only 193 of 1152 shotgun subclones yielded sequence containing STR102 repeats.
Sequence analysis of these contigs revealed a number of features (Figure 3a). One was a SIRE1 element (Laten and Morris 1993) inserted into a tract of STR102 repeats. The 3′ LTR of the SIRE1 element contained a sequence gap as it spanned two contigs within the scaffold. A SIRE1 solo LTR with 99.8% sequence identity to the 5′ LTR of the full element was also found inserted directly into a block of tandem repeats (Figure 3a). The SIRE1 element and the SIRE1 solo LTR were found in identical positions in two STR102 repeats. Several regions with homology to Calypso-like retroelements (Wright and Voytas 2002) were found scattered across the BAC (Figure 3a).
The 102-bp tandem repeat (named soybean tandem repeat 102, STR102) had 82.6% sequence identity (determined using BestFit of GCG) to the previously described 120-bp tandem repeat STR120 (Figure 3c, gi1147200) (Morgante et al. 1997). Two STR102 representatives were aligned to STR120 using ClustalW (Figure 3c). This analysis showed that the ∼102-bp monomers had regions of sequence identity with STR120 with two gaps (15 and 7 nt) in the alignment. Other tandem repeats (ranging from 5 to 191 bp with a minimum copy number of 5) were found within this BAC using the Tandem Repeat Finder program (Benson 1999), none of which were as frequent as STR102 but some of which can be seen as smaller blocks in the Dotter generated dot plot (Figure 3d) (Sonnhammer and Durbin 1995).
Dot-plot analysis of 076J21 showed that except for the SIRE1 element much of this BAC is duplicated internally (Figure 3d). For instance, the first ∼14 kb, before the first STR102 cluster, is duplicated several times from 48 kb to the end of the BAC. The LTRs of the SIRE1 element and the solo LTR are seen in the dot plot adjacent and within the second STR102 cluster (Figure 3d, arrows). All the Calypso 5-1-like elements shared sequence identity to the same region of the Calypso 5-1 element (4276-6089 of AF186186) although some of these were inverted relative to each other on the BAC (Figure 3d).
The assembled contigs for BAC 076J21 were used to query 1454 soybean GSSs derived from paired reads of clones with ∼3-kb inserts. These GSS sequences represent 1.29 Mb or ∼0.1% of the 1110 Mb soybean genome. RepeatMasker/Crossmatch (http://www.repeatmasker.org/) was used with STR102 and SIRE1 to estimate the frequency of each in the GSS data set as 0.7 and 0.6%, respectively. This is also shown in Figure 3a where the frequency of BLASTN hits of 076J21 to the GSS data set is plotted along the length of the BAC. Eight paired GSS sequences were almost entirely copies of STR102, indicating that these four clones may be composed primarily of STR102.
The 076J21 sequence was used to query the entire GenBank EST collection using an e-value cutoff of 4.0 × 10−4. A single soybean EST (gi22524207) had up to 98% sequence identity to the STR102 repeat. The SIRE1 element had sequence similarity to ESTs in the LTRs and one EST showed similarity to parts of the internal regions. Of the top 100 matches, 50 were derived from a soybean root hair subtracted cDNA library (gmrhRww). Fifteen potential genes were found on the BAC using FGENESH (http://www.softberry.com/berry.phtml) (Table 2). Three of the 15 (4–6) were derived from either the SIRE1 element or included STR102 repeats. Of the other 12, three of them (2, 9, and 13) had high levels of similarity to the retroelements Calypso 4-1 or 5-1. Two of the predicted genes had similarity to the Mdh1 genomic sequence; however, the Mdh1 sequence is >27 kb in length and the hits were not to the coding regions but rather to an upstream region that also has similarity to Calypso 4-1 retrotransposons. A similar situation was found for the hits to the SCB1 gene where the genomic sequence encompasses more than just the coding region, and the hits from the predicted genes from BAC 76J21 were to noncoding regions upstream of SCB1. Moreover, these 2 predicted genes (7 and 8) had significant TBLASTN hits to an LTR retrotransposon from pea (Neumann et al. 2003).
TABLE 2 .
Annotation of FGENESH prediction coding sequences on BAC 76J21
|
Feature |
Coordinates |
No. of hits to soybean ESTs (E < e−04) |
TBLASTNa GenBank (E < e−03) |
BLASTP UniProt (E < 0.03) |
Annotationb |
|---|---|---|---|---|---|
| 1 | 279..677 | 4 | — | — | Unknown |
| 2 | 1856..3554 | 13 | Calypso 4-1 | Gag/pol polyprotein | Retroelement |
| 3 | 6982..8874 | 10 | — | — | Unknown |
| 4 | 29856..34589 | 15 | SIRE1 | Gag-pol polyprotein | LTR retrotranposon |
| 5 | 34791..36620 | 2 | SIRE1 | Envelope-like protein | LTR retrotranposon |
| 6 | 41539..42707 | 3 | STR120 | — | Tandem |
| 7 | 47026..47694 | 9 | SCB1 | Hypothetical protein | LTR retrotranposon |
| 8 | 51974..53860 | 14 | SCB1 | Hypothetical protein | LTR retrotranposon |
| 9 | 54862..55980 | 4 | Calypso 4-1 | Gag/pol polyprotein | Retroelement |
| 10 | 57411..57809 | 6 | — | — | Unknown |
| 11 | 63592..64086 | 19 | AOX | — | Hypothetical |
| 12 | 66348..66722 | 2 | MDH1 | — | Retroelement |
| 13 | 70800..71918 | 4 | Calypso 4-1 | Gag/pol polyprotein | Retroelement |
| 14 | 73861..74259 | 6 | — | — | Unknown |
| 15 |
78107..78391 |
3 |
MDH1 |
— |
Retroelement |
Top hits are shown to the GenBank nucleotide database.
“Unknown” indicates EST matches with no GenBank or UniProt hits; “hypothetical” indicates EST hit and GenBank hit.
Three sequencing clones (Figure 3a) were chosen for further FISH analysis on both chromosomes and DNA fibers. Clone 2_P01, representing the 102-bp tandem repeat, had strong hybridization signals in heterochromatic regions (pericentromeric and other knob-like regions) on all 20 meiotic chromosomes (Figure 1, d and f). On extended genomic DNA fibers, this repeat was present in interrupted stretches of up to 435.6 kb (Figure 1g). The other two clones contained portions of either the SIRE1 element (1_L22) or a Calypso 5-1-like retroelement (1_E15). Neither of the retroelements had long fiber-FISH signals indicative of tandem repeats (data not shown); rather, the fiber-FISH signals were dispersed. On pachytene chromosomes subclones containing SIRE1 (1_L22) and Calypso 5-1 (1_E15) were pooled for FISH and were found to localize to pericentromeric heterochromatin and heterochromatic knobs on euchromatic arms (Figure 1, e and f).
Conservation and methylation status of sequences derived from 076J21:
Centromeric sequences from several other plant species have been isolated previously and, in the case of the cereals, a centromere-specific retrotransposon is conserved in both sequence and chromosomal locations across the cereal family (Aragon-Alcaide et al. 1996; Jiang et al. 1996a; Presting et al. 1998). Using a hybridization-based assay, we tested the conservation of both BACs (09M21 and 0076J21) and the three 076J21 subclones across a set of evolutionarily related legume species. When the two BACs were used as probes, they were conserved only in G. max and the closely related and sexually compatible G. soja, both of which are annuals (Figure 4, a and b). However, when the STR102 repeat (subclone 2_P01) was used as a probe on the blots containing the Glycine annuals and perennials, it hybridized to a fragment in G. latifolia (Figure 4c).
Figure 4.—
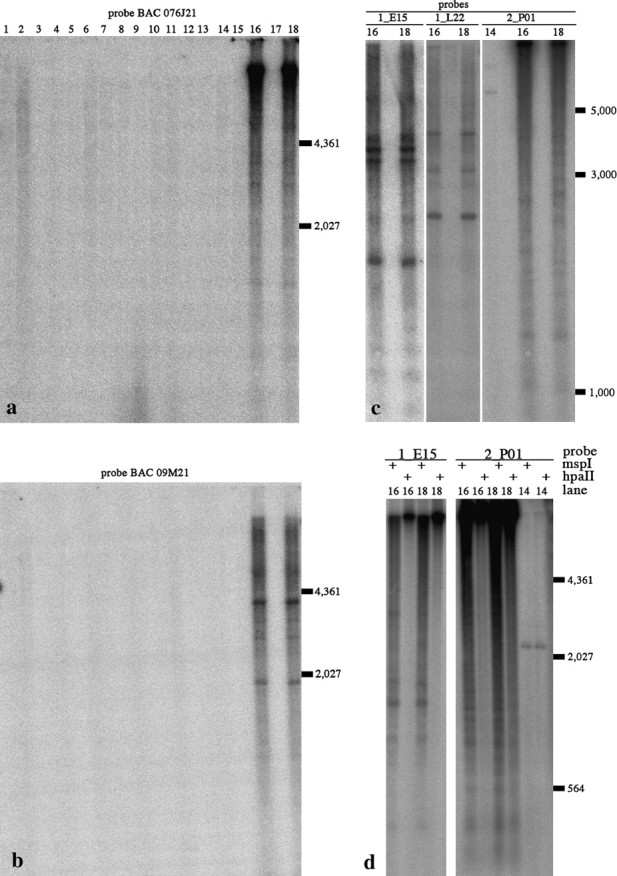
Southern analysis of conservation of BACs 076J21 and 09M21 and conservation and methylation of subclones of 076J21. Lane numbers for all a–d refer to the species in Table 1. (a) BAC 076J21 and (b) 09M21 probed against the Glycine species blot. (c) Subclones of 076J21 probed against the Glycine species blots (only lanes showing hybridization are shown). (d) Methylation of 076J21 subclones containing Calypso 5-1 (1E_15) and STR102 (2P_01) were tested by probing against genomic DNA digested with either MspI or HpaII.
The methylation status of the three subclones was tested using the methyl-cytosine-sensitive isoschizomer restriction enzymes MspI and HpaII. Both enzymes cut asymmetrically at 5′-CCGG-3′; however, when this sequence is CpG methylated, HpaII will not cut, whereas MspI will. In the case of Calypso 5-1 (1_E15), these sequences were methylated in both G. max and G. soja (Figure 4d). The STR102 repeat does not have a CCGG restriction site, but surrounding sequences do appear to be methylated on the basis of the hybridization of the SR102-containing clone 2_P01 to the blot (Figure 4d). The SIRE1 element (1_L22) did not appear to cut with either enzyme so its methylation status could not be determined using this approach.
DISCUSSION
Repetitive sequences can account for a major portion of eukaryotic genomes. Although often referred to as “junk” DNA, repetitive sequences are known to function in the organization of telomeres (Blackburn and Hall 1978) and centromeres (reviewed in Jiang et al. 2003) and may be involved in chromosome packaging, thereby regulating gene expression (Stam et al. 2002). Ribosomal clusters are another example of tandemly repeated but functional DNA. In soybean, estimates of the repetitive fraction range from 40 to 60% on the basis of DNA:DNA renaturation experiments (Goldberg 1978; Gurley et al. 1979). In maize, retroelements often insert within other retroelements, leading to “nested transposons” (SanMiguel et al. 1996) that separate “islands” of genic or low-copy sequences; in Arabidopsis, the majority of the repetitive sequences are localized to centromeric and pericentromeric regions of the genome (Arabidopsis Genome Initiative 2000). The genome of M. truncatula, a legume, has demarcated euchromatic/heterochromatic regions as shown by FISH mapping (Kulikova et al. 2001).
Two BACs anchored to MLG L were found to hybridize to the pericentromeric regions of all 20 pairs of soybean chromosomes. This indicated that sequences within these BACs are (1) repetitive within the soybean genome and (2) conserved in the chromosomal location. To follow up on this observation and to further characterize the physical makeup of the soybean genome, these BACs were molecularly analyzed using DNA sequencing, Southern analysis, and FISH to pachytene chromosomes and extended DNA fibers. These analyses allowed us to determine the distribution, DNA sequence composition, conservation, and methylation status of sequences in this BAC.
FISH of these BACs to pachytene chromosomes more finely determined the chromosome distribution of sequences from BAC 076J21. It was evident from DAPI staining that many of the chromosomal arms of soybean are euchromatic, confirming previous observations (Singh and Hymowitz 1988). This indicates that, for some chromosomes, the majority of the heterochromatin is likely to be confined to the pericentromeric regions. FISH analysis showed that BAC 076J21 and several of the subclones from this BAC localized to either side of the primary constriction (centromere). However, it is possible that there are homologous sequences within the centromeres that, due to chromosomal packaging, are unavailable as hybridization targets. A similar phenomenon, where centromeric sequences did not hybridize to FISH probes on meiotic chromosome preparations, was seen in potato (J. Jiang, personal communication).
The distribution of repeats on either side of a centromere was not even and sequences were occasionally found in heterochromatic regions outside of pericentromeric heterochromatin. In Arabidopsis, heterochromatic knobs containing pericentromeric sequences have been found physically disassociated from the centromeric regions, such as that seen on the short arm of chromosome 4 (Fransz et al. 2000). Although soybean pachytene chromosomes appear to be generally euchromatic, knob-like regions of heterochromatin are found in the euchromatic arms and some of the sequences in BAC 076J21 hybridize to these regions.
Sequence analysis of BAC 076J21 showed the presence of a 102-bp tandem repeat (STR102), fragments of Calypso-like elements, a SIRE1 element, and a SIRE1 solo LTR. Tandem repeats are a common motif of higher eukaryotic centromeric/pericentromeric regions. The 180-bp pAL1 repeat of Arabidopsis (Martinez-Zapater et al. 1986), the 155-bp CentO repeat of rice (Cheng et al. 2002), the 137-bp pSau3A9 repeat of sorghum, and the 156-bp CentC of maize (Ananiev et al. 1998) are examples of centromere-specific tandem repeats (also reviewed in Houben and Schubert 2003). Given the commonality of the tandem repeat feature at centromeric regions, it is thought that these approximately nucleosomal-length repeats may play a role in organizing centromere-specific nucleosomes (Nagaki et al. 2003; Black et al. 2004).
Very few tandem repeats have been reported for G. max apart from STR120 (Morgante et al. 1997); SB92, a 92-bp tandem repeat (Vahedian et al. 1995); and now STR102, a ∼102-bp repeat with 82.6% sequence similarity to STR120. FISH analysis of the STR102-containing clone 2_P01 showed that this repeat is almost exclusively located in heterochromatic regions that are either pericentromeric or knob-like regions embedded in euchromatic arms. Sequence analysis showed that there are at least two clusters of STR102 repeats within BAC 076J21, although digital mapping indicates that a third cluster is present that was not assembled into the sequence scaffold. This observation underscores the difficulty of sequencing and assembling sequences from repetitive regions.
The STR102 repeat was detected in clusters up to ∼435.6 kb in length, although longer arrays may exist. A similar organization of centromeric tandem repeats has been reported for rice (Cheng et al. 2002) and maize (Ananiev et al. 1998). The STR102 sequence was conserved in G. latifolia outside of G. max and G. soja; however, only one restriction fragment showed weak hybridization in G. latifolia, so it is possible that there was only limited sequence identity between STR102 and sequence(s) in G. latifolia. This is not unexpected since tandem repeats, such as the α-satellite of primates, have been found to evolve very rapidly (Waye and Willard 1989).
Retroelements are another common feature of eukaryotic genomes (reviewed in Bennetzen 1996). In maize, retroelements often insert within other retroelements leading to nested transposons (SanMiguel et al. 1996). A few retroelements have been described from the soybean genome, the most unusual of which is the SIRE1 element (Laten and Morris 1993), which falls into the copia/Ty1 family (Laten et al. 1998). Laten et al. (2003) found that the flanking sequences of 3 of 10 SIRE1 insertions were repetitive, belonging to either Ty3-gypsy or other repetitive families. In BAC 076J21, the SIRE1 element was inserted into a long array of STR102 repeats and BAC 09M21 also had sequences with significant sequence alignments to the SIRE1 element. This indicates that the SIRE1 element may preferentially insert into heterochromatic and/or pericentromeric regions; alternatively, insertions into gene-rich euchromatic regions may be selected against. This is further corroborated by our FISH results showing hybridization to obvious heterochromatic regions.
A SIRE1 solo LTR was also found in a tract of STR102 repeats with the same 5-bp insertion duplication as the SIRE1 element. There are two possible explanations for this. Two SIRE1 elements inserted into same tract of STR102 repeats with possible site-specific integration. Later one element was removed via unequal recombination (described below), leaving the solo LTR. Alternatively, one SIRE1 element inserted into a STR102 repeat followed by duplication of the inserted element; then one element was removed, possibly via unequal recombination, leaving the solo LTR. In Arabidopsis, for solo LTRs with intact direct repeats, intraelement unequal recombination was found to be the primary causative mechanism (Devos et al. 2002). Pairing and recombination among LTRs within an element would leave one LTR while removing the rest of the retrotransposon. The 5′ LTR from the SIRE1 is 99.8% identical over 1130 bases to the solo LTR, indicating that this is likely a recent insertion within the last 30,000 years (H. Laten, personal communication) and further confirming a previous suggestion that this element may still be active within the G. max genome (Laten et al. 2003).
The Calypso-like elements that are dispersed across 076J21 were fragments of either Calyspo 4-1 or Calyspo 5-1. The Calypso 5-1 fragments had homology to the Pol domain and the Calypso 4-1 fragments had homology to the first few kilobases of the elements that are not annotated. There does not appear to be a simple explanation as to how these fragments came to be.
Heterochromatin is generally methylated at cytosine residues, a hallmark of transcriptionally inactive regions (e.g., Soppe et al. 2002). There were, however, EST matches to sequences within this BAC, so we tested the methylation status of sequences in BAC 076J21 using methyl-sensitive and -insensitive restriction enzymes. STR102 (2_P01), the only sequence showing conservation outside of G. max and G. soja, did not have recognition sites for MspI/HpaII but flanking sequences were methylated, as was 1_E15, containing part of a Calypso 5-1 sequence. Thus, these sequences and/or flanking sequences are methylated in soybean, suggesting that they may be transcriptionally inert. Many of the EST matches, especially to the Calypso-like regions, were derived from a soybean root hair cDNA library, an observation that has no clear explanation.
Eukaryotic genomes protect themselves from retroelements by methylating the elements (Tompa et al. 2002). In cereals, however, transcripts can be found from transposons or retroelements (Vicient et al. 2001). In the case of centromeric repeats, it is hypothesized that strand-specific transcription of tandem repeats is the mechanism by which functional centromeres are epigenetically marked via an RNA-based mechanism, leading to the formation of heterochromatin (Dawe 2003; Martiennsen 2003; Topp et al. 2004). In Arabidopsis, it was recently shown that strand-specific methylation exists in centromeric heterochromatin (Song and Preuss 2003). One cDNA with sequence identity to STR102 was found; therefore, it is possible that transcripts from STR102 or related repeats are involved in the formation of epigenetic marks.
The distribution of SIRE1 and the other retroelements was clearly coincident with heterochromatic regions of the pachytene chromosomes. Thus, these repetitive sequences appear to be sequestered to repetitive regions of the genome and not dispersed throughout the euchromatic arms. The molecular organization of both tandem repeats and retroelements into specific chromosomal regions, as revealed by sequencing and FISH analysis, allows cursory insight into the organization of the soybean genome. The pachytene chromosomes have clearly defined euchromatin and heterochromatin and this work shows that the heterochromatic regions share repetitive elements such as STR102 and the Calypso and SIRE1 sequences. How these sequences function to organize DNA into functional chromosome units is still a mystery, but the organization suggests that sequencing of the euchromatic regions of the soybean genome may be a tractable approach to recover many of the genes of soybean.
We found two BAC clones, 09M21 and 076J21, from MLG L that are partially redundant and contain sequences found in the pericentromeric regions of all soybean chromosomes. One of the major sequence constituents of 076J21, tandem repeat STR102, was found at all or most major heterochromatic blocks in soybean. The repeat sequences present in 076J21 are not generally found outside of the two annuals G. max and G. soja, indicating that these are fast-evolving sequences.
Acknowledgments
We thank the Purdue University Agricultural Experiment Station and the United Soybean Board for generous support of this research, Jiming Jiang (University of Wisconsin-Madison) for advice and comments, and Laura Marek (Iowa State University) for the initial screening of the BAC library. We thank Jeff Doyle (Cornell University) for thoughtful advice in choosing various legume species for analysis of sequence conservation.
Footnotes
References
- Ananiev, E. V., R. L. Phillips and H. W. Rines, 1998. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95: 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Aragon-Alcaide, L., T. Miller, T. Schwarzacher, S. Reader and G. Moore, 1996. A cereal centromeric sequence. Chromosoma 105: 261–268. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 229–241. [Google Scholar]
- Bennetzen, J. L., 1996. The contributions of retroelements to plant genome organization, function and evolution. Trends Microbiol. 4: 347–353. [DOI] [PubMed] [Google Scholar]
- Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods et al., 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582. [DOI] [PubMed] [Google Scholar]
- Blackburn, E. H., and J. G. Hall, 1978. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 120: 33–53. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., F. Dong, T. Langdon, S. Ouyang, C. R. Buell et al., 2002. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14: 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh, D. S., S. Penuela, J. Mudge, R. L. Denny, H. Nordstrom et al., 1998. A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistance gene. Theor. Appl. Genet. 96: 196–202. [Google Scholar]
- Dawe, R. K., 2003. RNA interference, transposons and centromeres. Plant Cell 15: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K. M., J. K. M. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. Wendl and P. Green, 1998. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Fransz, P. F., S. Armstrong, J. H. de Jong, L. D. Parnell, C. van Drunen et al., 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367376. [DOI] [PubMed] [Google Scholar]
- Goldberg, R. B., 1978. DNA sequence organization in the soybean plant. Biochem. Genet. 16: 45–68. [DOI] [PubMed] [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Graham, M. A., L. F. Marek and R. C. Shoemaker, 2002. Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics 162: 1961–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley, W. B., A. G. Hepburn and J. L. Key, 1979. Sequence organization of the soybean genome. Biochim. Biophys. Acta 561: 167–183. [DOI] [PubMed] [Google Scholar]
- Houben, A., and I. Schubert, 2003. DNA and proteins of plant centromeres. Curr. Opin. Plant Biol. 6: 554–560. [DOI] [PubMed] [Google Scholar]
- Jackson, S. A., F. Dong and J. Jiang, 1999. Digital mapping of bacterial artificial chromosomes by fluorescence in situ hybridization. Plant J. 17: 581–587. [DOI] [PubMed] [Google Scholar]
- Jiang, J., S. Nasuda, F. Dong, C. W. Scherrer, S-S. Woo et al., 1996. a A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93: 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., S. H. Hulbert, B. S. Gill and D. C. Ward, 1996. b Interphase fluorescence in situ hybridization: a physical mapping strategy for plant species with large complex genomes. Mol. Gen. Genet. 252: 497–502. [DOI] [PubMed] [Google Scholar]
- Jiang, J., J. A. Birchler, W. A. Parrott and R. K. Dawe, 2003. A molecular view of plant centromeres. Trends Plant Sci. 8: 570–575. [DOI] [PubMed] [Google Scholar]
- Kulikova, O., G. Gualtieri, R. Geurts, D. J. Kim, D. Cook et al., 2001. Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J. 27: 49–58. [DOI] [PubMed] [Google Scholar]
- Laten, H. M., and R. O. Morris, 1993. SIRE-1, a long interspersed repetitive DNA element from soybean with sequence similarity to retrotransposons: initial characterization and partial sequence. Gene 134: 153–159. [DOI] [PubMed] [Google Scholar]
- Laten, H. M., A. Majumdar and E. A. Gaucher, 1998. SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc. Natl. Acad. Sci. USA 95: 6897–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laten, H. M., E. R. Havecker, L. M. Farmer and D. F. Voytas, 2003. SIRE1, an endogenous retrovirus family from Glycine max, is highly homogeneous and evolutionarily young. Mol. Biol. Evol. 20: 1222–1230. [DOI] [PubMed] [Google Scholar]
- Marek, L. F., and R. C. Shoemaker, 1997. BAC contig development by fingerprint analysis in soybean. Genome 40: 420–427. [DOI] [PubMed] [Google Scholar]
- Marek, L., J. Mudge, L. Darnielle, D. Grant, N. Hanson et al., 2001. Soybean genomic survey: BAC-end sequences near RFLP and SSR markers. Genome 44: 572–581. [PubMed] [Google Scholar]
- Martiennsen, R. A., 2003. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 35: 213–214. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater, J. M., M. A. Estelle and C. R. Somerville, 1986. A highly repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204: 417–423. [Google Scholar]
- Morgante, M., I. Jurman, L. Shi, T. Zhu, P. Keim et al., 1997. The STR120 satellite DNA of soybean: organization, evolution and chromosome specificity. Chromosome Res. 5: 363–373. [DOI] [PubMed] [Google Scholar]
- Mroczek, R. J., and R. K. Dawe, 2003. Distribution of retroelements in centromeres and neocentromeres of maize. Genetics 165: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., P. B. Talbert, C. X. Zhong, R. K. Dawe, S. Henikoff et al., 2003. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional element of Arabidopsis thaliana centromeres. Genetics 163: 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, P., D. Pozarkova and J. Macas, 2003. Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol. Biol. 53: 399–410. [DOI] [PubMed] [Google Scholar]
- Pagel, J., J. G. Walling, N. D. Young, R. C. Shoemaker and S. A. Jackson, 2004. Segmental duplications within the Glycine max genome revealed by fluorescence in situ hybridization of bacterial artificial chromosomes. Genome 47: 764–768. [DOI] [PubMed] [Google Scholar]
- Presting, G. G., L. Malysheva, J. Fuchs and I. Schuber, 1998. A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 16: 721–728. [DOI] [PubMed] [Google Scholar]
- Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice et al., 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., A. P. Tikhonov, Y-K. Jin, N. Motchoulskaia, D. Zakharov et al., 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P. J., W. Ramakrishna, J. L. Bennetzen, C. S. Busso and J. Dubcovsky, 2002. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A(m). Funct. Integr. Genomics 2: 70–80. [DOI] [PubMed] [Google Scholar]
- Schlueter, J. A., P. Dixon, C. Granger, D. Grant, L. Clark et al., 2004. Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876. [DOI] [PubMed] [Google Scholar]
- Shoemaker, R. C., K. Polzin, J. Labate, J. Specht, E. C. Brummer et al., 1996. Genome duplication in soybean (Glycine subgenus soja). Genetics 144: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., and T. Hymowitz, 1988. The genomic relationship between Glycine max and G. soja Sieb. and Zucc. as revealed by pachytene chromosome analysis. Theor. Appl. Genet. 76: 705–7011. [DOI] [PubMed] [Google Scholar]
- Song, L., and D. Preuss, 2003. Strand-biased DNA methylation associated with centromeric regions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 11133–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E. L. L., and R. Durbin, 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: 1–10. [DOI] [PubMed] [Google Scholar]
- Soppe, W. J. J., Z. Jasencakova, A. Houben, T. Kakutani, A. Meister et al., 2002. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21: 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., C. Belele, J. E. Dorweiler and V. L. Chandler, 2002. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16: 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa, R., C. M. McCallum, J. Delrow, J. G. Henikoff, B. van Steensel et al., 2002. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr. Biol. 12: 65–68. [DOI] [PubMed] [Google Scholar]
- Topp, C. N., C. X. Zhong and R. K. Dawe, 2004. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 101: 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedian, M., L. Shi, T. Zhu, R. Okimoto, K. Danna et al., 1995. Genomic organization and evolution of the soybean SB92 satellite sequence. Plant Mol. Biol. 29: 857–862. [DOI] [PubMed] [Google Scholar]
- Vicient, C. M., M. J. Jääskeläinen, R. Kalendar and A. H. Schulman, 2001. Active retrotransposons are a common feature of grass genomes. Plant Physiol. 125: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye, J. S., and H. F. Willard, 1989. Concerted evolution of alpha satellite DNA: evidence for species specificity and a general lack of sequence conservation among alphoid sequences of higher primates. Chromosoma 98: 273–279. [DOI] [PubMed] [Google Scholar]
- Wright, D. A., and D. F. Voytas, 2002. Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res. 12: 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., S. Sun, P. Nimmakayala, F. A. Santos, K. Meksem et al., 2004. A BAC- and BIBAC-based physical map of the soybean genome. Genome Res. 14: 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, X., P. F. Fransz, E. J. van Wennekes, P. Zabel, A. van Kammen et al., 1996. High-resolution mapping on pachytene chromosomes and extended DNA fibres by fluorescence in-situ hybridization. Plant. Mol. Biol. Rep. 14: 232–242. [Google Scholar]