Abstract
FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) are two genes that, unless plants are vernalized, greatly delay flowering time in Arabidopsis thaliana. Natural loss-of-function mutations in FRI cause the early flowering growth habits of many A. thaliana accessions. To quantify the variation among wild accessions due to FRI, and to identify additional genetic loci in wild accessions that influence flowering time, we surveyed the flowering times of 145 accessions in long-day photoperiods, with and without a 30-day vernalization treatment, and genotyped them for two common natural lesions in FRI. FRI is disrupted in at least 84 of the accessions, accounting for only ∼40% of the flowering-time variation in long days. During efforts to dissect the causes for variation that are independent of known dysfunctional FRI alleles, we found new loss-of-function alleles in FLC, as well as late-flowering alleles that do not map to FRI or FLC. An FLC nonsense mutation was found in the early flowering Van-0 accession, which has otherwise functional FRI. In contrast, Lz-0 flowers late because of high levels of FLC expression, even though it has a deletion in FRI. Finally, eXtreme array mapping identified genomic regions linked to the vernalization-independent, late-flowering habit of Bur-0, which has an alternatively spliced FLC allele that behaves as a null allele.
A plant's decision to initiate reproductive development is an important event that is controlled by the intersection of an endogenous program with environmental factors such as temperature and light. Biotic and abiotic factors affecting plant growth and survival are variable across habitats, requiring plants to specialize in sensing environmental stimuli and adapting their development accordingly. Presumably, natural selection has optimized this program to time the transition to flowering to provide optimal fitness in a given environment. Indeed the existence of latitudinal clines in many species, including Arabidopsis thaliana, is strongly indicative of adaptive variation in flowering (Mikola 1982; Hurme et al. 1997; Van Dijk et al. 1997; Stinchcombe et al. 2004) or light response (Maloof et al. 2001).
The study of floral induction in A. thaliana has long been approached through the isolation and characterization of mutants with altered flowering times. A large number of flowering-time genes have been identified and a scaffold of the underlying molecular network has been constructed (for review, see Mouradov et al. 2002; Simpson and Dean 2002; Searle and Coupland 2004; Sung and Amasino 2004). As may be expected, this network is complex. On the basis of an extensive body of physiological, genetic, and molecular studies, four main pathways that regulate the key floral identity genes have been outlined: photoperiod, vernalization, autonomous, and gibberellin. The photoperiod pathway utilizes photoreceptors in conjunction with the circadian clock to strongly accelerate flowering in the presence of long-day photoperiods. Flowering is also accelerated when plants with high levels of the floral inhibitor FLC are exposed to an extended period of winter-like temperatures. This process, called vernalization, causes epigenetic silencing of the FLC locus and thereby relieves repression of flowering. Independently of vernalization, FLC is also negatively regulated by the autonomous pathway, which was originally thought to function independently of the environment. Recently, however, it has been found that this pathway may also mediate response to ambient growth temperature (Blázquez et al. 2003). Finally, hormones of the gibberellin class promote flowering in the absence of positive cues from the photoperiod pathway.
The MADS-domain transcription factor encoded by FLC plays a central role in establishing the annual winter habit of many A. thaliana accessions. When active alleles of a second gene, FRI, are present, high levels of FLC potently inhibit the floral transition in a dosage-dependent manner until these plants have been vernalized (Michaels and Amasino 1999; Sheldon et al. 1999). In the wild, this combination of factors has been presumed to prevent germinated seedlings from flowering in the fall.
Conversion of winter annual to summer annual strains can result from natural variation at either FRI or FLC, as loss-of-function mutations in either gene eliminate the late-flowering phenotype. In accessions collected from the wild, variation at the FRI locus is common; to date, at least nine different lesions at FRI have been reported (Johanson et al. 2000; Le Corre et al. 2002; Gazzani et al. 2003). Two deletions first detected in the Columbia (Col) and Landsberg erecta (Ler) laboratory strains are frequently found in other accessions, leading to the conclusion that FRI is the predominant locus controlling flowering-time variation in native A. thaliana (Johanson et al. 2000; Le Corre et al. 2002). In addition, a smaller fraction of accessions seems to flower early because of weak alleles at the FLC locus (Gazzani et al. 2003; Michaels et al. 2003).
From the current framework, it appears that wild accessions of A. thaliana should fall into two distinct classes. The first group, which lacks FLC activity either because of a lesion in FRI or because of a weak FLC allele, should flower relatively early. The second group, possessing high levels of FLC activity, should flower very late even in long days, unless vernalized. Here, we have sought to estimate the amount of variation among wild accessions that is independent of FRI and to identify additional loci in wild accessions that influence flowering time. To this end, we have surveyed the flowering times of 145 single-seed descent accessions in long-day photoperiods, with and without a 30-day vernalization treatment, and genotyped these accessions for the Col-type and Ler-type FRI deletions.
We report substantial variation in flowering time that appears to be independent of FRI in accessions both with and without functional FRI alleles. While a large majority of the late-flowering accessions respond to vernalization, suggesting that much of the variation seen in this survey acts through FLC, there is evidence for late-flowering accessions that were not due to FLC expression. Furthermore, we characterized several accessions with interesting flowering-time phenotypes, given their FRI status to determine the genetic and molecular origins of their flowering-time phenotypes. Through genetic mapping and sequencing or genotyping loci known to affect flowering time, we identified novel loss-of-function FRI and FLC alleles as well as mapped late-flowering loci that are not allelic to FRI or FLC.
MATERIALS AND METHODS
Plant material:
A list of accessions and their phenotypes can be found at http://naturalvariation.org/werner. Most accessions were obtained from the Arabidopsis Biological Resource Center or Lehle Seeds. The LerK and CviK accessions are parents of the Ler/Cvi recombinant inbred line set in which the EDI allele of CRY2 was identified (El-Assal et al. 2001) and were kindly provided by Maarten Koornneef (Wageningen, The Netherlands). Combinations of functional and nonfunctional FRI and FLC alleles in Col (Lee et al. 1994b; Michaels and Amasino 2001) were kindly provided by Rick Amasino (Madison, WI). All lines were propagated prior to genotypic and phenotypic analysis.
Growth conditions:
Seeds were suspended in 1 ml of 0.1% phytagar (Invitrogen, San Diego) and imbibed at 4° in darkness for 4 days. Seeds were then sown onto presoaked Sunshine Mix no. 5 (McConkey, Garden Grove, CA) and thinned after 5 days. Plants were grown in a growth room at 22° under 16 hr of light provided by a 3:1 mixture of Cool-white and Gro-Lux (Sylvania) fluorescent bulbs, followed by 8 hr of darkness. For the long-day surveys, six plants for each line were raised in one pot, except for several control genotypes grown in three blocks of six plants. Over the course of the experiment, the pots were randomized across all flats several times and flats were rotated across and between shelves on a daily basis. Vernalization treatments were performed at the seedling stage, when 5-day-old plants were transferred from the growth room to incubators (Percival, Boone, IA) and kept under noninductive short-day photoperiods, consisting of 9 hr of light followed by 15 hr of darkness at 4°. After 30 days, plants were returned to the growth rooms. A replicate experiment with three blocks of six plants was performed for 31 accessions using newly harvested seeds. Plants used for RNA extractions were grown similarly, except at densities of 12–15 plants/pot.
For all F1, F2, and BC1 populations, seeds suspended in 0.1% phytagar were sown onto soil, allowed to germinate, and thinned (in an unbiased manner) to six seedlings per pot. Flats were rotated across and between shelves on a daily basis and pots were randomized frequently.
Flowering-time measurements:
Flowering time was measured both as total leaf number (TLN) and as days to flowering (DTF). Rosette leaf number and cauline leaf number on the main shoot were determined independently and added to yield TLN prior to formation of the first flower. DTF was recorded daily as the time from the date of sowing (and minus 30 days for vernalization treatment) until the first appearance of floral buds in the apex as seen by the unaided eye. Within a given genotype, we found a high correlation between TLN and DTF. Consequently, for some F1 and F2 experiments, only DTF was recorded to simplify data collection and allow better seed set for later analysis.
DNA sequencing:
For all genes, at least three independent PCR reactions with genomic DNA or cDNA as a template were pooled, gel purified, and used as templates for direct sequencing. The sequenced regions are indicated in Table 1.
TABLE 1 .
Sequenced regions in flowering-time genes
|
Gene (At ID) |
Genomic/cDNA |
GenBank accession no. |
Region (nt) |
|---|---|---|---|
| FCA (At4g16280) | Genomic | ATZ82992 | 1485–2454, 4317–8841 |
| FVE (At2g19520) | Genomic | AF498101 | 2337–5590 |
| FPA (At2g43410) | Genomic | AT2G43410.1 | 1–4593 |
| LD (At4g02560) | Genomic | AT4G02560 | 123–4828 |
| FRI (At4g00640) | Genomic | AF228499 | 40–3356 |
| FLD (At3g10390) | Genomic | AC011560 | 115506–118107 |
| AC009400 | 1517–2236 | ||
| FLK (At3g04610) | Genomic | AC011437 | 28984–33405 |
|
FLC (At5g10140) |
cDNA |
AF116527 |
29–802 |
RNA analysis:
RNA from 14-day-old seedlings grown in long-day periods was isolated using the RNeasy plant mini kit (QIAGEN, Chatsworth, CA). Ten micrograms of total RNA was separated in a 1% agarose gel and blotted onto Hybond-N+ membrane (Amersham Pharmacia Biotech). The membrane was hybridized with a random-labeled probe covering 38205 to 38535 of the FLC gene (GenBank accession no. AL356332) and hybridization signal was determined with a phosphorimager (Molecular Dynamics, Sunnyvale, CA). ImageQuant software (Molecular Dynamics) was used to quantify the signal strength. The blot was then stripped and rehybridized with a probe to 25S rRNA in a similar manner.
First-strand cDNAs were synthesized using the Reverse Transcription System (Promega, Madison, WI) starting with ∼1 μg of total RNA. RT-PCR for the FRI locus used primers JW159 (5′-AGG GCG TAG AGC ATT TAC-3′) and JW22 (5′-CTT GTG AGT CTC CAT ACA CTG-3′).
DNA genotyping:
A list of all PCR-based markers can be found at http://naturalvariation.org/werner.
eXtreme array mapping:
A total of 561 F2 individuals from the cross of Lz-0 with Col were grown in long-day periods and their flowering-time phenotypes were measured as DTF. Two leaves of similar size were collected from the 50 earliest and the 50 latest plants, yielding duplicate early and late pools. Similar amounts of tissue were collected from the parental lines. All tissue was flash frozen in liquid nitrogen and ground to a fine powder, and DNA was extracted according to a modified version of the standard CTAB protocol scaled up to 15 ml of 2× CTAB buffer (2% CTAB, 1.4 m NaCl, 20 mm EDTA, 100 mm TrisHCl, pH 8.0), followed by three rounds of phenol/chloroform extraction and ethanol precipitation. Three random-priming labeling reactions were prepared for each sample using the Bioprime labeling kit (Invitrogen) starting with ∼400 ng of genomic DNA and labeling overnight at room temperature. For each sample, the three reactions were pooled, ethanol precipitated, and ∼20 μg of DNA was hybridized to an Affymetrix ATH1 array as outlined in standard Affymetrix protocols for RNA. In total, 10 samples were prepared, hybridized, and analyzed: three replicates of Lz-0, three replicates of Col, and two replicates each for the early and late-flowering pools. Analysis of the hybridization data was similar to that described previously (Wolyn et al. 2004). For the Bur-0 × flc-3 F2 population (n = 330), the 65 earliest and the 65 latest plants, as measured by days to flowering, were pooled and processed similarly. A total of eight samples were prepared and hybridized: three replicates of Bur-0 and flc-3 and one sample each for the pools.
Lz-0 × Ler QTL mapping:
A total of 178 F2 plants from the cross of Lz-0 to Ler were grown in long days and genotyped for 32 markers distributed across all five chromosomes with an average distance of 15 cM, and a genetic map was determined using MapMaker/EXP 3.0 (Lander et al. 1987). Marker order was as expected from the physical locations of the markers in the Col reference sequence.
For QTL analysis presented in the text, composite interval mapping was performed with QTL Cartographer 1.16c (Basten et al. 2002) using model 6 with a window size of 4.00 cM and a 1.00-cM walking speed. Markers ciw1, nga172, FLC, and F5O24 were selected as background markers. The significance threshold, 3.06, was calculated as the 95th percentile of top LOD scores for 1000 permutations of the phenotypic data. Additional QTL mapping was done utilizing the bqtl package (http://hacuna.ucsd.edu; Borevitz et al. 2002) developed for the statistical package R (http://www.R-project.org; Ihaka and Gentleman 1996). Results from these analyses are available at http://naturalvariation.org/werner.
Bur-0 × flc-3 QTL mapping:
A random sample of 288 F2 plants from the same population that was analyzed by eXtreme array mapping (XAM) was genotyped for 16 markers located at sites identified by XAM. Ninty-six of the plants were also genotyped for two additional markers. As with the Lz/Ler population, these genotypes were used to construct a genetic map and QTL were identified using bqtl. The results of this analysis are available at http://naturalvariation.org/werner.
Estimates of QTL effects:
QTL effects for the Van-0 × Ler F2, Lz-0 × Ler F2, and Bur-0 × flc-3 (Col) F2 populations were calculated using the bqtl package. For the Van-0 × Ler F2 population, the final QTL model included additive and dominance main-effect terms for the markers FRI, HU13 (near FLC), and nga139 (near FLG; Alonso-Blanco et al. 1998) and the interaction terms FRI:HU13 and nga139:HU13. In the case of Lz-0 × Ler F2, we included additive and dominance main terms for ciw1, nga172, FLC, and F5O24, and the interaction terms FLC:F5O24 and ciw1:F5O24. Finally, the QTL model for the Bur-0 cross included additive and dominance main-effect terms for FKF1, FCA3, and BurFLC and an interaction term, FKF1:BurFLC.
RESULTS
Association of FRI deletions with flowering-time effects:
We measured the flowering times of 145 single-seed descent accessions in long-day photoperiods and genotyped them for the Col and Ler FRI deletions. As observed in previous surveys (Johanson et al. 2000; Le Corre et al. 2002; Stinchcombe et al. 2004), the FRI deletions first described in the Col and Ler laboratory strains occur with considerable frequency among accessions, with 34 having the Ler-type and 46 the Col-type deletion (Figure 1). No accession has both deletions, as expected due to their very tight linkage (253 bp). The effect of these deletions on flowering was obvious, with the mean flowering times of the Ler-type lines and Col-type lines being 21.1 ± 3.7 (standard error) leaves (P < 7 × 10−8) and 26.5 ± 3.4 (standard error) leaves (P < 2 × 10−12) earlier, respectively, than that of the remaining 58 accessions that flowered under our growth conditions. The presence or absence of these deletions, along with three other rare potential loss-of-function alleles in FRI (described below), account for 40% of the variation in flowering time among our accessions.
Figure 1.—
Frequency distributions of flowering-time means for 145 accessions grown in long days. (A) Sixty-five accessions lacking Col- and Ler-type FRI deletions. Seven lines did not flower during the course of the experiment (asterisk). (B) Forty-six accessions having the Col-type FRI deletion. (C) Thirty-four accessions having the Ler-type FRI deletion.
Early flowering in lines lacking FRI deletions:
Even though the 65 accessions lacking the Col- and Ler-type deletions in FRI flowered on average much later than those with these deletions, these accessions still showed a considerable range of flowering times (Figure 1A). Eleven accessions flowered with <20 leaves, while 7 had still not flowered after 115 days, when these accessions all had >90 leaves.
Given the substantial number of different FRI lesions discovered from natural populations to date (Gazzani et al. 2003; Johanson et al. 2000; Le Corre et al. 2002), a likely cause for the early flowering is that at least some of the 11 early lines harbored alternate mutations in FRI. Indeed, two of these lines are Cvi accessions, which have recently been shown to contain a premature stop codon in FRI (Gazzani et al. 2003). In the remaining 9 early lines, we sequenced 3.3 kb of the FRI gene encompassing the entire coding region, plus 534 bp upstream and 471 bp downstream of it. An apparent disruption in the FRI coding sequence is evident in only two lines. Accession An-1 has a 99-bp deletion combined with a 31-bp insertion at the start of exon 3. This indel is likely identical to “Indel2” found in the BUI accession (Le Corre et al. 2002). Accession Or-0 was found to have a novel 1-bp deletion in exon 2 of FRI, causing a frameshift in the coding sequence that leads to a premature stop codon.
While we found variation at the nucleotide and amino acid level in the remaining seven early accessions [Da(1)-12, Dra-0, Est, Shahdara, Van-0, Wa-1, Wil-1], none had an obvious disruption of the FRI coding sequence. Furthermore, we could detect FRI mRNA by RT-PCR in the six accessions that we examined (data not shown; Wil-1 not tested), suggesting that they may have subtle defects in FRI expression or may carry alleles at other loci that suppress FRI activity.
A new FLC loss-of-function allele in Van-0:
Since FRI acts entirely through FLC to control flowering (Michaels and Amasino 2001), accessions with a functional FRI allele could be early flowering because of defects in FLC. In fact, two of these seven early flowering accessions, Da(1)-12 and Shahdara, have FLC alleles that show attenuated responses to FRI (Gazzani et al. 2003; Michaels et al. 2003).
One of the early accessions that lacked an obvious FRI lesion, and which we analyzed in more detail, is Van-0. The F1 progeny derived from a cross of Van-0 to Ler, which has nonfunctional FRI and a weak FLC allele, flowered later than either parent, suggesting an interaction between at least two loci that delay flowering. The transgression observed in the F1 progeny is even greater when Van-0 is crossed to Col (J. Borevitz, unpublished data), which also lacks functional FRI. Since the FLC allele from Col is notably stronger than that of Ler (Koornneef et al. 1994; Lee et al. 1994b), we reasoned that variation at FLC, along with the observed polymorphism at FRI, may be partially responsible for the late flowering of F1 hybrids derived from Van-0. Another potential locus is the FLG QTL that interacts with the FLF QTL, which is very likely allelic to FLC, first identified in the Cvi × Ler recombinant inbred lines (RILs) (Alonso-Blanco et al. 1998).
An F2 population from the cross of Van-0 to Ler was genotyped for three markers, the Ler deletion at FRI, a marker tightly linked to FLC, and another 15 cM south on chromosome 5, near FLG. The Ler allele at FLC had a major additive effect (10 leaves increase) that interacted with both Van-0 alleles at FRI and FLG to increase the phenotype, together explaining 81% of the variance in flowering time (supplementary material at http://naturalvariation.org/werner). Figure 2 shows that all plants homozygous for the Van-0 FLC region were early flowering, while other genotypes were quite variable. Backcross analysis of F1 plants crossed to Ler revealed FRI variation dependent on FLC copy number controlling flowering time, while a backcross to Van-0 revealed that only the FLC-linked marker had a significant effect (supplementary material at http://naturalvariation.org/werner). This suggests that the Van-0 allele of FLC is even less active than the weak Ler allele and may be a null.
Figure 2.—
Frequency distribution of total leaf number for an F2 population derived from a cross of Van-0 with Ler, grown in long days. Shaded bars indicate individuals homozygous for the Van-0 allele of HU13, a marker close to FLC. Flowering-time ranges and means of the parents and F1 hybrid are denoted by arrows and horizontal lines, respectively.
Sequencing of the FLC gene from Van-0 revealed a nucleotide substitution in exon 6, creating a premature stop codon at position 158 of the open reading frame. Termination of translation at this site would produce a protein containing the MADS box, I-box, and K-box, but lacking the final 39 amino acids. Reverse transcription followed by PCR showed that Van-0 produces, in approximately equal proportions, a normal length FLC transcript with the nonsense mutation, as well as an alternatively spliced transcript lacking exon 6, which is apparently the consequence of nonsense-associated altered splicing (Wang et al. 2002). As exon 6 is 42 nucleotides long, translation of this transcript is predicted to yield an FLC protein with an internal deletion of amino acids 150–163. This nonsense mutation in FLC and the behavior of the QTL linked to FLC make FLC a strong candidate gene for causing the early flowering of Van-0.
Late flowering in lines with FRI deletions:
There was also significant variation in the accessions having nonfunctional FRI alleles (Figure 1, B and C). Of the 80 lines harboring a Ler- or Col-type deletion in FRI, under our conditions 24 flowered with significantly more than 20 total leaves, with several flowering as late as FRI-Sf2 FLC-Col, a line with the late-flowering FRI allele from Sf-2 in the Columbia background (Lee et al. 1994b). For 19 of these late-flowering accessions and for 10 early flowering accessions lacking the Col and Ler deletions, we confirmed the flowering phenotypes in a second experiment containing more individuals per line (r = 0.923 with first experiment).
To evaluate both the dominance of the late-flowering phenotypes and the effect of adding functional FRI and FLC alleles in F1 hybrid backgrounds, three of these accessions were crossed to four lines containing different homozygous combinations of active and inactive FRI and FLC alleles in the Columbia background. Inactive alleles were the fast-neutron flc-3 allele and the FRI deletion allele from Col (fri-Col). Active alleles were FLC from Col (FLC-Col) and FRI introgressed from the accession Sf-2 (FRI-Sf2) (Lee et al. 1994b; Michaels and Amasino 1999; Michaels and Amasino 2001). Early flowering plants were obtained when the three accessions, Er-0, Rak-2, Wu-0, were crossed to fri-Col flc-3 and fri-Col FLC-Col, revealing that their late-flowering phenotypes are largely recessive (Table 2). In contrast, adding a functional copy of FRI, through crossing to FRI-Sf2 flc-3, resulted in late flowering, with the F1 progeny from the cross to FRI-Sf2 FLC-Col being even later flowering, proving the effect of the FRI deletions and the absence of dominant suppressors. In addition, since introduction of functional FRI alone in the FRI-Sf2 flc-3 hybrids caused late flowering, the accessions in question have functional FLC alleles, perhaps of varying strength.
TABLE 2 .
Flowering-time analysis of late-flowering accessions
|
Flowering timea (mean ± SE) |
|||||
|---|---|---|---|---|---|
| Accession |
Parent |
× flc-3 Col F1 |
× Col F1 |
× FRI-Sf2 flc-3 Col F1 |
× FRI-Sf2 FLC-Col F1 |
| Wu-0 | 23.6 ± 0.43 | 18.5 ± 0.60 | 18.8 ± 0.31 | 29.0 ± 0.85 | 57.1 ± 3.74 |
| Er-0 | 30.7 ± 0.68 | 20.6 ± 0.96 | 19.9 ± 0.13 | 36.6 ± 0.94 | >90b |
| Rak-2 | 36.6 ± 1.29 | 19.3 ± 0.31 | 21.3 ± 0.61 | 57.3 ± 3.41 | >90b |
| Bur-0 |
38.9 ± 0.23 |
24.1 ± 0.13 |
>67b |
22.1 ± 0.35 |
74.8 ± 4.18 |
Flowering time was measured as days to flowering in long days without vernalization.
Some or all of the plants had not flowered after 90 days.
n = 7 or 8 for all genotypes. Comparisons of parents with the corresponding F1 populations are significant at P < 0.05 (t-test).
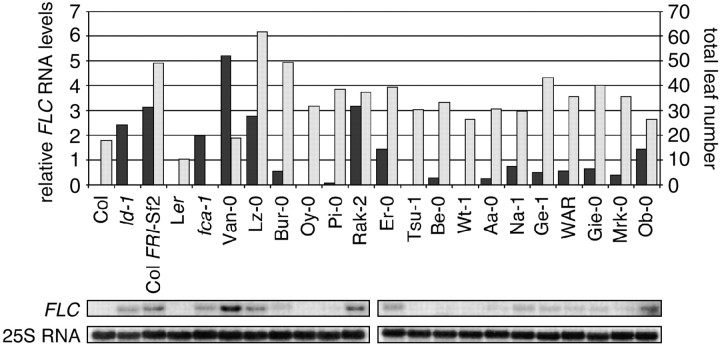
It is plausible that lines lacking FRI activity may still have significant FLC expression due to the presence of alternate activators of FLC such as FLG or ART1 (Grbic and Bleecker 1996; Alonso-Blanco et al. 1998; Poduska et al. 2003). Consequently, we examined FLC expression in 15 of these accessions by RNA blot analysis (Figure 3). These late-flowering lines showed considerable variation in FLC levels, with modest correlation between FLC levels and flowering time (r = 0.55). Be-0, Oy-0, Pi-0, and Wt-1 had very little or no detectable FLC expression, while Er-0, Na-1, Ge-1, Gie-0, Lz-0, Ob-0, and Rak-2 had substantial FLC levels, despite the presence of nonfunctional FRI alleles.
Figure 3.—
(Top) Comparison of relative FLC RNA levels (solid bars) with flowering times (shaded bars) in late-flowering accessions with deletions in FRI. The five leftmost strains are controls; flowering times were not determined for ld-1 (in Col background) and fca-1 (in Ler background), which are late-flowering mutants known to have elevated FLC levels. Also shown is Bur-0, which does not have a deletion in FRI. (Bottom) RNA blots. FLC levels are expressed relative to 25S rRNA.
Flowering-time QTL in Lz-0:
Lz-0 was the latest flowering of all accessions with a FRI deletion (62 leaves) and had high levels of FLC RNA (Figure 3). This lateness was largely eliminated by vernalization (25 leaves after a 30-day vernalization treatment), suggesting that the late flowering in absence of vernalization is due, at least in part, to FLC activation. In agreement with this notion, FLC RNA levels in Lz-0 were much lower after 30 days of vernalization (data not shown). When Lz-0 was crossed to Col and Ler, the resulting F1 hybrids were early (data not shown), indicating that activation of FLC in Lz-0 is suppressed by dominant alleles present in Col and Ler. This behavior is very much reminiscent of loss-of-function mutations in autonomous pathway genes such as FCA, FLD, FLK, FPA, FVE, FY, or LUMINIDEPENDENS (LD) (Koornneef et al. 1991; Lee et al. 1994a; Sanda and Amasino 1996; Nelson et al. 2000; Lim et al. 2004; Mockler et al. 2004). To examine whether the Lz-0 behavior is associated with a major mutation in any of these genes, we sequenced the entire coding regions including small introns for FCA, FLD, FPA, FVE, FLK, and LD. Although nonsynonymous changes were found, there were no obvious loss-of-function mutations.
To identify loci responsible for the FRI-independent activation of FLC in Lz-0, we next examined 178 F2 plants derived from the cross of Lz-0 to Ler. These plants showed a wide range of flowering times, with many individuals flowering earlier or later than either parent (Figure 4A). A QTL analysis using 32 markers spaced at ∼15-cM intervals throughout the genome failed to detect any major QTL near the autonomous pathway genes FVE, FPA, FLK, FLD, LD, or FCA. Suggestive peaks were found at the bottom of chromosome 1, near marker ciw1, and at the top of chromosome 3, in the vicinity of FLK and FLD, respectively (Figure 4B).
Figure 4.—
Flowering-time QTL in crosses of Lz-0 with Ler and Col. (A) Flowering-time distribution in an F2 population derived from a cross of Lz-0 with Ler, grown in long days. Parental means and ranges are denoted by arrows and horizontal lines, respectively. (B) Flowering-time QTL maps for chromosomes 1, 3, and 5 on the same population genotyped for 32 markers spanning the genome. QTL likelihoods were calculated with QTL Cartographer (see materials and methods). The dashed line corresponds to a P = 0.05 threshold set by permutations (LOD 3.06). No evidence for QTL was apparent on chromosomes 2 and 4. Solid diamonds indicate marker locations. (C) Flowering times of 561 F2 individuals derived from a cross of Lz-0 with Columbia, grown in long days. Tissue from the 50 earliest and the 50 latest individuals (solid bars) was used for XAM. Ranges and mean values of parental lines are indicated by horizontal lines and arrows, respectively. (D) XAM results for four comparisons between two independent replicates from the earliest 50 plants and two independent replicates from the latest 50 plants. Positive allele frequency indicates bias to Col alleles, negative toward Lz-0 alleles. Roman numerals indicate chromosomes.
The likelihood of QTL was significant for much of chromosome 5. Background markers placed at FLC (4.4 cM) and 11.5 cM south of FLC (F5O24) could resolve two peaks. One QTL within this region is very likely to be FLC, given the known weakness of the Ler FLC allele and the high levels of FLC RNA present in Lz-0. The peak south of FLC is in the same region as known natural alleles that interact with FLC, such as ART1, FLG, and the QTL that we detected in the Van/Ler cross (Alonso-Blanco et al. 1998; Poduska et al. 2003). Genome-wide testing for epistasis suggested interactions that further delayed flowering between Lz-0 alleles at markers near ciw1 and F5O24 as well as between Lz-0 alleles at markers near F5O24 and FLC (supplementary data, QTL effect table at http://naturalvariation.org/werner).
XAM on an F2 population derived from the cross of Lz-0 with Col grown in long days (Figure 4C) provided a complementary analysis of the genomic regions responsible for the lateness of Lz-0 (Wolyn et al. 2004). As a first step, labeled Lz-0 and Col genomic DNA was hybridized to Affymetrix ATH1 arrays (Borevitz et al. 2003), and 15,000 single-feature polymorphisms (SFPs) were identified as differences in hybridization intensity between the two strains [20% false discovery rate (FDR) by permutations and ∼30% FDR by sequence analysis]. Clusters of SFPs revealed 286 potential deletions. When probes prepared from DNA pools of the 50 earliest and the 50 latest segregating F2 plants (earliest and latest 8.9% each) were hybridized independently to ATH1 arrays, allele frequencies for each SFP could be estimated. Allele frequencies of SFPs linked to QTL will deviate in opposite directions between early and late pools while unlinked loci will have intermediate allele frequencies, i.e., equal contributions of parental alleles in both pools.
Significant allele frequency differences caused by QTL were detected in three regions of the genome: the bottom of chromosome 1, a large part of chromosome 4, and the top of chromosome 5 (Figure 4D). In the late-flowering pool, Lz-0 alleles were enriched at the bottom of chromosome 1 at ∼19 Mb, colocalizing with a suggestive QTL peak seen in the cross with Ler. A similar overlap with the Ler QTL results was seen for QTL on chromosome 5. However, unlike the results from the Ler cross, allelic variation was observed at the top of chromosome 4. Here, Lz-0 alleles were also enriched in the late-flowering pool. Although FRI is located in this region, both Lz-0 and Columbia contain lesions at the FRI locus, suggesting that this QTL corresponds to another gene(s).
Variation in vernalization response:
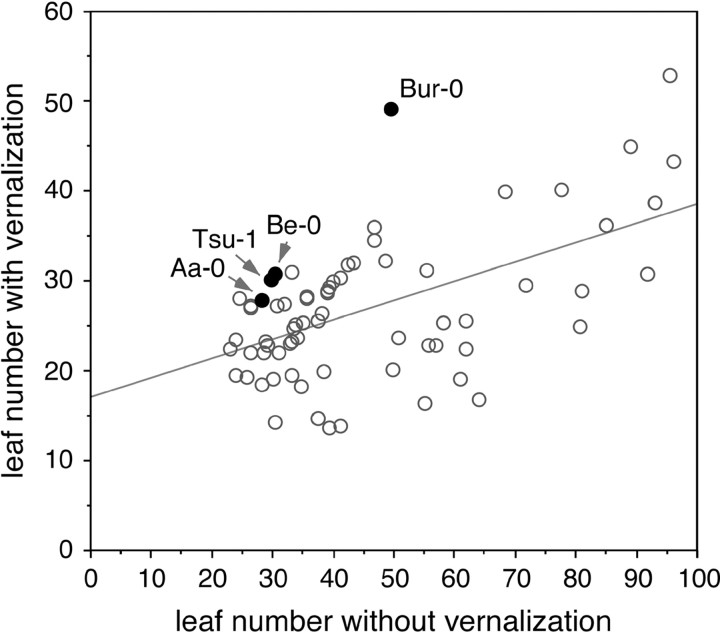
As exemplified in mutants with defects in the photoperiod pathway, A. thaliana has the potential for a late-flowering, vernalization-insensitive growth habit. To date, however, most late-flowering accessions have been found to be vernalization responsive and therefore are assumed to have high levels of FLC causing the late flowering in the absence of vernalization. To determine whether there are exceptions to this general rule, we measured the flowering times of 78 late-flowering accessions grown in long days after a 30-day vernalization treatment, which is considered a nearly saturating vernalization treatment for most late-flowering, vernalization-responsive genotypes (Lee and Amasino 1995).
For the vast majority of lines that flowered with >22 leaves without vernalization or did not flower at all during the course of the experiment, the 30-day treatment substantially reduced the number of leaves produced prior to flowering. However, there was considerable variation in vernalization sensitivity, ranging from no effect to >70% reduction in total leaf number (Figure 5). Specifically, four accessions, Aa-0, Be-0, Bur-0, Tsu-1, that flowered with >30 total leaves showed little response to vernalization, with less than a 10% decrease in leaf number. In a previous study, Tsu-0, another accession from Tsu (Japan), showed a similar behavior (Nordborg and Bergelson 1999).
Figure 5.—
Response of late-flowering accessions to 30-day vernalization treatment. Only the 70 accessions having >22 total leaves and flowering in long days are included. Bur-0 stands out as displaying no response to vernalization.
Flowering-time QTL in Bur-0:
The most dramatic behavior among the late-flowering, vernalization-insensitive group was shown by Bur-0, which flowered with ∼50 leaves regardless of vernalization. Unlike strong photoperiod pathway mutants, such as co and gi (Koornneef et al. 1991), Bur-0 flowered later in short days than in long days, demonstrating that it can still respond to day length (data not shown). When crossed to fri-Col flc-3 and FRI-Sf2 flc-3, the F1 hybrids were early flowering in long days (Table 2). In contrast, very-late-flowering plants were obtained from the cross of Bur-0 with Col, which contains an active FLC allele. Taken together, these results indicate that Bur-0 has an FLC allele that does not respond to FRI and that its recessive late-flowering phenotype may be independent of FLC.
FLC transcript in Bur-0 was detected by both RNA gel-blot analysis (Figure 3) and RT-PCR (data not shown), although the transcript size appeared larger than that found in other accessions. When we cloned the FLC cDNA from Bur-0, we found that it contained 64 bp of intron sequence immediately upstream of exon 7. The use of an alternate 3′ splice site appeared to be the result of a mutation of an invariant G in the final position of intron 6. The inserted sequence causes a frameshift, leading to the addition of four new codons followed by a premature stop codon. The result is an FLC open reading frame that, similar to that of Van-0, would encode the MADS-, I-, and K-boxes while lacking the C-terminal 33 amino acid residues. On the basis of the failure of the Bur-0 FLC allele to respond to FRI and the similarity among the proteins encoded by the Van-0 and Bur-0 FLC transcripts, we reason that the Bur-0 FLC allele may also be a null.
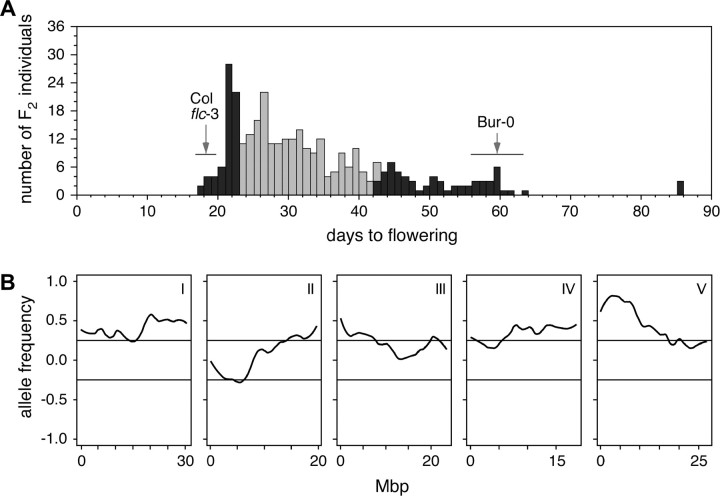
To identify QTL affecting flowering time in Bur-0 aside from FLC, we again turned to XAM to analyze a population of 330 F2 plants from the cross of Bur-0 with flc-3 (Col background) grown in long days (Figure 6A), where any QTL that we detect are likely to be FLC independent. A total of 15,000 SFPs were identified between Bur and Col (7% FDR by permutations and 30% FDR by sequence analysis) that could be clustered into 253 potential deletions (Borevitz et al. 2003). There was severe segregation distortion on the bottom of chromosome 1, when the average allele frequency of pools was compared to the midparent genotype (see http://naturalvariation.org/werner). However, because this segregation distortion was seen in both late and early pools, it does not affect the mapping results.
Figure 6.—
Mapping of genomic regions that delay flowering in Bur-0. (A) Flowering times of 330 F2 individuals derived from a cross of Bur-0 with flc-3 (Col background), grown in long days. Tissue from the 65 earliest and the 65 latest individuals (solid bars) was used for eXtreme array mapping. Ranges and mean values of parental lines are indicated by horizontal lines and arrows, respectively. (B) XAM results for earliest 65 plants. Positive allele frequency indicates bias to Col flc-3 alleles, negative toward Bur-0 alleles. Roman numerals indicate chromosomes.
As shown in Figure 6B, XAM suggests the presence of long-day flowering time loci on the bottom of chromosome 1 and on the top of chromosome 5. The calculated peak for the QTL on chromosome 1 was at ∼21 Mb, while the QTL on chromosome 5 peaked at 4.6 Mb. Sites with less drastic deviations from neutral allele frequency appeared at the top of chromosome 2 and the middle of chromosome 4. We also individually genotyped a large number of plants from the entire F2 population used for XAM, using molecular markers surrounding the predicted QTL. This analysis confirmed the position of QTL predicted by XAM. As seen with XAM, an extreme deficiency in Col homozygotes (7% observed, 25% expected, χ2 test, P < 1 × 10−16) was observed on the bottom of chromosome 1.
The flowering time QTL on chromosome 1 and chromosome 5 were semidominant, with single Bur-0 alleles delaying flowering by ∼6 and 10 days, respectively, relative to single Col alleles. Interestingly, these loci showed a significant interaction with both Bur-0 alleles, further delaying flowering. Furthermore, even the chromosomal regions showing only slight biases in allele frequency proved to significantly affect flowering time in this population. The Bur-0 allele at the chromosome 4 locus was estimated to delay flowering by 4 days/allele, while Col homozygotes at the chromosome 2 locus flowered ∼3 days later than did Bur-0 homozygotes.
DISCUSSION
Previous studies of natural variation in flowering time in A. thaliana either have focused on a small number of accessions (e.g., Gazzani et al. 2003) or did not include genetic analyses (e.g., Stinchcombe et al. 2004). Here, we examined variation of flowering time in a large collection of single-seed descent accessions available from stock centers. We have confirmed and extended previous observations regarding the frequency and magnitude of effect of two natural FRI deletions. Furthermore, we not only have estimated the extent of variation likely to be due to genetic loci other than FRI, but also have carried out initial genetic and molecular analyses of accessions that have interesting flowering phenotypes with respect to their genotype at FRI.
With the identification of the Or-0 fri allele, there are now at least 10 different mutations known to disrupt the FRI coding region (Johanson et al. 2000; Le Corre et al. 2002 ; Gazzani et al. 2003). At least 84 of the 145 accessions that we surveyed carry FRI loss-of-function alleles, with the Col and Ler deletions being, by far, the most prevalent. At a minimum, the presence or absence of natural fri alleles are responsible for 40% of the variation in long-day flowering time seen in our experiment.
Some of the FRI-independent flowering-time variation is likely due to allelic variation at FLC itself. Ler, C24, Da(1)-12, and Shahdara all have FLC alleles often described as “weak” due to their attenuated responses to active FRI alleles (Koornneef et al. 1994; Lee et al. 1994b; Sanda and Amasino 1996; Gazzani et al. 2003; Michaels et al. 2003). In the case of Ler, the reduced sensitivity to FRI is the result of a transposon insertion in the first intron, leading to decreased steady-state levels of FLC mRNA. Similarly, a larger and unrelated insertion was found in intron 1 of the Da(1)-12 FLC allele and may be responsible for its behavior (Michaels et al. 2003).
In our work, we identified additional early flowering accessions, Dra-0, Est, Wa-1, and Wil-1, that appear to have intact FRI coding sequences and, except for Wil-1 (not tested), express the FRI transcript, making them good candidates for having defects in FLC. Gazzani et al. (2003) have analyzed another accession from Wilna (Lithuania), Wil-2, and found that its early flowering may be the result of either a nonfunctional FRI allele caused by changes outside the coding region or a closely linked dominant suppressor of FRI activity. Wil-1 may be early flowering for the same reason.
In contrast to other natural FLC alleles that have been characterized as weak, the Van-0 and Bur-0 FLC alleles affect the coding region and appear to be incapable of delaying flowering. All FLC transcripts that we found in Van-0 and Bur-0 were expressed at reasonably high levels and code for FLC proteins with disrupted C termini, suggesting that these amino acids are essential for FLC function. In Van-0, the defect in FLC is very likely the source of its early flowering phenotype, while the effect of the inactive FLC allele in Bur-0 is masked by the presence of other late-flowering loci.
Another possible source of flowering-time variation in nonvernalized plants is through FRI-independent modulation of FLC expression levels. To search for alternate regulators of FLC in natural populations, we sought accessions that flowered late and were vernalization responsive, but lacked FRI activity. Principal among the accessions that we uncovered with this phenotype was Lz-0.
QTL mapping in the Lz/Ler population identified at least two linked QTL on the top of chromosome 5, one at FLC and one south of FLC, near marker F5O24. A flowering-time QTL not only is expected at FLC, due to the known phenotype of the Ler allele, but also is consistent with FLC mediating the vernalization-sensitive late-flowering phenotype of Lz-0. Other late-flowering loci from Lz-0 detected in the cross would then be expected to interact with the Lz-0 FLC allele. Indeed, we found interaction effects between Lz-0 alleles for QTL at marker F5O24 and FLC. However, the statistical support for this interaction was weak, which may well be a consequence of the limited recombination between these linked QTL. The QTL linked to F5O24 is near ART1 and FLG, known or suspected natural modifiers of FLC (Alonso-Blanco et al. 1998; Poduska et al. 2003). We also identified an FLC-interacting locus in this region in the cross between Van-0 and Ler. However, ART1 and FLG, as well as the Van-0 allele in this region, seem to act in a dominant or semidominant fashion compared to those of Ler, while the Lz-0 gene appears to be recessive.
XAM identified several additional regions, near markers ciw1 and F5O24, as well as FLC as associated with late flowering in the cross of Lz-0 with Col. Relative to Ler, Col has a strong FLC allele capable of responding to FRI and to mutations in autonomous pathway genes. Consequently, if allelic variation at FLC underlies the effect of this QTL at FLC, it would suggest that the Lz-0 FLC allele may be stronger than the Col allele or may show a particular interaction with other loci. Alternatively, enrichment for Lz-0 alleles at FLC could be a consequence of linkage between FLC and loci from Lz-0 that activate its expression or that delay flowering independently of FLC.
Contrary to the ample natural variation acting through FLC, little variation has been assigned to the genes of the photoperiod pathway, with the notable exception of the EDI amino acid substitution in CRY2 (El-Assal et al. 2001), which has been found only in accessions originating from the Cape Verde Islands. It is possible that variation in this pathway does exist in the wild, but that the effects are more subtle and therefore generally masked by the dramatic variation conferred by the FRI/FLC pathway. Consistent with this, several small-effect QTL in the Cvi/Ler RIL population were detected as significant only after vernalization, although it is possible that these QTL may not have effects in the absence of vernalization (Alonso-Blanco et al. 1998).
We took an alternative approach to examining natural variation in the photoperiod pathway by identifying late-flowering accessions with limited FLC activity and little response to vernalization. One such accession is Bur-0. Since we found that Bur-0 has a strong loss-of-function allele at FLC, we used a mapping cross with a Col line that carries a null allele, flc-3. Thus, any flowering-time loci that we detect should be FLC independent. XAM identified four regions of the genome as responsible for flowering-time differences between Bur-0 and Col flc-3. Despite substantial segregation distortion, the effects of all four regions could be confirmed by genotyping the population with individual molecular markers.
Interestingly, Bur-0 has a short circadian period (Michael et al. 2003), which may be causally related to its flowering-time phenotype. The confidence interval for the QTL on chromosome 1 includes the genomic location of the clock gene and the putative blue-light photoreceptor FKF1 (Nelson et al. 2000; Imaizumi et al. 2003), while the XAM QTL on chromosome 5 peaks at 4.6 Mb, which is ∼1.4 Mb south of FLC, near CONSTANS (CO), the major output of the photoperiod pathway (Putterill et al. 1995). Flowering-time QTL at the bottom of chromosome 1 or at the top of chromosome 5, but distinct from FLC, also have been found in other mapping crosses (Clarke et al. 1995).
Early flowering has arisen independently many times in natural A. thaliana populations, demonstrated first by numerous lesions in FRI and now by a growing collection of weak and possibly null FLC alleles. However, in contrast to the Col- and Ler-type deletions in FRI, these disruptions in FLC are relatively rare. The Bur- and Ler-type FLC alleles were found in only one and four additional strains, respectively, while the FLC-Van allele was unique among the 145 accessions that we surveyed here. For the other loci mapped in this work, determining the prevalence and effect among accessions awaits their molecular identification.
Acknowledgments
We thank the National Science Foundation (NSF)-funded Arabidopsis Biological Resource Center at Ohio State University, Maarten Koornneef and Rick Amasino for seed stocks, and Tsegaye Dabi for technical assistance. We thank Chris Schwartz and Norman Warthmann for comments and critical reading of the manuscript. Support for the joint program in quantitative genetics in the Weigel and Chory laboratories was provided by a Helen Hay Whitney Fellowship (J.O.B.), a NSF Predoctoral Fellowship (J.D.W.), funds from the Howard Hughes Medical Institute (HHMI) to J.C. and the Max Planck Society to D.W., a grant from Torrey Mesa Research Institute/Syngenta to D.W., and a grant from the National Institutes of Health (GM62932) to J.C. and D.W. J.C. is an Investigator of the HHMI, and D.W. is a Director of the Max Planck Institute.
Footnotes
Sequences data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AY781906, AY785055, AY849982, AY849983, AY849984, AY849985, AY849986, AY849987, AY849988, AY849989, AY849990, AY849991, AY849992, AY849993, AY849994, AY849995, AY849996, AY849997, AY849998, AY849999, AY850000, AY850001, AY850002, AY854371, AY854372, and AH014566.
Communicating editor: V. Sundaresan
References
- Alonso-Blanco, C., S. E. El-Assal, G. Coupland and M. Koornneef, 1998. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basten, C. J., B. S. Weir and Z-B. Zeng, 2002 QTL Cartographer, Version 1.16. Department of Statistics, North Carolina State University, Raleigh, NC.
- Blázquez, M. A., J. H. Ahn and D. Weigel, 2003. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33: 168–171. [DOI] [PubMed] [Google Scholar]
- Borevitz, J. O., J. N. Maloof, J. Lutes, T. Dabi, J. L. Redfern et al., 2002. Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160: 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J. O., D. Liang, D. Plouffe, H.-S. Chang, T. Zhu et al., 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J. H., R. Mithen, J. K. M. Brown and C. Dean, 1995. QTL analysis of flowering time in Arabidopsis thaliana. Mol. Gen. Genet. 248: 278–286. [DOI] [PubMed] [Google Scholar]
- El-Assal, S. E.-D., C. Alonso-Blanco, A. J. Peeters, V. Raz and M. Koornneef, 2001. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29: 435–440. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., A. R. Gendall, C. Lister and C. Dean, 2003. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132: 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic, V., and A. B. Bleecker, 1996. An altered body plan is conferred on Arabidopsis plants carrying dominant alleles of two genes. Development 122: 2395–2403. [DOI] [PubMed] [Google Scholar]
- Hurme, P., T. Repo, O. Savolainen and T. Pääkkönen, 1997. Climatic adaptation of bud set and frost hardiness in Scots pine (Pinus sylvestris). Can. J. For. Res. 27: 716–723. [Google Scholar]
- Ihaka, R., and R. Gentleman, 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5: 299–314. [Google Scholar]
- Imaizumi, T., H. G. Tran, T. E. Swartz, W. R. Briggs and S. A. Kay, 2003. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306. [DOI] [PubMed] [Google Scholar]
- Johanson, U., J. West, C. Lister, S. Michaels, R. Amasino et al., 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., C. J. Hanhart and J. H. van der Veen, 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., H. Blankestijn-de Vries, C. Hanhart, W. Soppe and T. Peeters, 1994. The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta phenotype. Plant J. 6: 911–919. [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Day et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Le Corre, V., F. Roux and X. Reboud, 2002. DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol. Biol. Evol. 19: 1261–1271. [DOI] [PubMed] [Google Scholar]
- Lee, I., and R. M. Amasino, 1995. Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108: 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., M. J. Aukerman, S. L. Gore, K. N. Lohman, S. D. Michaels et al., 1994. a Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., S. D. Michaels, A. S. Masshardt and R. M. Amasino, 1994. b The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6: 903–909. [Google Scholar]
- Lim, M. H., J. Kim, Y. S. Kim, K. S. Chung, Y. H. Seo et al., 2004. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof, J. N., J. O. Borevitz, T. Dabi, J. Lutes, R. B. Nehring et al., 2001. Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29: 441–446. [DOI] [PubMed] [Google Scholar]
- Michael, T. P., P. A. Salome, H. J. Yu, T. R. Spencer, E. L. Sharp et al., 2003. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053. [DOI] [PubMed] [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 2001. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S. D., Y. He, K. C. Scortecci and R. M. Amasino, 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikola, J., 1982. Bud-set phenology as an indicator of climatic adaptation of Scots pine in Finland. Silva Fenn. 16: 178–184. [Google Scholar]
- Mockler, T. C., X. Yu, D. Shalitin, D. Parikh, T. P. Michael et al., 2004. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc. Natl. Acad. Sci. USA 101: 12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov, A., F. Cremer and G. Coupland, 2002. Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14: S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. C., J. Lasswell, L. E. Rogg, M. A. Cohen and B. Bartel, 2000. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., and J. Bergelson, 1999. The effect of seed and rosette cold treatment on germination and flowering time in some Arabidopsis thaliana (Brassicaceae) ecotypes. Am. J. Bot. 86: 470. [PubMed] [Google Scholar]
- Poduska, B., T. Humphrey, A. Redweik and V. Grbic, 2003. The synergistic activation of FLOWERING LOCUS C by FRIGIDA and a new flowering gene AERIAL ROSETTE 1 underlies a novel morphology in Arabidopsis. Genetics 163: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., F. Robson, K. Lee, R. Simon and G. Coupland, 1995. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Sanda, S. L., and R. M. Amasino, 1996. Ecotype-specific expression of a flowering mutant phenotype in Arabidopsis thaliana. Plant Physiol. 111: 641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, I., and G. Coupland, 2004. Induction of flowering by seasonal changes in photoperiod. EMBO J. 23: 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C. C., J. E. Burn, P. P. Perez, J. Metzger, J. A. Edwards et al., 1999. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G. G., and C. Dean, 2002. Arabidopsis: The Rosetta stone of flowering time? Science 296: 285–289. [DOI] [PubMed] [Google Scholar]
- Stinchcombe, J. R., C. Weinig, M. Ungerer, K. M. Olsen, C. Mays et al., 2004. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. USA 101: 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S., and R. M. Amasino, 2004. Vernalization and epigenetics: how plants remember winter. Curr. Opin. Plant Biol. 7: 4–10. [DOI] [PubMed] [Google Scholar]
- Van Dijk, H., P. Boutry, H. McCombie and P. Vernet, 1997. Flowering time in wild beet (Beta vulgaris ssp. maritima) along a latitudinal cline. Acta Oecol. 18: 47–60. [Google Scholar]
- Wang, J., Y. F. Chang, J. I. Hamilton and M. F. Wilkinson, 2002. Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol. Cell 10: 951–957. [DOI] [PubMed] [Google Scholar]
- Wolyn, D. J., J. O. Borevitz, O. Loudet, C. Schwartz, J. Maloof et al., 2004. Light-response quantitative trait loci identified with composite interval and eXtreme array mapping in Arabidopsis thaliana. Genetics 167: 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]