Abstract
The value of molecular markers and pedigree records, separately or in combination, to assist in the management of conserved populations has been tested. The general strategy for managing the population was to optimize contributions of parents to the next generation for minimizing the global weighted coancestry. Strategies differed in the type of information used to compute global coancestries, the number and type of evaluated individuals, and the system of mating. Genealogical information proved to be very useful (at least for 10 generations of management) to arrange individuals' contributions via the minimization of global coancestry. In fact, the level of expected heterozygosity after 10 generations yielded by this strategy was 88–100% of the maximum possible improvement obtained if the genotype for all loci was known. Marker information was of very limited value if used alone. The amount and degree of polymorphism of markers to be used to compute molecular coancestry had to be high to mimic the performance of the strategy relying on pedigree, especially in the short term (for example, >10 markers per chromosome with 10 alleles each were needed if only the parents' genotype was available). When both sources of information are combined to calculate the coancestry conditional on markers, clear increases in effective population size (Ne) were found, but observed diversity levels (either gene or allelic diversity) in the early generations were quite similar to the ones obtained with pedigree alone. The advantage of including molecular information is greater when information is available on a greater number of individuals (offspring and parents vs. parents only). However, for realistic situations (i.e., large genomes) the benefits of using information on offspring are small. The same conclusions were reached when comparing the use of the different types of information (genealogical or/and molecular) to perform minimum coancestry matings.
THE maintenance of high levels of genetic variability and low levels of inbreeding is a major objective in conservation programs. Genetic variation is a prerequisite for populations to be able to face future environmental changes and to ensure long-term response to selection, either natural or artificial, for traits of economic or cultural interest (Frankham et al. 2003). Also, inbreeding levels should be kept as low as possible to avoid deleterious effects on fitness-related traits, which could compromise the viability of the populations.
The classical criterion used to quantify genetic variability has been the expected heterozygosity (Nei 1973), usually called gene diversity (GD). GD represents the expected proportion of heterozygotes if the population were in Hardy-Weinberg equilibrium and is directly related to the amount of additive genetic variance for quantitative traits (Falconer and Mackay 1996). From an evolutionary perspective, another important measure of variability is allelic diversity (AD), or allelic richness (i.e., the number of different alleles at a particular locus, or the average over loci, present in the population). High levels of AD are essential for the long-term evolutionary potential of populations because the limit of selection response is determined by the initial number of alleles (assuming that mutation is negligible), regardless of the allelic frequencies (James 1971; Hill and Rasbash 1986).
Loss of alleles in small populations, as in those under conservation programs, is mainly driven by genetic drift. Moreover, the increase of inbreeding under random mating is also a function of population size. Inbreeding refers to the probability of identity by descent (IBD) in a locus. In simulation studies, like the present one, this probability can be calculated by counting if we assign different alleles to all individuals in the base population.
The magnitude of the effect of genetic drift under different management strategies is really dependent on the effective population size (Ne; Falconer and Mackay 1996), instead of on the census size. Usually, Ne is calculated through the increase of inbreeding in the population as Ne = 1/2ΔF, where ΔF is the rate of inbreeding. When management is based on genealogical information, ΔF soon reaches an asymptotic value (Falconer and Mackay 1996; Wang 1997). Therefore, the effective population size (Ne) has been often used as a measure of the long-term performance of the population regarding both diversity and inbreeding. However, when decisions are made only on the basis of marker information, Ne loses usefulness as it does not reach a constant value, but increases as generations go by (Toro et al. 1999). This effect is not observed when both sources of information (pedigree and molecular markers) are used.
There is a consensus on the optimal way to manage GD when the pedigree of the population is available (Ballou and Lacy 1995; Caballero and Toro 2000; Fernández et al. 2003). In this scenario, the best strategy is to optimize contributions of parents (i.e., number of offspring that each individual leaves to the next generation) by minimizing the global coancestry weighted by those contributions. Furthermore, under random mating, this strategy also implies the maximization of effective population size (Ne) (Caballero and Toro 2000, 2002). In a parallel way, when only molecular marker (rather than genealogical) information is available, the optimal strategy for maintaining GD is to minimize the global molecular coancestry, as defined below (Toro et al. 1999). When both genealogical and molecular information is available, it can be combined to calculate the coancestry conditional on markers (Toro et al. 1999; Wang 2001). In this way, markers can help to ascertain the global “realized” coancestry from the “expected” coancestry provided by the pedigree.
In ex situ conservation programs, space resources are limited. One possible procedure is to generate only the individuals that are going to be kept. Consequently, decisions on contributions have to be made on the basis of parents' information. Another possibility is to generate a large number of offspring that will exceed the maximum number that can be kept, and some of them have to be discarded. Notwithstanding, molecular information on the surplus offspring could be used together with parental information to help in breeding decisions. This is more likely to be done with highly prolific species. Under the last scenario, Toro et al. (1999) and Wang (2001) shown that the use of coancestry conditional on markers to decide the selected offspring to be kept as breeders could yield effective population sizes >40% larger than those obtained using only pedigree coancestry (Wang 2001). Both studies consider that all parents contributing to the next generation had the same number of offspring. However, differential contributions of parents have proven to be very efficient for managing the rate of inbreeding (Fernández and Toro 1999; Villanueva et al. 2004). Also, the studies of Toro et al. (1999) and Wang (2001) focused on the comparison between different management strategies for Ne and, therefore, they referred to a time horizon where those parameters had reached asymptotic values. They ignored the consequences of each strategy on the levels of GD and inbreeding at shorter time horizons, which are determinants of the adaptation ability of the population and of the inbreeding depression in fitness-related traits.
Nonrandom mating systems have proven to be efficient for increasing Ne (Caballero et al. 1996) and therefore for maintaining genetic variability and controlling inbreeding levels. In particular, minimum coancestry matings, which minimize the average pairwise coancestry between couples (Toro et al. 1988), have proven to be, in some cases, effective in reducing F-levels when only pedigree information is used in artificial selection (Sonesson and Meuwissen 2000, 2002) and conservation programs (Fernández and Caballero 2001; Sonesson and Meuwissen 2001). However, nonrandom mating systems have not been evaluated when molecular information is used to compute coancestries.
The objective of this study was to evaluate the efficiency of the use of molecular markers and pedigree information (separately or in combination) on the maintenance of genetic diversity and the control of inbreeding in conserved populations. Both sources of information were considered for optimizing contributions of parents and for optimizing matings between selected parents.
METHODS
Population and genetic models
Populations of constant census size N = 18 (Nm = 9 males and Nf = 9 females) or N = 27 (Nm = 9 and Nf = 18) were modeled through stochastic computer simulations.
The genome of individuals consisted of 1 or 20 chromosomes. Chromosome length was 1 M. Each chromosome carried 100 evenly spaced loci that were used to evaluate genetic diversity parameters. A random number of crossovers (Poisson distributed with mean one) were assumed in randomly chosen places without interference when obtaining gametes.
All individuals in the base population were assumed to be unrelated and not inbred. Therefore, all base population individuals carried two different alleles at each locus and GD and AD were at their maximum values (1 − 1/2N and 2N, respectively). In most scenarios management strategies started in the base population. However, in some simulations, five unmanaged generations (with random contributions and matings) were performed prior to the application of any management strategy. These simulations aimed to evaluate the effect of different amounts of diversity present in the population when the conservation program starts on the relative performance of the strategies investigated. They also represent more realistic scenarios as, in practice, relationships differ between individuals.
In addition to the 100 multiallelic loci, 1–100 evenly distributed markers were simulated per chromosome. Each marker position coincided with the position of one of the multiallelic loci. The number of alleles per marker ranged from 2 (modeling the typing of low polymorphic markers such as SNPs) to 10 (e.g., microsatellites). In generation zero (where the population starts to be managed), marker alleles were assigned at random with the same probability.
Management strategies
As a reference for comparison, unmanaged populations (random contributions and random mating; R) were simulated for each value of N.
For the rest of the cases, the general strategy for managing the population was to maximize the expected heterozygosity (GD). This was achieved by minimizing the global weighted coancestry, calculated as
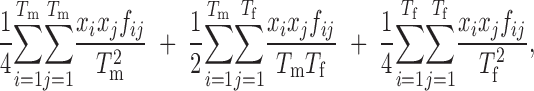 |
where xi is the contribution from individual i, fij is the coancestry between individuals i and j (computed in different ways as described below) and Tm and Tf are, respectively, the numbers of males and females evaluated. Several restrictions were imposed in the optimization: (i) only integer nonnegative solutions were allowed; (ii) the sum of all contributions equaled twice the total number of individuals evaluated (Tm + Tf); and (iii) half of the contributions arose from males and half from females. Optimal solutions for contributions were obtained via a simulated annealing algorithm (Kirkpatrick et al. 1983).
Management strategies were applied for 10 discrete generations (in addition to the 5 unmanaged generations in some cases). These strategies differed in the number and type of evaluated individuals, in the type of information used to compute global coancestries, and in the system of mating.
Evaluated individuals:
Two different scenarios were considered.
Parents genotyped:
In the first scenario, the decisions about the optimal individual contributions to the next generation were based on information on potential parents. Thus, the number of evaluated individuals (i.e., the number of individuals included in the optimization) was equal to the number of individuals kept in the population as breeders (Nm = Tm = 9 males and Nf = Tf = 9 or 18 females).
Offspring genotyped:
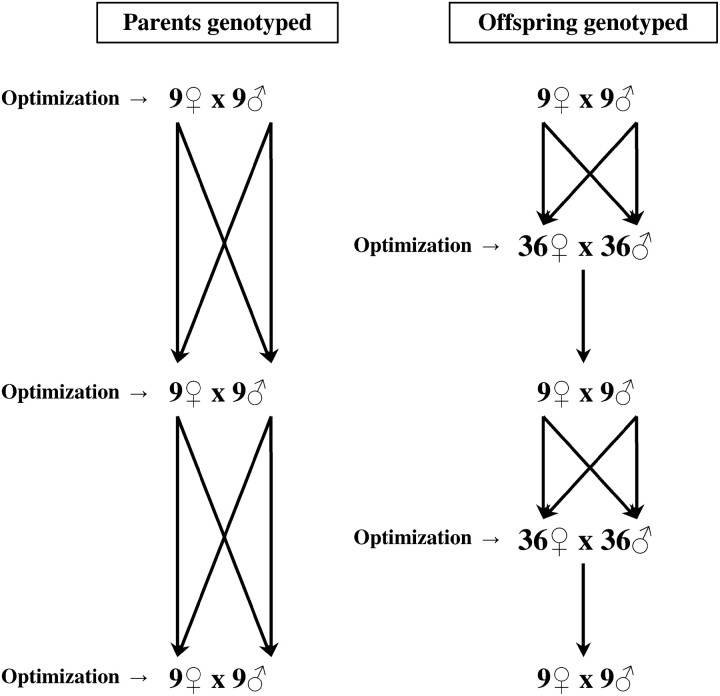
In the second scenario, available parents produced several offspring that were genotyped. Then, the individuals to keep as breeders for the next generation were decided on the basis of offspring information (and the rest of the offspring were discarded). The total number of evaluated individuals was 72 (36 of each sex) and therefore Tm = Tf = 36. This case corresponds to the situation where more individuals than needed are born. To make the results comparable, it should be noted that the number of individuals kept in the population (N, i.e., the number of selected/contributing individuals) was forced to be the same as in the first scenario. To achieve this, an additional restriction was imposed in the optimization, allowing a maximum number of 9 males and 9 or 18 females to contribute to the next generation (the rest of the evaluated individuals had zero contributions). The number of offspring per parent was not fixed, but was also optimized at the same time. Figure 1 shows a scheme of both scenarios for the case of Nm = Nf = 9).
Figure 1.—
Scheme of the two simulated scenarios depending on the number and type of the individuals evaluated (genotyped).
The efficiency of the second scenario was expected to be higher than that of the first scenario, as a larger number of evaluated individuals were available, and higher than that in the work by Toro et al. (1999) and Wang (2001) as selected individuals with lower mean coancestry produced more offspring to be evaluated than did those highly related with the rest of the population (it would be not very likely to select many offspring from that individual).
Information used for computing coancestry:
Different strategies were evaluated and named according to the type of coancestry used in the optimization.
Pedigree (fP):
Coancestries were calculated from the genealogy only, including unmanaged generations in the scenarios where they were simulated. This represented the expected IBD for the whole genome.
Molecular (fM):
Coancestries were calculated from marker information only. Molecular coancestry between two individuals is defined in a similar way to Malecot's definition but referring to identity by state (IBS), which is the probability that two alleles, taken at random from the same locus in two individuals, are equal. Values were averaged across marker loci.
Conditional on markers (fPM):
Coancestries were calculated by combining molecular and genealogical information using the method proposed by Pong-Wong et al. (2001). The IBD was estimated every 5 cM (i.e., at 20 positions in each chromosome), and it was averaged across positions. Preliminary simulations computing coancestry at 100 positions per chromosome produced the same results in the levels of genetic diversity (data not shown). Using coancestry computed only at 20 positions, however, reduced the computation time considerably.
Genomic (fG):
Coancestries were calculated from information on all positions in the genome. This situation corresponds to scenarios where the genotype for all loci of the genome is known and, therefore, it establishes the upper theoretical limit of efficiency for any strategy. As all individuals carried two different alleles in each locus in the base population, fG represented the real IBD.
Mating systems:
The performance of the different strategies was evaluated by (i) optimizing the contributions of parents to the next generation and mating the parents at random and (ii) combining the optimization of contributions with minimum coancestry matings. In the latter situation, the type of information (i.e., the type of coancestry) used in both optimizations (“selection” and mating) was the same. A particular case was simulated where selection decisions were based only on pedigree information but mating decisions were based on both pedigree and molecular information. The a priori advantage of this strategy (relative to the strategy using pedigree and marker information jointly in both selection and mating decisions) is the lower number of individuals to be genotyped for the markers. A simulated annealing algorithm (Kirkpatrick et al. 1983) was also used to optimize matings.
Parameters evaluated
The expected heterozygosity (GD), AD, and inbreeding level (F, the proportion of homozygous loci observed in the population) were calculated each generation for the breeding individuals using all loci, and they were averaged over 100 (for genomes of 1 chromosome) or 50 (for genomes of 20 chromosomes) replicates. The effective population size (Ne) was calculated as Ne = 1/2ΔF, where ΔF was the average rate of inbreeding, ΔF = (Ft+1 − Ft)/(1 − Ft), from generations t = 5 to t = 10. The latter was not calculated when the decision criteria were the molecular coancestry (fM) or the genomic coancestry (fG), because in these situations Ne does not reach a constant, asymptotic value, as was stated in the Introduction.
RESULTS
Random mating:
Table 1 shows the expected heterozygosity (GD, averaged over all nonmarker loci across the genome) at generation 10 and the effective population size (Ne) yielded by each management strategy for Nm = Nf = 9 and different combinations of number of chromosomes (c), number of markers typed per chromosome (m), and number of alleles per marker (a). Results presented correspond to the case where the starting population was constituted by unrelated and noninbred individuals. Scenarios with differential relationships between individuals at the beginning of the conservation program and those with larger census sizes (Nm = 9 and Nf = 18) produced very similar trends to those presented in Table 1 and are, therefore, not shown.
TABLE 1 .
Genome-wide expected heterozygosity (GD, in percentage) at generation 10 and effective population size (Ne)
| Parents genotyped |
Offspring genotyped |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
fM |
fPM |
fM |
fPM |
|||||||||||||||
| c | a | R | fP | fG | m = 1 | m = 5 | m = 10 | m = 1 | m = 5 | m = 10 | fG | m = 1 | m = 5 | m = 10 | m = 1 | m = 5 | m = 10 | |
| GD | 1 | 2 | 73.93 | 84.09 | 87.22 | 71.72 | 69.53 | 68.51 | 84.01 | 84.91 | 85.09 | 95.11 | 74.90 | 74.04 | 76.45 | 84.82 | 88.67 | 90.46 |
| 10 | 73.99 | 82.75 | 84.45 | 84.44 | 86.58 | 86.84 | 78.12 | 87.84 | 90.35 | 87.75 | 91.82 | 92.71 | ||||||
| 20 | 2 | 84.09 | 68.08 | 77.97 | 80.39 | 84.00 | 83.98 | 84.00 | 86.46 | 74.99 | 81.89 | 83.15 | 83.52 | 84.26 | 84.82 | |||
| 10 | 80.28 | 83.24 | 83.76 | 84.02 | 83.99 | 84.12 | 83.35 | 85.27 | 85.72 | 84.17 | 85.55 | 85.94 | ||||||
| Ne | 1 | 2 | 18.61 | 35.67 | 33.14 | 39.10 | 49.97 | 38.02 | 48.69 | 69.31 | ||||||||
| 10 | 41.66 | 45.14 | 56.49 | 45.01 | 103.31 | 181.83 | ||||||||||||
| 20 | 2 | 35.27 | 34.94 | 35.66 | 33.57 | 33.37 | 39.91 | |||||||||||
| 10 | 34.03 | 34.87 | 34.15 | 36.03 | 44.28 | 47.19 | ||||||||||||
Population with Nm = Nf = 9 under random mating is shown. Management strategies: R, random; fP, pedigree coancestry; fG, genomic coancestry; fM, molecular coancestry; and fPM, coancestry conditional on markers. c, number of chromosomes; a, number of alleles per marker; m, number of markers per chromosome. Standard errors range from 0.04 to 0.52 for GD and from 0.47 to 4.78 for Ne.
The upper limit of efficiency (measured as the level of GD maintained), provided by minimizing fG, was lower for large than for small genomes. Similarly, when coancestry was computed using both pedigree and molecular markers (i.e., fPM), lower levels of GD were observed for genomes of 20 chromosomes. However, rather paradoxically, the opposite trend was observed in some situations for strategies relying only on molecular information (i.e., fM; for example, a = 10, m = 1).
The efficiency in the short and medium term of using only pedigree information when optimizing contributions is clear from the values of GD maintained in the population after 10 generations (Table 1). In fact, the values of GD obtained with this strategy were 88–100% of the maximum attainable increase (i.e., obtained by minimizing fG). For large genomes (i.e., c = 20), the pedigree-based strategy achieved nearly the highest possible diversity (i.e., the same as fG), and consequently adding molecular information provided little or no extra benefit. Note that, when dealing with genealogies alone, no improvement is expected from using offspring information as pedigree relationships are equal for all individuals within the same family.
When only molecular information (fM) from parents was assumed to be available, the number of markers needed to reach the same levels of GD as with genealogical information only (i.e., fP) was very high. Differences between fM and fP were more evident with biallelic markers, but even with a = 10, 5–10 markers per Morgan were required for fM to give levels of GD similar to those obtained with fP (Table 1). The levels of GD obtained when using fM improved when the offspring was genotyped (i.e., when 72 individuals were evaluated), but still a considerable number of markers per morgan (≥5) were required for some schemes to outperform the pedigree-based method.
Unexpectedly, in some cases when only parents were genotyped, the levels of GD obtained through the exclusive use of markers were even lower than the levels achieved in unmanaged populations. Moreover, we found another counterintuitive behavior of markers when the number of these was scarce and/or their degree of polymorphism was low. In such situations, increases in the number of markers (or alleles per marker) led to lower levels of maintained genetic diversity (e.g., c = 1 and a = 2 with only parents genotyped). Figure 2 shows the levels of GD kept in a population with Nm = Nf = 9, after one or five generations of management, when different numbers of markers and alleles per marker are used to calculate molecular coancestry. It is clear that, for some combinations of m and a, increasing the number of markers or their degree of polymorphism was counterproductive, as the larger the number of markers (or the number of alleles per marker) used, the lower the expected heterozygosity maintained (even up to 10 markers in the case of biallelic ones). This performance was more evident for small genomes (Table 1) and in the short term (Figure 2), but the effect could last as long as 10 generations in extreme cases (Table 1).
Figure 2.—
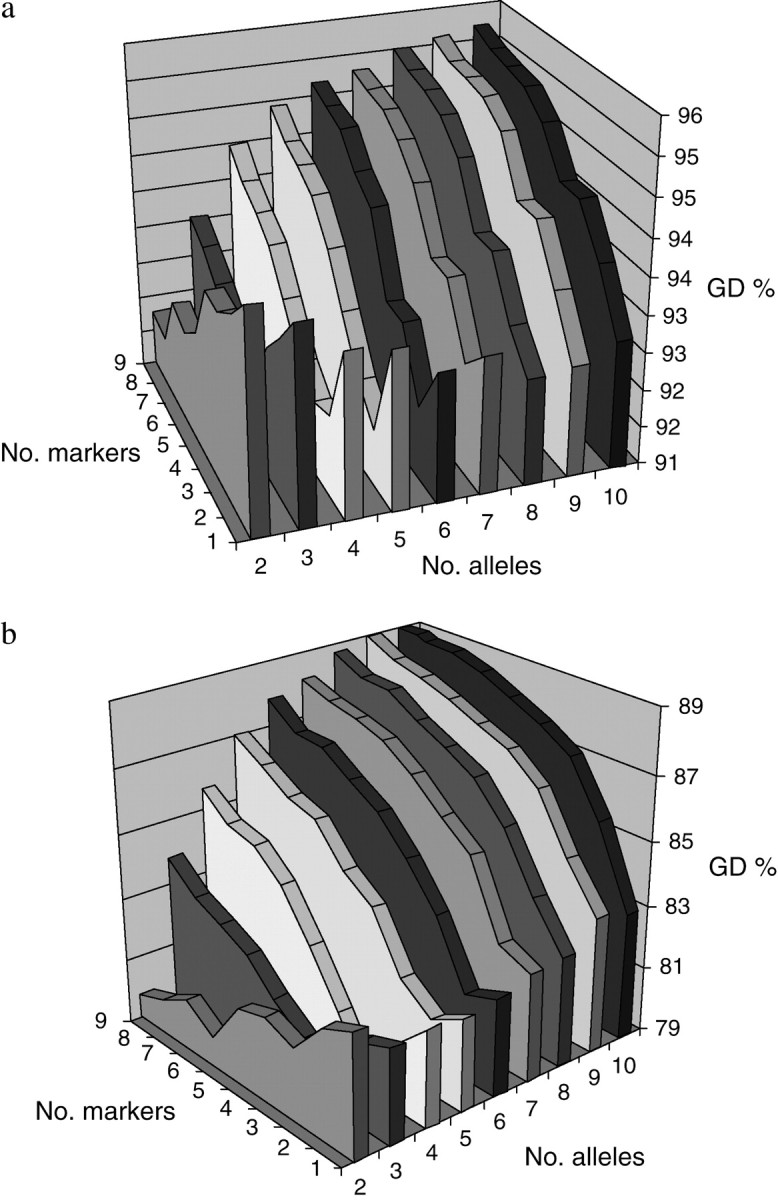
Genome-wide expected heterozygosity (GD %) maintained by minimizing molecular coancestry for different numbers of markers and alleles per marker. Only parents were genotyped. The genome length was 1 M, Nm = Nf = 9, and matings were at random. (a) After one generation of management. (b) after five generations of management.
When information on offspring's markers is available, the performance of the strategy using fM improved and GD levels after 10 generations were higher than those for unmanaged populations even with only one biallelic marker per chromosome.
As mentioned above, the levels of GD obtained by minimizing fP were close to the maximum expectations (i.e., by minimizing fG), leaving, thus, a narrow margin of improvement for molecular information. The inclusion of marker information into the management strategy, via the coancestry conditional on markers (i.e., fPM), hardly gave extra gains, if any, relative to using pedigree alone when the only available information is that from parents (Table 1). However, for small genomes and using offspring information, important increases in Ne were observed when molecular information was combined with pedigree information (relative to the Ne obtained by using fP). For large genomes (c = 20) the values obtained for Ne with fPM were not significantly different from those obtained with fP when decisions were made on parents' genotype or on offspring's genotype with little marker information (i.e., a = 2 and m = 1 or 5).
AD and inbreeding (F) showed a parallel behavior to that of GD (Table 2). Most of the increase in AD and most of the decrease in F relative to unmanaged populations were due to the use of genealogical information, and little improvement was observed when including molecular information, especially for large genomes. If the genome was small (c = 1) and offspring information was used, greater advantages were obtained via the minimization of fPM.
TABLE 2 .
Genome-wide allelic diversity (AD, in percentage) and inbreeding coefficients (F, in percentage) at generation 10
| Parents genotyped |
Offspring genotyped |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
fM |
fPM |
fM |
fPM |
|||||||||||||||
| c | a | R | fP | fG | m = 1 | m = 5 | m = 10 | m = 1 | m = 5 | m = 10 | fG | m = 1 | m = 5 | m = 10 | m = 1 | m = 5 | m = 10 | |
| AD | 1 | 2 | 17.07 | 26.15 | 28.67 | 15.61 | 13.74 | 12.6 | 26.08 | 26.78 | 27.01 | 57.37 | 17.38 | 16.39 | 17.35 | 27.93 | 35.85 | 41.06 |
| 10 | 16.17 | 21.98 | 23.98 | 21.61 | 28.38 | 28.32 | 19.14 | 30.42 | 37.09 | 34.80 | 46.86 | 49.55 | ||||||
| 20 | 2 | 25.89 | 13.35 | 19.12 | 21.42 | 26.07 | 26.09 | 26.07 | 28.53 | 17.24 | 22.95 | 24.32 | 25.35 | 26.22 | 26.82 | |||
| 10 | 21.34 | 24.70 | 25.35 | 26.06 | 25.99 | 25.99 | 24.47 | 26.88 | 27.48 | 26.13 | 27.62 | 28.02 | ||||||
| F | 1 | 2 | 21.86 | 11.88 | 7.38 | 23.57 | 24.77 | 25.00 | 12.01 | 11.08 | 9.66 | 2.17 | 20.56 | 21.53 | 18.53 | 11.21 | 7.85 | 6.09 |
| 10 | 19.63 | 12.02 | 9.59 | 10.71 | 9.53 | 8.27 | 15.86 | 7.74 | 5.36 | 8.26 | 4.83 | 3.51 | ||||||
| 20 | 2 | 11.89 | 26.52 | 17.10 | 14.87 | 11.97 | 12.08 | 11.95 | 9.20 | 20.04 | 13.70 | 12.39 | 12.53 | 11.86 | 10.89 | |||
| 10 | 14.84 | 12.42 | 11.80 | 12.07 | 11.77 | 11.85 | 12.29 | 10.35 | 9.93 | 11.47 | 10.02 | 9.62 | ||||||
Population with Nm = Nf = 9 under random mating is shown. Management criteri: R, random; fP, pedigree coancestry; fG, genomic coancestry; fM, molecular coancestry; and fPM, coancestry conditional on markers. c, number of chromosomes; a, number of alleles per marker; M, number of markers per chromosome. Standard errors range from 0.04 to 0.42 for allelic diversity and from 0.05 to 0.25 for inbreeding.
Optimized mating:
Table 3 shows the inbreeding coefficient at generation 10 when contributions and matings were both optimized. Two situations are presented: one (more theoretical, to illustrate upper limits of performance) with 100 markers in just 1 chromosome and another one (more practical) with 20 chromosomes and 5 markers on each. The inbreeding obtained in unmanaged populations (R) is also shown for comparison. The levels of GD obtained when the mating scheme was also managed are not shown because, as expected, they were the same as those found with random mating. It can be proven that, once contributions have been decided, the global coancestry in the next generation is independent of the mating design.
TABLE 3 .
Inbreeding coefficient (F, in percentage) at generation 10 under minimum coancestry mating
| Parents genotyped |
Offspring genotyped |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| c | m | a | R | fP, fPa | fM, fM | fPM, fPM | fP, fPM | fM, fM | fPM, fPM | fP, fPM |
| 1 | 100 | 2 | 26.05 | 9.83 | 5.49 | 2.84 | 5.81 | 3.06 | 0.70 | 6.55 |
| 10 | 3.08 | 2.85 | 6.33 | 0.48 | 0.63 | 6.54 | ||||
| 20 | 5 | 2 | 14.96 | 10.33 | 9.82 | 11.64 | 8.98 | 10.06 | ||
| 10 | 10.07 | 9.45 | 9.52 | 7.67 | 7.43 | 9.55 | ||||
Population with Nm = Nf = 9 under random mating is shown. Management criteria: R, random; fP, pedigree coancestry; fM, molecular coancestry; and fPM, coancestry conditional on markers. c, number of chromosomes; m, number of markers per chromosome; a, number of alleles per marker. Standard errors range from 0.06 to 0.30.
The first element in column headings is the criterion used to determine contributions and the second element is the one used to arranged matings.
The good performance of the pedigree-based strategy and the limited ability of marker-based strategies to improve the former were again clear in the more realistic situation (large genomes and few markers genotyped). In general, the lowest inbreeding was achieved when both pedigree and molecular information were used to decide both contributions and matings, and offspring genotype was available. The main finding when comparing random vs. nonrandom mating (Table 2 vs. Table 3) is that the effect of avoiding mating between relatives is small in practical scenarios. In fact, the reduction in levels of F at generation 10 is only ∼2–3%.
DISCUSSION
This article has investigated the efficiency of molecular markers and pedigree records, separately or in combination, to assist in the management of conserved populations. The results have shown that genealogical information proves to be a very powerful tool for maintaining genetic diversity and low levels of inbreeding via the minimization of global pedigree coancestry, at least for the period of time considered (10 generations). In fact, levels of expected heterozygosity yields by such a strategy were 88–100% of the maximum possible levels obtained if all loci in the genome were genotyped (i.e., the levels obtained by minimizing fG). The minimization of fP was equally efficient for maintaining allelic diversity and this agrees with previous results of Fernández et al. (2004) showing that strategies directed to maintaining GD are also efficient in maintaining AD.
On the other hand, the exclusive use of marker information was of limited value for the maintenance of genetic diversity. The amount and degree of polymorphism of markers to be used to compute molecular coancestry had to be very high to mimic the performance of the strategy relying on pedigree coancestry in the short-term and still moderate in the long-term horizon, especially for large genomes. Moreover, we found an unexpected behavior of markers. When the “quality” of molecular information was low (i.e., the number of markers and/or the number of alleles per marker was low), increasing the amount of information could lead to decreased levels of genetic diversity in the population. The reason for this performance could be the following. In generation zero, marker alleles were assigned at random and, thus, there was no direct relationship between the real (i.e., genomic) coancestry and the molecular coancestry. However, just by chance, some individuals could be less/more marker related with the rest of the population (i.e., lower/higher average fM) and they would be erroneously favored/penalized. This was more likely with an intermediate number of alleles than with low (high) polymorphic markers. Therefore, going from very low to intermediate polymorphic markers led to more wrong decisions and, thus, to lower levels of genetic diversity maintained. As the number of generations increased, real relationships between molecular coancestry and coancestry at positions near the markers were established and, therefore, decisions based on markers became more effective. The greater the number of alleles, the sooner these relationships were generated. In nonequilibrium situations, the performance of molecular-based methods would depend on the particular degree of disequilibrium and the way it is generated. A similar argument can be invoked to explain the observation of decreased genetic diversity maintained, in some situations, when increasing the number of markers for a given number of alleles per marker. In this case, the number of different haplotypes is the key parameter, playing the role of the number of alleles in the previous explanation.
The other paradoxical result related to the minimization of fM (i.e., better performance for large genomes in some situations) is also a consequence of markers being in linkage equilibrium with other loci in the base population. Although diversity in the markers follows the logical trend, behavior in the rest of the genome depends on how fast disequilibrium is generated, which is a function of the number of markers and their degree of polymorphism.
Finally, another fact should be pointed out relative to the use of molecular information alone. When minimization of fM (or fG) is the chosen strategy, Ne is not useful as a measure of efficiency, because it does not reach an asymptotic value but increases with time. For example, with Ns = Nd = 9, 72 genotyped offspring, c = 1, m = 5, and a = 10, estimates are equal to 58.36, 78.31, and 92.67 if we averaged ΔF to calculate Ne for generations 5–10, 10–15, or 15–20, respectively. This happens as alleles become fixed in some positions and the number of markers to be jointly optimized decreases. Therefore, we cannot make predictions on the future performance of the population base on a particular value of Ne.
When only parents are genotyped, the inclusion of molecular information together with genealogical data (fPM) in the management of contributions showed limited value for improving the levels of diversity (either GD or AD) and the levels of inbreeding in the first 10 generations (Tables 1 and 2). With small genomes (c = 1) we obtained greater Ne when minimizing fPM was the chosen strategy, implying some benefits could be found in the long-term horizon (Table 1). However, this advantage disappears for larger, and more realistic, genomes (c = 20).
If the genotype for a number of offspring was available, there was a greater improvement in Ne by minimizing fPM relative to the values reached with fP, especially for small genomes (Table 1). These values, for combinations with a similar number of chromosomes and number of markers per chromosome, were in the range of those found by Toro et al. (1999) and Wang (2001). Therefore, results presented in this article are in agreement with those from previous studies, regarding long-term performance of strategies, although no test of significance can be made due to the lack of standard errors for Ne of the Toro et al. (1999) and Wang (2001) data.
When comparisons between management methods are made on the basis of levels of AD maintained in the population, conclusions are similar to those observed for GD (i.e., very good performance of pedigree-based strategies and little improvement from the inclusion of marker information, fPM, except for small genomes when offspring genotype is available). These results are in agreement with the work by Fernández et al. (2004), which showed that strategies directed to the maintenance of GD (minimization of weighted global coancestry) are also effective in the maintenance of AD.
Marker information was also of relatively low value for optimizing matings among selected individuals to decrease inbreeding levels, at least for realistic scenarios. Previous studies (Fernández and Caballero 2001) have shown that, when using exclusively pedigree information for conservation purposes, the effect of the mating design on the inbreeding levels is minimal, once contributions have been optimized. In selection schemes (Sonesson and Meuwissen 2000, 2002) improvements can be larger, but it depends on the structure of the population, the selection pressure, and the restriction on inbreeding imposed. In the present study, when marker information was used separately or in combination with pedigree information, little improvements in F levels were obtained by managing the matings, which was not surprising since levels of inbreeding at generation 10 were very similar when optimizing contributions using fP or fPM and matings were at random. From a practical point of view, a comparison of interest is that between strategies that use fPM to optimize both contributions and matings or to optimize only matings. With the former, slightly lower levels of inbreeding were generally obtained, but the costs involved in the program were higher since a larger number of individuals needed to be genotyped.
As a general conclusion, managers of a conservation program should be advised to critically evaluate the convenience of including molecular information into the management design, because the cost of molecular techniques is still high and markers will not be very abundant except for domestic species. The results from this study suggest that, for lowly prolific species and thus basing decisions only on breeders' data, it would be more efficient to use genealogical information in the management, if such information is available. Obviously, if we lack pedigree, it is better to use molecular information to manage the population than leave it unmanaged, except for very unrealistic scenarios. When more offspring than needed can be generated and genotyped, the advantage of using molecular information can be larger, especially when combined with genealogical data on a species with small genomes. However, in realistic situations (i.e., species with large genomes and a limited number of available markers), probably it would be more efficient to allocate the available resources to the enlargement of the population or to a better control of pedigree and restrict the use of markers to more specific tasks such as solving pedigree uncertainties. Notwithstanding, these considerations should be studied for each particular case.
Acknowledgments
This work was supported by grants BMC2003-03022 (Ministerio de Ciencia y Tecnología and Fondos Feder) and RZ03-010 (Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria). Jesús Fernández was supported by a Programa Ramón y Cajal contract. Beatriz Villanueva acknowledges financial support from the Secretaría de Estado de Educación y Universidades (Ministerio de Educación, Cultura y Deporte, Spain) and from the Scottish Executive Environment and Rural Affairs Department (United Kingdom). Ricardo Pong-Wong acknowledges financial support from The Biotechnogy and Biological Research Council.
References
- Ballou, J. D., and R. C. Lacy, 1995 Identifying genetically important individuals for management of genetic variation in pedigreed populations, pp 76–111 in Population Management for Survival and Recovery, edited by J. D. Ballou, M. Gilpin and T. J. Foose. Columbia University Press, New York.
- Caballero, A., and M. A. Toro, 2000. Interrelations between effective population size and other pedigree tools for the management of conserved populations. Genet. Res. 75 331–343. [DOI] [PubMed] [Google Scholar]
- Caballero, A., and M. A. Toro, 2002. Analysis of genetic diversity for the management of conserved subdivided populations. Conserv. Genet. 3 289–299. [Google Scholar]
- Caballero, A., E. Santiago and M. A. Toro, 1996. Systems of mating to reduce inbreeding in selected populations. Anim. Sci. 62 431–442. [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996 An Introduction to Quantitative Genetics, Ed. 4. Longman, Harlow, UK.
- Fernández, J., and A. Caballero, 2001. A comparison of management strategies for conservation with regard to population fitness. Conserv. Genet. 2 121–131. [Google Scholar]
- Fernández, J., and M. A. Toro, 1999. The use of mathematical programming to control inbreeding in selection schemes. J. Anim. Breed. Genet. 116 447–466. [Google Scholar]
- Fernández, J., M. A. Toro and A. Caballero, 2003. Fixed contributions designs vs. minization of global coancestry to control inbreeding in small populations. Genetics 165 885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, J., M. A. Toro and A. Caballero, 2004. Managing individuals' contributions to maximize the allelic diversity maintained in small, conserved populations. Conserv. Biol. 18 1358–1367. [Google Scholar]
- Frankham, R., J. D. Ballou and D. A. Briscoe, 2003 Introduction to Conservation Genetics. Cambridge University Press, Cambridge, UK.
- Hill, W. G., and J. Rasbash, 1986. Models of long term artificial selection in finite populations. Genet. Res. 48 41–50. [DOI] [PubMed] [Google Scholar]
- James, J. W., 1971. The founder effect and response to artificial selection. Genet. Res. 12 249–266. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, S., C. D. Gelatt and M. P. Vecchi, 1983. Optimization by simulated annealing. Science 220 671–680. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pong-Wong, R., A. W. George, J. A. Woolliams and C. S. Haley, 2001. A simple and rapid method for calculating identity-by-descent matrices using multiple markers. Genet. Sel. Evol. 33 453–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonesson, A. K., and T. H. E. Meuwissen, 2000. Mating schemes for optimum contribution selection with constrained rates of inbreeding. Genet. Sel. Evol. 32 231–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonesson, A. K., and T. H. E. Meuwissen, 2001. Minimization of rate of inbreeding for small populations with overlapping generations. Genet. Res. 77 285–292. [DOI] [PubMed] [Google Scholar]
- Sonesson, A. K., and T. H. E. Meuwissen, 2002. Non-random mating for selection with restricted rates of inbreeding and overlapping generations. Genet. Sel. Evol. 34 23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro, M. A., B. Nieto and C. Salgado, 1988. A note on minimization of inbreeding in small-scale selection programmes. Livest. Prod. Sci. 20 317–323. [Google Scholar]
- Toro, M. A., L. Silió, M. C. Rodríguez, J. Rodrigáñez and J. Fernández, 1999. Optimal use of genetic markers in conservation programmes. Genet. Sel. Evol. 31 255–261. [Google Scholar]
- Villanueva, B., R. Pong-Wong, J. A. Woolliams and S. Avendaño, 2004 Managing genetic resources in commercial breeding populations, pp. 113–131 in Farm Animal Genetics Resources, edited by G. Simm, B. Villanueva, K. D. Sinclair and S. Townsend. BSAS Occasional Pub. 30, Nottingham University Press, Nottingham, UK.
- Wang, J., 1997. More efficient breeding systems for controlling inbreeding and effective size in animal populations. Heredity 79 591–599. [DOI] [PubMed] [Google Scholar]
- Wang, J., 2001. Optimal marker-assisted selection to increase the effective size of small populations. Genetics 157 867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]