Abstract
Mushrooms, such as Coprinus cinereus, possess large families of pheromones and G-protein-coupled receptors that are sequestered at the B mating-type locus and whose function is to confer vast numbers of different mating types. This ability results from complex patterns of cognate and noncognate pheromone/receptor pairings, which potentially offer a unique insight into the molecular interaction between receptor and ligand. In this study we have identified many more members of these families by molecular analysis of strains collected worldwide. There are three groups of genes at each B locus. We have identified two alleles of group 1, five alleles of group 2, and seven alleles of group 3, encoding in total 14 different receptors and 29 different pheromones. The specificity of many newly identified alleles was determined by transformation analysis. One striking finding was that receptors fall into groups based on sequence homology but these do not correspond to the groups defined by position, indicating that complex evolutionary processes gave rise to the B loci. While additional allelic versions may occur in nature, the number of B specificities possible by combination of the alleles that we describe is 70, close to previous estimates based on population analysis.
THE role of pheromone signaling in fungal mating has largely been elucidated from studies of the budding yeast Saccharomyces cerevisiae. This fungus has just two mating types and pheromones are secreted to act as chemoattractants for identifying compatible mating partners. Binding of a pheromone to an appropriate receptor on the cell surface triggers an intracellular G-protein-linked MAP kinase cascade that results in changes in growth direction to permit cell fusion, which is then followed immediately by nuclear fusion (reviewed by Kurjan 1993). Once cells are diploid, pheromone signaling ceases.
Pheromone signaling also plays an essential role in mating in basidiomycete fungi (reviewed by Casselton and Olesnicky 1998), but in mushroom species such as Coprinus cinereus, unlike S. cerevisiae, there is no evidence that pheromones have a role in mate attraction (Olesnicky et al. 1999). Cell fusion is mating type independent, and pheromone signaling is activated only after cells with compatible mating types fuse. The recognition of a compatible mating depends on acquisition of a complement of genes that encode pheromones that activate receptors in the same cellular compartment. For this reason, the genes that encode the pheromones and receptors were first identified as mating-type determinants and map to what has been designated the B mating-type locus. A second mating-type locus, A, encodes the subunits of a transcription factor belonging to the homeodomain family (Kües et al. 1994a) that is necessary, together with pheromone signaling, to promote the initial stages of sexual development. Compatible A genes encode versions of the proteins that can heterodimerize following cell fusion, an interaction that is analogous to that between the MATa1 and MATα2 mating-type proteins in mated cells of S. cerevisiae (reviewed by Johnson 1995).
In contrast to S. cerevisiae, compatible cell fusion in C. cinereus is not followed immediately by nuclear fusion, but by an extended vegetative phase in which the nuclei from each mate remain paired in each cell, a phase known as the dikaryophase or dikaryon. The B mating-type genes have long been known to regulate critical steps in the initiation and maintenance of the dikaryophase (Swiezynski and Day 1960), steps that we now recognize as being dependent on pheromone signaling. Initially, compatible B genes promote nuclear exchange and migration following compatible cell fusion; subsequently, they act together with the A genes to regulate a complex tip cell division, which involves the formation of a structure known as the clamp connection.
The chance of random cell fusion resulting in mating is enhanced by the fact that the mating-type genes of C. cinereus are multiallelic and allelic variation is sufficient to generate some 12,000 different mating specificities (Raper 1966), most of which would be cross-compatible. There are an estimated 79 versions of the B locus, the subject of this report. We have shown previously that each B locus is complex and contains a tandem array of three groups of paralogous genes (O'Shea et al. 1998; Halsall et al. 2000). Each group of genes comprises one or more pheromone genes and a single receptor gene. Variation in only one group of alleles between two mating partners generates different B mating specificities. Thus the large numbers of different versions of the B locus derive from different allelic combinations of the three groups of genes.
C. cinereus pheromone receptors are traditionally classified under the rhodopsin-like superfamily of seven-transmembrane domain G-protein coupled-receptors (GPCRs). Fungal GPCRs constitute the class D subfamily, and C. cinereus GPCRs specifically belong to the S. cerevisiae Ste3p-like receptor subgroup (http://www.gpcr.org/7tm/). The lipopeptide pheromones belong to the same family as the S. cerevisiae a-factor, which are distinguished by having precursors with a C-terminal CaaX motif, where C is cysteine, a is aliphatic, and X is one of several amino acids (Caldwell et al. 1995). Post-translational processing includes farnesylation of the cysteine residue, removal of the three terminal amino acids, carboxymethylation of the resulting carboxy-terminal cysteine, and removal of the amino-terminal precursor region (Chen et al. 1997).
Although GPCRs have evolved different specificities to respond to a variety of ligands and have very different primary structure, they are believed to have similar tertiary structure. In mushroom species, like C. cinereus, evolution has created a family of functionally redundant receptors and corresponding ligands but sequence variation permits them to display highly specific interactions. A single receptor may be activated by several different pheromones and each pheromone may activate several different receptors. However, previous data indicate that only pheromones and receptors encoded by genes within the same group can activate each other (Halsall et al. 2000). At present we do not understand how the specificity of receptor-ligand recognition is achieved but by analyzing the diversity of naturally occurring worldwide strains of C. cinereus this study has aimed to (1) isolate sufficient pheromone and pheromone receptor genes to account for the predicted numbers of B specificities; (2) identify putative key residues involved in a compatible interaction; and (3) understand the evolutionary process that has generated such a complex redundant family of proteins.
MATERIALS AND METHODS
C. cinereus strains and growth conditions:
We have continued to use the name C. cinereus in this report since this is the name by which this model species is best known, but recent phylogeny analysis has led to the new name Coprinopsis cinerea (Redhead et al. 2001). Genomic DNAs were isolated from the following strains: 68 (A2B1), LCO12 (A2B3 trp-3), TC4 (A5B5), H5 (A5B6), S337 (A7B8), D5-12 (A12 B12), ZBW601 (A40B40), JV6 (A42B42), OK130 (A43B43 ade-8), ScotF2 (A44B44), ScotF1 (A45B45), ScotE11 (A47B47), and KF2#1 (A91B92). Host strains for transformation were: 218 (A3B1 trp-1.1:1.6), LCO12 (A2B3 trp-3), HT8 (A5B5 trp-1.2), RS74 (A5B6 trp-1.1:1.6), LN118 (A42 B42 trp-1.1:1.6), AT8 (A43 B43 trp-3 ade-8), and NL1 (A44B44 trp-1.1:1.6). Strains used in mating assays were: 68 (A2B1), PR94226 (A6B3 ade-5 cho-1), TC10 (A6B5), J6.5-5 (A43B42), J6.5-4 (A42B43), JL58 (A6B42), and NL1R (A43B44). These strains have a worldwide origin: B1, B3, B5, B6, B44, B45, and B47 come from United Kingdom collections; B8, B12, B43, and B92 come from Japanese collections; B40 is from Czechoslovakia; and B42 is from Java. All cultures were grown at 37°. Cultures for genomic DNA extraction were grown in liquid yeast, malt, glucose medium (Rao and Niederpruem 1969) in petri dishes for 5 days, and DNA was isolated by the method of Dellaporta et al. (1983). For general culture and mating tests the complete medium of Lewis (1961) was used, supplemented with 100 mg/liter l-tryptophan. Transformation was based on the method of Casselton and de la Fuente (1989). Plasmids or PCR products containing pheromone genes were cotransformed with either plasmid pCc1001 containing the C. cinereus trp-1 gene (Binninger et al. 1987) or plasmids pDB3 or pDB6 containing the C. cinereus trp-3 gene (Burrows 1991). trp+ transformants were isolated to minimal medium (Shahriari and Casselton 1974). Receptor activation was assessed by the method of O'Shea et al. (1998). This involved crossing a minimum of 25 transformants to a tester strain having a different A mating specificity but the same B mating specificity as the host strain. If the introduced pheromone gene activated the pheromone response pathway in the host, this resulted in a compatible mating interaction with the formation of the characteristic dikaryotic mycelium.
Isolation of new B alleles:
Cosmid libraries constructed from genomic DNA of strains OK130 (A43B43 ade-8, May et al. 1991) and E117.9 (A6m-1B5 ade-8, Kües et al. 1994b) yielded the complete set of genes comprising the B43 and B5 specificities, respectively. The libraries were screened for clones hybridizing to sequences flanking the B locus either in our own laboratory (B5) or in that of M. E. Zolan (B43). To amplify long fragments of DNA containing unknown alleles flanked by known sequences, long range (LR)-PCR (Herculase polymerase, Stratagene, La Jolla, CA) was used according to manufacturer's instructions. The amplified PCR products were purified by agarose gel electrophoresis (QIAquick gel extraction kit, QIAGEN, Chatsworth, CA) and cloned into pGEM-T Easy vectors (Promega, Madison, WI). DNA sequencing was carried out on an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) by the Department of Biochemistry, University of Oxford. Plasmid subclones containing small fragments of DNA were first end sequenced with universal primers and completed by primer walking. Long fragments of isolated and subcloned DNA of B43 were sequenced using the transposon insertion-based method GPS-LS linker scanning system (New England BioLabs, Hitchin, Herts, UK).
Southern blot analyses:
These were carried out using standard procedures (Sambrook et al. 1989) Probes derived from B3, B6, and B42 were described by Halsall et al. (2000). Additional probes were derived from B1 (rcb2), B44 (rcb2), B43 (rcb2 and rcb3), B45 (rcb3), and B47 (rcb3) using primers listed in Table 1 to amplify the appropriate sequences.
TABLE 1 .
List of oligonucleotide primers used for PCR amplification
| Name | Sequence (5′–3′) | Application |
|---|---|---|
| PB31 | GGCAGGTCTAAAGGTAGCCACG | LR of group 3 (from msf 1.2) |
| PMR1 | AAAAGCCCTTGGTGGTGAAGCC | LR of B44, B1 group 2 (from B6 phb1.2R) |
| PMR2 | ACACCAAACCTGAGCGACAATC | LR of B44, B1 group 2 (from B3 Rcb3F) |
| PMR3 | CTCCATTCCGATATCACGTTCT | B1-RCB2-end-F |
| PMR4 | CAACAGAATCGTTCAACAAAACTC | B44-RCB2-end-F |
| PMR5 | GTATGGGCGCCTTACGTTT | phb3.23 primer 1 |
| PMR6 | AGACAAAGGCGATCGGATG | phb3.23 primer 2 |
| PMR7 | TTTGGACCATTGTTCGCTTT | phb3.13 primer 1 |
| PMR8 | CACAGAAGGGACTACGCACA | phb3.13primer 2 |
| PMR9 | TACCCTAACGTCCACGATGA | rcb33primer 1 |
| PMR10 | AAGGGGTCGTGGGCTAGTAT | rcb33primer 2 |
| PMR11 | GGCGAGTTGCTTATCTGTGG | phb13primer 1 |
| PMR12 | CTCTGTCATTCGCCTTGCAC | phb13primer 2 |
| PMR13 | CTCAAGAACATGGCGTCGAT | rcb13primer 1 |
| PMR14 | GCTGAAGTCGTCAAGACGTG | rcb13primer 2 |
| PMR15 | TAAGGAGTCAGCCCGAAGAA | phb3.242primer 1 |
| PMR16 | TTAAACGCTTTCCACAATGG | phb3.242primer 2 |
| PMR17 | TCAGCACAAGAGCAGGAAGA | phb3.142primer 1 |
| PMR18 | CGAATTCAGACGACGCTCAT | phb3.142primer 2 |
| PMR19 | ACCCTCAAAACCTCCAGATG | rcb342primer 1 |
| PMR20 | GTGAGCCAGAAACCAGAGGA | rcb342primer 2 |
| PMR21 | GCACAGACCGACCAGTAAGC | phb2.242primer 1 |
| PMR22 | CGGAACAACCTAAAGACGGTA | phb2.242primer 2 |
| PMR23 | TTGGAAGGGCACAAAAGCTA | phb2.142primer 1 |
| PMR24 | TAGCTGCAAAGGGATTGAGC | phb2.142primer 2 |
| PMR25 | TACCCTCAATGGACACGATG | rcb242primer 1 |
| PMR26 | AGCTAGCTGTGACGGTCGTT | rcb242primer 2 |
| PMR27 | AAGCGATAACGCACCAGAAG | phb3.26primer 1 |
| PMR28 | CAGTCGACAATCGCTACGAA | phb3.26primer 2 |
| PMR29 | GCATACGTACGGCAAGGAGT | phb3.16primer 1 |
| PMR30 | CGATTTTGACCACACATCCA | phb3.16primer 2 |
| PMR31 | TCTTCTCCCTCCTCACCTTG | rcb36primer 1 |
| PMR32 | GTGAGCCAGAAACCAGAGGA | rcb36primer 2 |
| PMR33 | TGCCACTCGAAGAGAGACAA | phb2.26primer 1 |
| PMR34 | ATGGCCGGATGTATAAAACG | phb2.26primer 2 |
| PMR35 | CTTCATGTCGAAGTTGTTTCG | phb2.16primer 1 |
| PMR36 | CGCGTACGGTCCTAATCG | phb2.16primer 2 |
| PMR37 | GACGCTTGGGGCGGACGAT | rcb26primer 1 |
| PMR38 | AAGCTTAGTAAGAGGACATGAGTCC | rcb26primer 2 |
| PMR39 | CTCGAGTTCCTTGGGTTGGT | phb1.26primer 1 |
| PMR40 | AAGGGCTCGGATAGAGCTTC | phb1.26primer 2 |
| PMR41 | CCGTTCCAAGTTTTGACGTT | phb1.16primer 1 |
| PMR42 | GCGAACTCACTGTTGCTTCA | phb1.16primer 2 |
| PMR43 | CCCCGACGGCCTTGTACTGTAGC | rcb16primer 1 |
| PMR44 | CTCGCTCTGCTCCCGGACC | rcb16primer 2 |
| PMR45 | GTTGAGATCAAGGCGAACGAT | phb2.244primer 1 |
| PMR46 | CCGCAATTCTCGGTATAAAGC | phb2.244primer 2 |
| PMR47 | GGTTTATGTTGAGGCGGACTAC | phb2.144primer 1 |
| PMR48 | TACTGACTACAATGCGGACTCTG | phb2.144primer 2 |
| PMR49 | ATCTTAAAACTGTCATCTGCCACA | rcb244primer 1 |
| PMR50 | CAGGGCAAATGAGAAAGATAGAAG | rcb244primer 2 |
| PMR51 | GTTTCCACGAAACACGCAAAC | phb2.21primer 1 |
| PMR52 | AAGTCTGTCAAGCAATGTTAGCC | phb2.21primer 2 |
| PMR53 | GCAGATGGCAGGATCTTTGT | phb2.11primer 1 |
| PMR54 | AGGGGTTAGACGATCCAGGTAT | phb2.11primer 2 |
| PMR55 | TAAAGACCTGATTCTGCTTCAAGG | rcb21primer 1 |
| PMR56 | AGGGCAAATGAGAAAGAAAGAAGT | rcb21primer 2 |
| PMR57 | CACAGTCTTAAGCATTCTCAGTCAA | phb3.343primer 1 |
| PMR58 | ATTATCGAGGTTTGCTTTGCTCT | phb3.343primer 2 |
| PMR59 | ATTGATAATTAAGGCGACCTTCTG | phb3.243primer 1 |
| PMR60 | ACGGACTCTAAGATCGATGTTCTC | phb3.243primer 2 |
| PMR61 | TGGTTTGCATACAAAGGTATTGTC | phb3.143primer 1 |
| PMR62 | TAACATATGGAGATGGTTGGGTAG | phb3.143primer 2 |
| PMR63 | ACATCTTCTCCTCTCCTCCTTTTC | rcb343primer 1 |
| PMR64 | GCTGGTACTAAGTTAATGTCATCGAG | rcb343primer 2 |
| PMR65 | CAAATTGCTGCATTGAATAGAGAG | phb2.343primer 1 |
| PMR66 | GAAGCTCTTCGTCATAATCCTGTT | phb2.343primer 2 |
| PMR67 | TGAGCTCAGTGAGTGTTGAAAGA | phb2.243primer 1 |
| PMR68 | ATTCGAGTCTAAGGAAAGGAGTGA | phb2.243primer 2 |
| PMR69 | GGATCCTGTTAACCGACGTTAT | phb2.143primer 1 |
| PMR70 | GATGACTTGCGTCTGAGTGAACTA | phb2.143primer 2 |
| PMR71 | ATGCTAAATATGCTCAAATACTGTGC | rcb243primer 1 |
| PMR72 | AAGTCATGAAAAGATCGTGTGTAAAG | rcb243primer 2 |
| PMR73 | TAATCCGAGCGATAAAACACAGTA | phb35primer 1 |
| PMR74 | AGTTGGGAGTAAGATGGCTTACAA | phb35primer 2 |
| PMR75 | GACCTCTCTGCTTTTCACGTTATAC | rcb35 primer 1 |
| PMR76 | ACTGTCCAGAGAAAATTGTCAGACTT | rcb35primer 2 |
| PMR77 | ATCTCTCACTCGTCCACCAAAC | mfs1.1 primer 1 |
| PMR78 | GGTCCTACGTCCATCCTAATGAC | mfs1.1 primer 2 |
| PMR79 | GCTGGTAAAACGATAACACGATT | mfs1.2 primer 1 |
| PMR80 | TAAGACTGTATGGGTCCACAACC | mfs1.2 primer 2 |
Plasmid and PCR strategies:
Routine cloning was in pBluescript II, pBC SK+, Litmus 28, and plasmid amplification was in Escherichia coli strain DH5α. To amplify pheromone and receptor genes for transformation, we used either pfu polymerase or the TaqPlus Precision PCR system (Stratagene). Oligonucleotides were synthesized to order by GIBCO (Gaithersburg, MD), Life Technologies (Paisley, Scotland), or MWG-BIOTECH AG (Germany). The oligonucleotides used in this study for PCR amplification are described in Table 1.
Bioinformatic tools:
Sequence analysis was performed with the Staden package software (v2002.2). Sequence assembly was obtained with the programs Pregap4 and Gap4. GPCR-encoding sequences were identified by BLASTx searches in the National Center for Biotechnology Information database, and potential pheromone genes were identified by the signature terminal CaaX motif in predicted open reading frames (Gene Jockey, Taylor 1996). Introns were predicted by a combination of BLAST search and visual identification of the conserved splice junctions. Initial multiple alignments of nucleotide or protein sequences were performed with ClustalX 1.81 (Thompson et al. 1994), with the following default alignment parameters: Gonnet series protein weight matrix, 10 gap opening, and 0.20 gap extension. Alignments were manually edited with the Sequence Alignment Editor Se-Al (v2.0a11; Rambaut, A; http://evolve.zoo.ox.ac.uk/) and formatted in MacBoxshade.
Phylogeny analysis:
Multiple sequence alignments were produced using ClustalW (Thompson et al. 1994) or ALIEN via the Multiple Alignment General Interface (MAGI) at the UK Human Genome Mapping Project Bioinformatics Resource Centre (HGMP-RC; Rysavy et al. 1992). Peptide alignments were optimized through MAGI using RASCAL (Thompson et al. 2003). Resulting GPCR genomic DNA and peptide data sets were evaluated by a range of phylogeny algorithms: “distance” using the Phylogeny Interface Environment at HGMP-RC and DNAdist or PROTdist (Felsenstein 1993); “parsimony” using PAUP* (v 4.0b10; Swofford 2000); and “maximum likelihood” using TreePuzzle (v5.0; Schmidt et al. 2000) at HGMP-RC with quartet puzzling (Strimmer and Von Haeseler 1996). Substitution models were either JTT (Jones et al. 1992) for peptide analysis or HKY (Hasegawa et al. 1985) for nucleic acids. Outgroup peptides composed S. cerevisiae Ste3p (P06783) alone or together with proteins from other basidiomycete species: Ustilago maydis Pra1, Pra2 (P31302, P31303); Schizophyllum commune Bar1, Bbr1, Bbr2 (P87022, P78741, AF148501); and Pleurotus djamor Ste3.3 (AAS46748). The C. cinereus Rcb26, Rcb36, Rcb142, Rcb242, and Rcb342 have been deposited previously (Y11081, Y11080, AF186383, AF186384, AF186385).
Sequence data accession numbers:
GenBank accession numbers for C. cinereus genes are as follows: rcb13 AY172107; phb13 AY172109; rcb33 AY172108; phb3.13 AY172110; phb3.23 AY172111; rcb244 AY393904; phb2.244 AY393913; phb2.144 AY393912; rcb21 AY393903; phb2.21 AY393911; phb2.11 AY393910; rcb343 AY393906; phb3.343 AY393919; phb3.243 AY393918; phb3.143 AY393917; rcb243 AY393905; phb2.343 AY393916; phb2.243 AY393915; phb2.143 AY393914; phb35 AY393920; rcb35 AY393907; rcb347 AY393909; phb3.347 AY393925; phb3.247 AY393924; phb3.147 AY393923; rcb345 AY393908; phb3.245 AY393922; and phb3.145 AY393921. The peptide multiple sequence file used in phylogenetic analyses is available as the EMBL-Align database accession ALIGN000818.
RESULTS
Identifying allelic variation in uncharacterized B loci:
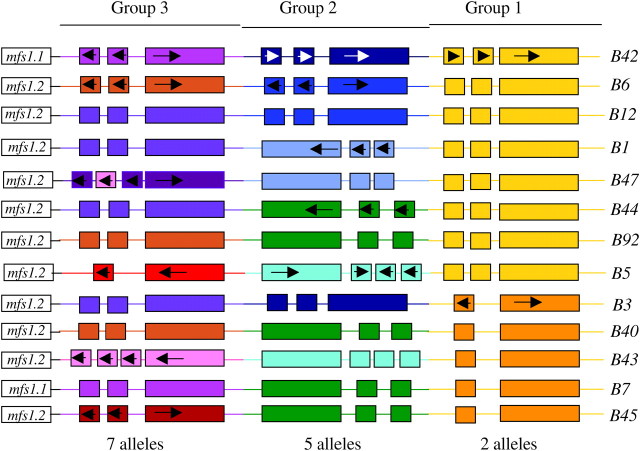
We have previously characterized three variants of the B locus, B3, B6, and B42 (O'Shea et al. 1998; Halsall et al. 2000; Milner 2000), each of which derives its unique specificity from a particular combination of alleles of three tandemly arranged groups of genes, which we refer to as groups 1, 2, and 3. This organization is illustrated in Figure 1 where the B3, B6, and B42 loci are compared with 10 other loci that we have characterized in this study. We found that B6 and B42 are homoallelic for the group 1 genes and B3 and B42 are homoallelic for the group 2 genes, and all three loci are heteroallelic for group 3 genes. Importantly, we found that sequence variation between alleles and the sequences within which the genes are embedded is such that they failed to cross-hybridize in Southern analyses. Positive hybridization has thus been used to identify homoalleles in uncharacterized B loci. This approach was used for a preliminary analysis of six strains in our collection that exhibited different B mating specificities (B1, B5, B40, B41, B43, and B44) and identified shared alleles in all of them (Halsall et al. 2000). This screen indicated that there were likely to be only two alleles of the group 1 genes, both of which we had sequenced. Since classical population studies had predicted that there are some 79 different B mating specificities in nature, we assumed that there must be many more alleles of group 2 and group 3 genes.
Figure 1.—
Allele constitution of 13 B mating specificities of C. cinereus. The different alleles of the three groups of genes together with allele-specific flanking sequences are differentiated by color. Pheromone genes are represented by short boxes and receptor genes by long boxes. mfs1.1 and mfs1.2 are alternative alleles of a conserved gene flanking the B locus. Arrows indicate the direction of pheromone and receptor gene transcription.
In this study we set out to identify more of the sequence diversity of C. cinereus pheromones and receptors by completing the analysis of the six partially characterized strains described by Halsall et al. (2000) and by acquiring five new strains from Japan and the United Kingdom that exhibited other B specificities (B7, B12, 45, B47, and B92) and potentially had new B alleles. The designation of the B mating specificity has been arbitrary in different wild collections so we confirmed differences by mating tests with our standard strains. In doing this, we noted that, contrary to our previous classification, B40 and B41 have the same specificity, so we eliminated B41 from this current analysis. We have thus examined a total of 13 strains. Southern analyses were used to confirm our previous data and to screen the new strains for homoalleles of genes in B3, B6, and B42 (Table 2). Also we performed PCR on a panel of genomic DNAs derived from all strains using primers designed to amplify known receptor genes. Where product of expected length was obtained, end sequencing confirmed correct allele assignment (Table 2). As new alleles were identified by DNA sequencing (rcb2 from B1, B43, and B44 and rcb3 from B43, B45, and B47), Southern analyses and PCR/end sequencing were used to identify other loci that shared them.
TABLE 2 .
PCR and Southern blot analyses used to identify sharedrcb alleles in 13 strains ofC. cinereus with differentB specificities
| Hybridizationa/PCR amplification with strains of different B specificity |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primer pair | Expected product (kb) |
B1 | B3 | B5 | B6 | B7 | B12 | B40 | B42 | B43 | B44 | B45 | B47 | B92 |
| rcb33 | 1.6 | +a | +a | − | − | − | +a | − | − | − | +a | − | − | −a |
| rcb35 | 1.8 | − | − | + | − | − | − | − | − | − | − | − | − | − |
| rcb36 | 1.6 | − | − | − | +a | − | − | +a | − | − | − | − | − | +a |
| rcb342 | 1.7 | − | − | − | − | +a | −a | − | +a | − | − | − | − | −a |
| rcb343 | 1.8 | − | − | − | − | − | − | − | − | + | − | − | − | − |
| rcb21 | 1.5 | + | − | − | − | − | − | − | − | − | − | − | + | − |
| rcb26 | 2.6 | − | − | − | +a | − | +a | − | − | − | − | − | − | +/−a |
| rcb242 | 1.5 | − | +a | − | − | − | − | − | +a | − | − | − | − | − |
| rcb243 | 2.0 | − | − | + | − | − | − | − | − | + | − | − | − | − |
| rcb244 | 1.5 | − | +/− | − | − | + | − | + | − | − | + | + | − | + |
| rcb13 | 2.1 | − | +a | − | − | +a | − | +a | − | +a | − | +a | − | − |
| rcb142 | 2.6 | +a | − | +a | +a | − | +a | − | +a | − | +a | − | +a | +a |
+, PCR amplification of a fragment of expected size confirmed as rcb by end sequencing; −, no PCR amplification; +/−, PCR amplification of a fragment of expected size but confirmed as not rcb by end sequencing.
Hybridization in Southern blot analyses.
Each group of genes comprises a receptor gene and one to three pheromone genes and we have adopted the following conventions to assign designations. The receptor genes are designated rcb1, rcb2, or rcb3, which indicates that they derive from group 1, group 2, and group 3, respectively. Pheromone genes are similarly designated phb1, phb2, and phb3 to indicate the group, followed by 1, 2, or 3 to distinguish the individual gene (see Figure 2). All genes are given a superscript number to indicate the B specificity from which they were isolated.
Figure 2.—
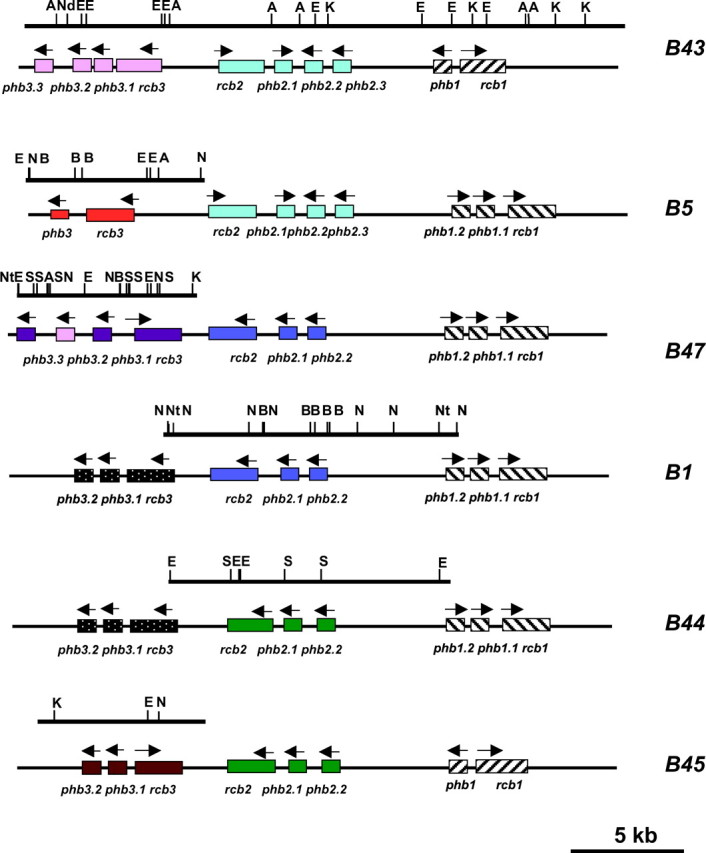
Identification of new group 3 and group 2 alleles. New alleles of the group 2 and group 3 genes at the B mating-type locus of C. cinereus are differentiated by colors; previously sequenced genes are presented in black and white motifs. Restriction sites are represented by A, AflII; B, BamHI; E, EcoRI; K, KpnI; N, NcoI; Nd, NdeI; Nt, NotI; and S, SacI.
Group 1 contains the minimum allelic diversity with only two alleles:
The Southern analyses summarized in Table 2 clearly indicated that there were only two allelic variants of the group 1 genes in the 13 genomic DNAs tested. Primers designed to rcb13 amplified a fragment with the expected sequence from B3, B7, B40, B43, and B45 and primers designed to rcb16 amplified a fragment with the expected sequence from B1, B5, B12, B42, B44, B47, and B92. Hence among the strains that we have looked at, group 1, with just two alleles, contains the minimum allelic diversity possible.
Identifying group 2 and group 3 alleles:
Our previous analysis identified two group 2 alleles and three group 3 alleles. Southern analysis and PCR/end sequencing unambiguously identified the homoallele of rcb26 in B12 and rcb242 in B3. In the analysis of group 3, the primers designed to rcb33 amplified additional products to the expected 1.6-kb fragment from B1, B12, and B44 genomic DNA but in each case the 1.6-kb fragment was confirmed as the homoallele of rcb33 by sequencing. Also Southern data for two of the new strains were ambiguous with respect to the group 3 alleles present in B12 and B92 since hybridization was to more than one probe (Table 2). The reasons for this were unclear but could relate to some of the normally allele-specific flanking sequences included in the probe rather than to the genes themselves. PCR/end sequencing, however, showed that B92 contained the homoallele of rcb36 and B12 contained the homoallele of rcb33 (Table 2).
To identify new alleles of group 2 and group 3 genes, we adopted two cloning strategies; the first is a strategy based on LR-PCR that we had previously shown to be able to isolate long fragments spanning just a single group of unknown genes (Halsall et al. 2000) and the second is isolation of clones from cosmid libraries. Once a new allele was identified, the PCR/end sequencing technique was used to test whether homoalleles of this new allele were present in any of the other partially uncharacterized B loci.
LR-PCR was used to amplify the group 2 genes of B1 and B44 since their group 1 and group 3 genes were homoallelic to known alleles. To minimize the size of the fragment to be amplified, primers were designed to the group 1 pheromone gene phb1.2, which is close to the group 1/group 2 boundary, and to the 3′-end of the group 3 rcb33 receptor, which is close to the group 3/group 2 boundary. To amplify group 3 genes, we designed primers to the group 2 sequence close to the group 3/group 2 boundary (from pheromone or receptor, depending on the orientation) and to the mfs-1 gene that flanks the B locus, which encodes a member of the major facilitator superfamily of putative membrane transporters. Two alleles of the mfs-1 gene encode almost identical proteins but have very different DNA sequences (Halsall et al. 2000). By using appropriate primers we were able to show which allele was present in each of our 13 strains and to design primers accordingly.
We amplified fragments of 9.5 and 7.5 kb, respectively, from B1 and B44 and, by sequencing, identified two new group 2 alleles (Figure 2). These each comprised a receptor gene and two pheromone genes. Primers to B1 rcb21 identified a homoallele in B47, whereas primers to B44 rcb244 identified homoalleles in B7, B40, B45, and B92 (Table 2). Using primers designed from the 5′-end of mfs-1.2 and the 3′-end of rcb244 or rcb21, we amplified a 6.85-kb fragment from B45 and a 7.44-kb fragment from B47, respectively, and sequencing identified new group 3 alleles in both. We were able to characterize the remaining unidentified alleles, the group 2 and group 3 alleles from B5 and B43, by sequencing clones containing the entire B5 or B43 loci isolated from cosmid libraries (see materials and methods). Both loci were homoallelic for the group 2 genes but had different alleles of the group 3 genes. Figure 2 summarizes the new sequences obtained in this study, which are indicated by the restriction maps above the diagram of the genes.
Our completed analysis of 13 B specificities identified five alleles of the group 2 genes, seven alleles of the group 3 genes, and two alleles of the group 1 genes. The allele combinations found in the 13 B specificities analyzed are summarized in Figure 1. If we assume that the three groups of genes are indeed functionally independent and that all allele combinations can be generated in nature, this level of allelic variation is sufficient to generate 70 unique B mating specificities.
The number of pheromone genes within any one group ranges from one to three (Figure 1), far fewer than found in the corresponding groups in the only other mushroom species for which data are available, S. commune, where there may be as many as eight (Fowler et al. 2004). The orientations of the genes differ from one allele to another. Remarkably, phb3.247 is identical in sequence to phb3.343. We checked the promoter region and could find no differences in sequence that would indicate that it is a pseudogene in either background. This is the only instance where we have found different alleles containing identical genes.
Receptors within the three groups have a complex origin:
A question of considerable interest with respect to the evolution of the B locus is whether or not the three subgroups of receptor genes have evolved independently: this is key to understanding how receptor/pheromone specificity is determined. Are the members of each group derived from an early duplication event with subsequent diversification giving rise to subfamilies of closely related genes and proteins, or has there been shuffling of genes between groups with appropriate sequence changes occurring to enable group specificity?
We have approached this question by comparing the predicted amino acid sequences of the receptors. Table 3 gives the percentage identity (top half of table) and similarity (bottom half of table) of the proteins. Sequence identity ranges from 18 to 81% and, significantly, we see this range between proteins within the same group as well as between proteins in different groups. Although six very similar proteins are encoded by the group 3 alleles present in B5, B6, B42, B43, B45, and B47, all having >60% identity with each other, these show only ∼20% identity to the seventh member of the group present in B3. This latter receptor is most similar in sequence to a group 2 receptor present in B43, the two sharing 68% identity. Of the five group 2 receptors, only two, found in B1 and B44, show >45% identity. The two group 1 receptors are also very dissimilar, only 18% identity, and each resembles more closely receptors in the other groups.
TABLE 3 .
Comparison of group 1, group 2, and group 3 receptor protein sequences
| Group 3 |
Group 2 |
Group 1 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rcb35 | Rcb36 | Rcb342 | Rcb343 | Rcb345 | Rcb347 | Rcb33 | Rcb21 | Rcb26 | Rcb242 | Rcb243 | Rcb244 | Rcb13 | Rcb142 | |
| Rcb35 | — | 77 | 67 | 78 | 70 | 62 | 23 | 38 | 33 | 24 | 22 | 55 | 31 | 20 |
| Rcb35 | 83 | — | 79 | 74 | 81 | 73 | 24 | 40 | 34 | 25 | 21 | 41 | 31 | 21 |
| Rcb35 | 76 | 85 | — | 69 | 74 | 69 | 21 | 40 | 34 | 24 | 20 | 40 | 31 | 21 |
| Rcb343 | 85 | 81 | 77 | — | 73 | 68 | 22 | 38 | 34 | 24 | 22 | 40 | 30 | 21 |
| Rcb345 | 77 | 86 | 81 | 80 | — | 81 | 23 | 39 | 32 | 24 | 21 | 40 | 19 | 21 |
| Rcb347 | 69 | 72 | 74 | 73 | 86 | — | 20 | 33 | 29 | 21 | 19 | 35 | 27 | 19 |
| Rcb33 | 34 | 35 | 34 | 34 | 33 | 30 | — | 23 | 20 | 34 | 69 | 23 | 19 | 33 |
| Rcb21 | 53 | 54 | 53 | 52 | 52 | 46 | 37 | — | 31 | 27 | 22 | 81 | 26 | 20 |
| Rcb26 | 46 | 46 | 46 | 47 | 45 | 41 | 34 | 42 | — | 22 | 20 | 33 | 47 | 19 |
| Rcb242 | 39 | 40 | 39 | 38 | 38 | 34 | 47 | 40 | 32 | — | 33 | 23 | 30 | 32 |
| Rcb243 | 34 | 33 | 33 | 34 | 33 | 30 | 76 | 33 | 31 | 45 | — | 23 | 18 | 31 |
| Rcb244 | 40 | 55 | 53 | 54 | 54 | 49 | 37 | 88 | 45 | 43 | 35 | — | 30 | 21 |
| Rcb13 | 40 | 42 | 41 | 40 | 30 | 37 | 31 | 36 | 59 | 40 | 30 | 40 | — | 19 |
| Rcb142 | 32 | 33 | 32 | 31 | 33 | 29 | 42 | 33 | 30 | 42 | 41 | 34 | 29 | — |
Matrix summarizes percentage identity (upper diagonal) and percentage similarity (lower diagonal). Matrix diagonals are separated by dashes. Values >60% are shown in italics.
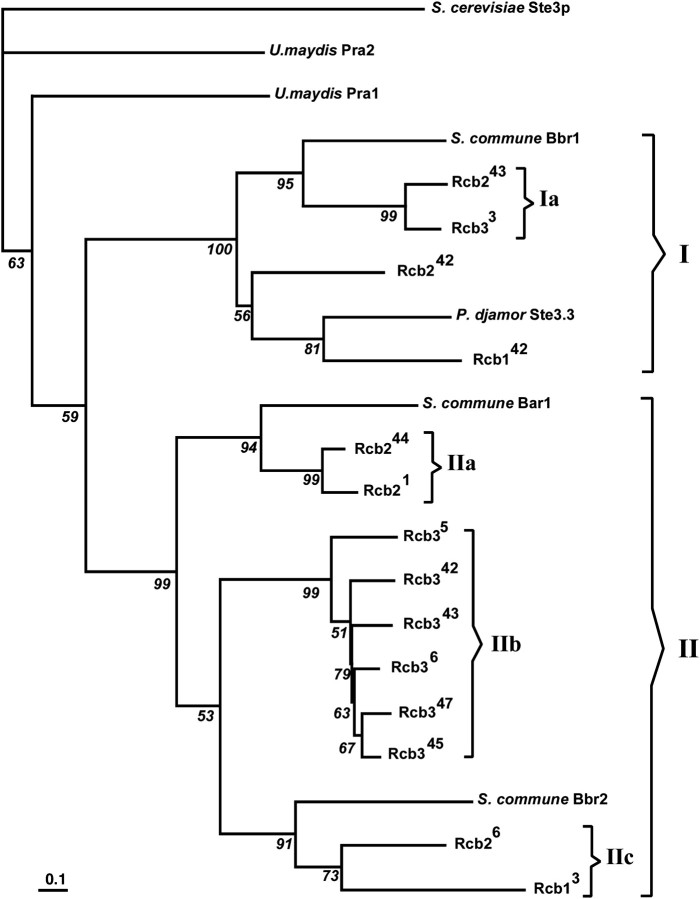
The phylogenetic analysis (presented in Figure 3), strongly supported by all tree-building algorithms (distance, parsimony, and maximum likelihood), revealed two major clusters that were distinct from the basal outgroup species (S. cerevisae Ste3p, U. maydis Pra1 and Pra2). Four of the C. cinereus receptors (Rcb243, Rcb33, Rcb242, Rcb142) group in cluster I, together with the orthologous S. commune Bbr1 (Bβ) and P. djamor Ste3.3 proteins, whereas the other 10 C. cinereus receptors group in cluster II, together with S. commune Bar1 (Bα) and Bbr2 (Bβ) proteins. These two major clusters and their subgroups proved robust, independent of basidiomycete outgroup inclusion, and were revealed by all analyses. Four C. cinereus subgroups were also evident: Ia (Rcb243, Rcb33; 99% support), IIa (Rcb244, Rcb21; 99%), IIb (Rcb35, Rcb342, Rcb343, Rcb36, Rcb347, Rcb345; 99%), and IIc (Rcb26, Rcb13; 73%). These phylogenetic subgroups were also evident in gDNA tree topologies (data not presented).
Figure 3.—
Quartet puzzling tree with maximum-likelihood branch lengths for C. cinereus receptor proteins. Branch lengths were calculated using the JTT model for substitution with uniform rate heterogeneity (log L, −17850.83). Bar indicates the number of substitutions per site. Branch support values (percentages) for 25,000 puzzling steps are shown in italics. Nominated outgroup was S. cerevisiae Ste3p.
Our analysis suggests that an early duplication and diversification of the progenitor of the C. cinereus family resulted in at least two evolutionary groups. Two major clusters and four subgroups indicate that the paralogous C. cinereus rcb genes are polyphyletic and diverge through several lineages. Significantly, members of all three of the C. cinereus receptor groups that we have defined as functionally different by their position within the B locus can be found in each of the two major clusters seen in the phylogenetic tree. Although there is obviously a common origin for six of the group 3 receptor genes in cluster IIb, reflected by the similar amino acid sequences of their predicted proteins, the third member of this subfamily, Rcb33, is found in cluster I, and its predicted protein most closely resembles receptors in group 1 and group 2. We conclude, therefore, that gene shuffling has played a major role in the evolution of the present functional groups. Although only three S. commune receptors were used in this analysis, it would appear that similar events have occurred in the evolution of the Bα and Bβ families. Available Bα receptor sequences are very similar and group together in any phylogenetic analysis (James et al. 2004) and thus resemble the subgroup of similar Rcb3 proteins found in C. cinereus, but the two Bβ proteins are found in different clusters with Bbr2 being much closer in sequence to the Bα proteins.
Predicted pheromone sequences:
We have identified and sequenced 17 new pheromone genes, which, together with those described previously, constitute 29 sequences: 3 from group 1, 11 from group 2, and 14 from group 3 (Figure 4). The new genes were identified by searching for open reading frames adjacent to receptors with the characteristic C-terminal CaaX motif. The full precursor sequences are compared in Figure 4 and are arranged according to the group from which the alleles derive. There is considerable length variation of the precursors, ranging from 48 to 85 amino acids, but in all we see a conserved two-residue charged motif (ER in 24 of the 28 sequences and DR/QR/ED in the others), which we previously predicted to be at the amino terminus of the mature peptide pheromone (Casselton and Olesnicky 1998; Olesnicky et al. 1999). In Figure 4 the predicted mature pheromone sequences are shown in larger type and range in length from 12 to 15 amino acids.
Figure 4.—

Alignment of pheromone precursor sequences. The conserved ER/ED/ QR doublet considered to be at the N terminus of the mature pheromone is in boldface type as is a doublet of amino acids preceding the terminal cysteine residue. The C-terminal CaaX motif is underlined and the amino acids removed by C-terminal processing are shown in smaller type. Sequences obtained from O'Shea et al. (1998), Halsall et al. (2000), and Milner (2002) are indicated by (*). Pheromones are classified according to the pair of amino acids found at positions −1 and −2 with respect to the carboxy-terminal cysteine residue (the subterminal doublet); rr, both are aromatic; ar, an aliphatic and aromatic pair; aa, both are aliphatic.
The amino-terminal regions of the pheromone precursors that are removed during maturation are highly divergent even when the mature peptide sequence is identical, as in the case of the two group 2 pheromones present in B1, Phb2.11 and Phb2.21. Hence alignment is not greatly informative of pheromone origin except in the case of Phb3.247 and Phb3.343, which apparently derive from a recent shuffling event. The processing machinery must recognize conserved structural features of the precursor molecule, which may include, in addition to the conserved dipeptide, a proline that is often present at position −4 and/or an aspartate/asparagine at position −2 relative to the putative cleavage site (Figure 4).
The pheromone-receptor interaction is highly specific because it ensures that B-regulated development can be activated only by the complement brought together by compatible mating partners. We have assumed that the genes within each of the three groups at the B locus are functionally independent (O'Shea et al. 1998; Halsall et al. 2000); thus we predict that pheromones must be able to distinguish among the different receptors encoded by alleles within the same group as well as those encoded by genes in other groups. We have looked, therefore, for likely determinants of specificity in the pheromone sequences. In vitro mutation studies indicated that the subterminal doublet adjacent to the C-terminal cysteine residue plays a role in specificity determination; by reversing the order of these two amino acids we were able to switch the allele specificity of a group 3 pheromone (Olesnicky et al. 1999). Moreover, the identity of the subterminal doublet in the pheromones of S. commune was one of the defining features in the grouping presented by Fowler et al. (2004). In C. cinereus, as in S. commune, this doublet is often common between several pheromones within the same group, so it cannot be the only determinant of allelic specificity but could have a more critical role in determining group specificity. As can be seen in Table 4, group 2 pheromones represent a homogenous group in which the C-terminal doublet is always an aliphatic/aromatic (ar) pair and, in all but two instances, this is AF. Group 3 pheromones fall into two distinct subgroups; all seven alleles encode pheromones with a C-terminal aromatic (rr) pair (FW, WF, or WW), but four of the alleles also encode a quite different pheromone with an aliphatic (aa) pair (GQ, GA, or TL). The alternative alleles of the group 1 genes encode pheromones that are quite unrelated to each other; Phb1.142 and Phb1.242 resemble group 2 pheromones in having the aliphatic/aromatic AF C-terminal doublet whereas Phb1.13 resembles a group 3 pheromone in having a TA aliphatic/aliphatic doublet. The C-terminal doublet cannot be an indicator of group specificity for these three pheromones but more likely reflects the fact that the receptors that they activate have a common origin with those in either group 2 or group 3, respectively.
TABLE 4 .
Functional analysis of group 1, group 2, and group 3 pheromones
| Group | Pheromone | Subterminal doubleta |
Hosts in which receptor is activatedb |
Hosts in which receptor is not activatedb |
Receptor specificityc |
|---|---|---|---|---|---|
| I | Phb1.142 | ar | B3 | (B42) | Group 1, two classes: |
| Phb1.242 | ar | B3 | (B42) | (1) Rcb13 ar activated; and | |
| Phb1.13 | aa | B42 | (B3) | (2) Rcb142 aa activated | |
| II | Phb2.11 | ar | B6, B42, B43, B44 | (B1) | Group 2, one class: |
| Phb2.21 | ar | B6, B42, B43, B44 | (B1) | all receptors ar activated | |
| Phb2.16 | ar | B1, B42, B43, B44 | (B6) | ||
| Phb2.26 | ar | B1, B42, B43, B44 | (B6) | ||
| Phb2.142 | ar | B1, B6, B43, B44 | (B42) | ||
| Phb2.242 | ar | B1, B6, B43, B44 | (B42) | ||
| Phb2.243 | ar | B1, B6, B42, B44 | (B43) | ||
| Phb2.243 | ar | B1, B6, B42, B44 | (B43) | ||
| Phb2.144 | ar | B1, B6, B42, B43 | (B44) | ||
| Phb2.244 | ar | B1, B6, B42, B43 | (B44) | ||
| III | Phb3.13 | rr | B42, B43 | (B3), B5, B6 | Group 3, four classes: |
| Phb3.23 | rr | B42, B43 | (B3), B5, B6 | (1) Rcb343 aa and rr activated; | |
| Phb3.15 | rr | B3, B6, B42, B43 | (B5) | (2) Rcb35 aa activated; | |
| Phb3.16 | aa | B3, B5, B43 | (B6), B42 | (3) Rcb33 activated by all aa and | |
| Phb3.26 | rr | B42, B43 | (B6) B3, B5 | some rr; and | |
| Phb 3.142 | aa | B3, B5, B43 | (B42), B6 | (4) Rcb36 and Rcb342 activated | |
| Phb 3.242 | rr | B6, B43 | (B42), B3, B5 | by some aa and some rr | |
| Phb 3.143 | rr | B3, B6, B42 | (B43), B5 | ||
| Phb 3.243 | rr | B3, B6, B42 | (B43), B5 | ||
| Phb 3.343 | aa | B3, B5, B6, B42 | (B43) |
Pheromones are classified on the basis of a subterminal doublet comprising an aliphatic amino acid pair (aa), an aromatic amino acid pair (rr), or an aliphatic aromatic pair (ar).
Pheromone genes were tested by transformation for ability to activate the pheromone response in hosts with different allelic versions of the receptor genes. B specificities in parentheses are the self-hosts from which the pheromone gene was isolated.
Receptors are classified according to the subterminal doublet of the pheromone that activates them. It is assumed that receptors can be activated only by pheromones from the same group.
Functional analysis of pheromone specificity:
In S. commune, a B-null stain in which the entire Bα and Bβ gene complexes have been deleted by mutation has been identified (Fowler et al. 2001), enabling single pairs of receptors and pheromones to be tested in vivo for compatibility. Unfortunately, no such B-null strain exists in C. cinereus; instead, compatibility can be tested only in host strains of known receptor constitution. To do this, pheromone genes were amplified by PCR and introduced into hosts of different specificity by cotransformation. Approximately 25 transformants were isolated and crossed to tester strains carrying the same B specificity as the transformation host and different A specificity. When the pheromone activated a compatible receptor in the host, this resulted in a successful mating as indicated by formation of a dikaryon (see materials and methods). We performed this analysis for a range of pheromones, introducing them into host strains that contained receptors that we predicted they would activate. All the group 2 pheromones activated mating in the non-self hosts tested; therefore, on the basis of the assumption that pheromones activate only receptors within the same group, all group 2 pheromones can activate all non-self group 2 receptors. In contrast, several group 3 pheromones failed to activate mating in non-self hosts but, in all but one of the cases tested, the full allelic complement of pheromones was sufficient to activate the other specificity; for example, Phb3.242 fails to activate B3 and B5 but activates B6, and Phb3.142 activates B3 and B5 but not B6. This resembles the situation in S. commune where pheromones activate only subsets of non-self receptors (see Fowler et al. 2004). Above we classified the group 3 pheromones into two types on the basis of their aa or aromatic (rr) subterminal doublets, since these residues are known from mutagenesis to determine receptor specificity in at least one case (Olesnicky et al. 1999). We were therefore interested to observe whether group 3 receptors fell into classes according to whether they interacted with aa or with rr pheromones. Again following the assumption that pheromones activate only same-class receptors, our transformation suggested this was true only for Rcb35, which was activated only by aa pheromones. The other group 3 specificities tested were activated by both aa and rr pheromones.
DISCUSSION
Organization of the B locus:
In this study we have made a molecular analysis of 10 previously uncharacterized loci of C. cinereus that confer different B mating specificities. The organization of each locus is similar with three groups of multiallelic genes encoding a pheromone receptor and one to three pheromones. Our transformation data, used to test many of the pheromones that we have identified, are consistent with these three groups of genes being functionally independent, such that pheromones activate only receptors within the same group.
The complexity of the homobasidiomycete B locus and the role of functionally redundant genes in generating large numbers of mating specificities was first evident from classical recombination studies with the mushroom S. commune. Here it was found that the B genes resided at two closely linked loci designated Bα and Bβ (see Raper 1966). Nine different alleles of each locus were identified in world samples, sufficient to generate 81 unique αβ combinations and thus B mating specificities in nature (Raper et al. 1960; Parag and Koltin 1971; Stamberg and Koltin 1973). In fact the number is <81 because not all αβ combinations can be generated by recombination (Koltin and Raper 1967; Fowler et al. 2004). Each locus contains a pheromone receptor and from three to eight pheromone genes (Wendland et al. 1995; Vaillancourt et al. 1997; Fowler et al. 2001, 2004); thus each locus is equivalent to one of the three groups of genes at the B locus of C. cinereus. The complex B locus of C. cinereus is essentially equivalent to three tightly linked loci that have no homologous sequences separating them. During evolution, however, these groups of genes have been recombined to generate different allele combinations that generate the different B mating specificities in the population. By having three groups of functionally redundant genes rather than two, large numbers of specificities can be generated from relatively few alleles. C. cinereus and S. commune have predicted similar numbers of B mating specificities, but preliminary studies on P. djamor suggest that this fungus may have as many as 231. Like C. cinereus, P. djamor has three groups of B genes and, like S. commune, on the basis of recombination analysis at least two reside at separable loci (James et al. 2004).
In total we have identified seven alleles of the group 3 genes, five alleles of the group 2 genes, and two alleles of the group 1 genes, sufficient to generate 70 of the 79 predicted specificities. There may be more alleles in the wild that remain to be identified. C. cinereus is a saprophytic fungus of horse dung and our finding of so many shared alleles in strains isolated from widely separate locations such as England and Japan may be attributed to the movement of domesticated animals, a point made by May et al. (1999) when analyzing A locus variation.
Although we can detect no homologous sequence that would permit recombination between the groups of genes, recombination within the B locus may play some role in maintaining variability in nature. Where two B loci share alleles of the group 2 genes, as in the case of B3 and B42 or B44 and B45 (Figure 1), this shared sequence of some 7 kb would permit a low frequency of recombination of different group 1 and group 3 genes and could generate nonparental B mating specificities. Early attempts to detect recombination between B genes of C. cinereus were unsuccessful (Haylock 1978; P. Day, personal communication) but these experiments were carried out with B5 and B6 strains, which share group 1 genes, making recombination undetectable, and between B3 and B6 strains, which share no genes, making recombination impossible.
Evolution of the complex B locus:
The homobasidiomycetes are unique among fungi in having evolved large families of receptors and pheromones for mate recognition. Although the pheromones and receptors play a similar role in mating-type determination in hemi-basidiomycetes such as U. maydis, there are only two versions of the corresponding mating-type locus. Pheromone signaling is essential for mate chemoattraction and recognition and induces the formation of mating filaments that promote compatible cell fusions. After cell fusion, as in homobasidiomycetes, pheromone signaling continues to be important for maintenance of the dikaryophase (Hartmann et al. 1996; Urban et al. 1996; Müller et al. 2003). Without mate attraction, it is obviously advantageous in homobasidiomycetes to have large numbers of mating types to increase the chances of a random hyphal fusion leading to dikaryosis and sexual reproduction. Twenty mating specificities for each of A and B would be sufficient for 90% outbreeding potential in the population (Koltin et al. 1972). Remarkably, gene diversification and locus duplication have led to far higher numbers than these, and in many homobasidiomycetes outbreeding potential approaches 100%.
Our phylogeny analysis based on receptor sequence suggests a complex origin for the three groups of C. cinereus receptor genes and their cognate pheromones. The C. cinereus genes can be separated into two major clusters indicative of an early duplication of an ancestral gene and subsequent sequence diversification. Significantly, each cluster contains representative members of all three groups of genes, indicating that they have not evolved independently and that recombination events have moved receptors of different lineage between groups. To maintain group specificity, this would have required sequence changes that prevented a receptor from being activated by pheromones from another group and from acquiring responsiveness to pheromones within the new group. Fowler et al. (2001) have proposed a simple mutation model for the generation of new receptor specificities and Kothe et al. (2003) have proposed that they arise by recombination. Whatever the origin, there has been subsequent strong selection for sequence diversification because the integrity of the B locus is maintained by the very dissimilar DNA sequences of both homologous and paralogous genes and the sequences within which they are embedded.
The sequence identity between receptors ranges between 18 and 81%, and this similarity is unlinked to pheromone specificity; for example, Rcb26 and Rcb243 are only 20% identical but are both activated by non-self group 2 pheromones, whereas Rcb35 and Rcb343 are 78% identical but discriminate among numerous pheromones, such as Phb3.26. This suggests that pheromone specificity evolves faster than the rate of diversification of the whole receptor sequence and is consistent with only a small group of residues in the receptor forming the pocket of interaction with the pheromone. The selective pressure on the remainder of the receptor is presumably to maintain overall signaling function (3D structure, interaction with G-protein, etc.), and therefore the similarity between receptors can be related to time since diversification from a common ancestor. There is a striking pattern of receptors with >60% identity, which presumably derives from the most recent duplication events. Group 3 shows evidence of recent expansion, with six of seven receptors >60% identical. In contrast, receptors of groups 1 and 2 are <40% identical with the exception of Rcb21 and Rcb244 (81% identical). Rcb243 is characterized by similarity to Rcb33, perhaps as a result of the most recent intergroup gene-shuffling event.
Pheromone specificity determinants—multiple sequence positions within mature pheromones dictate receptor specificity:
Our working model for the structural basis of pheromone-receptor interaction is that all are variations on a theme, having in common a primary mode of interaction that involves recognition of structural features common to all pheromones. These may be obvious, such as the ER motif or the terminal modified cysteine, or may be secondary structural features not immediately apparent from the primary sequence. This idea is supported by the relatedness of all the receptors to Ste3p of S. cerevisiae and by observed common features within the pheromones. In addition, we envisage a secondary recognition process, involving residues presented by the primary interaction into a variable binding site within the receptor that is able to differentiate amino acids at multiple different positions of the pheromone. This may define whether the pheromone is able to bind or, alternatively, whether binding and activation of the receptor may be separable; i.e., there may be a wide spectrum of pheromones able to bind a receptor, only a subset of which activates it. Whether pheromones are able to bind but not activate receptors has not been tested so far.
Multiple positions within mature pheromone sequences are likely to dictate receptor specificity. The data presented in this study and elsewhere are consistent with this conclusion (Olesnicky et al. 1999, 2000; Fowler et al. 2001; Gola and Kothe (2003). We previously highlighted Phb3.242, which contains the sequence WF as the subterminal doublet (i.e., positions −1 and −2 relative to the terminal cysteine) (Brown and Casselton 2001). The specificity of this pheromone can be switched to be similar to Phb3.26 (i.e., able to activate Rcb36) by mutagenesis, reversing the subterminal doublet from WF to FW, as occurs in Phb3.26. However, Phb3.23 also contains WF but it fails to activate Rcb36; it differs from Phb3.242 at positions −4 and −6. Therefore we can say that positions −1, −2, and −4 or −6 act coordinately together in conferring ability of phb2.342 to activate Rcb36. Fowler et al. (2004) similarly compared mature sequences of two S. commune pheromones, Bbp2(7) and Bbp2(4), which differ at the same positions (−4 and −6) but which activate different receptors (β3 and β9, or α8 and β, respectively). The authors showed by mutagenesis that the amino acid at position −4 confers this receptor selectivity, being aliphatic [glycine in Bbp2(7)] or aromatic [phenylalanine in Bbp2(4)]. The two C. cinereus pheromones at position −4 also have either aliphatic or aromatic residues: proline in Phb3.23 and leucine in Phb3.242. This is an important observation, because it suggests that pheromone identity might be encoded at common positions in the mature sequence and by common mechanisms often involving aromatic residues across the homobasidiomycete family. Other examples are Phb2.143 and Phb2.242, which differ at only a single position (−7, alanine and serine, respectively) but discriminate appropriately between Rcb242 and Rcb243. The sequence information on this large and interdependent family of receptors and ligands, combined with the specificity relationships that we have presented in this study, will be a powerful resource in combination with future modeling and mutagenesis studies in elucidating the molecular details of the receptor-ligand interaction.
Many of our conclusions about receptor/pheromone specificity depend on the assumption of functional independence of the groups, i.e., that pheromones activate only receptors in the same group. However, Fowler et al. (2004) have recently shown that in particular specificities of S. commune, pheromones from Bβ can activate receptors in the other group, Bα. To prevent recombination from generating self-compatible receptor/pheromone combinations in the same haploid individual, the Bα-Bβ intervening region is short and contains sequence features that prevent homologous recombination. If the analogous situation occurs in C. cinereus, presumably in one of the combinations of group 2 and 3 that does not occur in any of the strains analyzed (it seems even less likely to be true for group 1 with only two alleles), this would complicate the interpretation of pheromone/receptor specificity from the transformation analyses. We believe this situation in C. cinereus is less likely due to the smaller number of pheromones associated with each receptor. In S. commune, pheromones have evolved to have limited range of activity toward different receptors, whereas in C. cinereus, there appears to have been greater selection for cross-specificity in each group, to maximize potential for non-self recognition. In the only example in S. commune where the sequences of the cross-group receptors are known, Bar8 and Bbr1, these show 88% identity (Fowler et al. 2004), which is significantly greater than that of the most similar cross-group receptors in C. cinereus, Rcb243 and Rcb33, which are only 69% identical. Cross-group specificity may need to be tested directly, ideally using a B-null host strain of C. cinereus. However, our extensive attempts to generate such a strain by directed mutation have been unsuccessful so far.
Acknowledgments
We are particularly grateful to Takashi Kamada for sending us strains with new mating specificities from Japan and to Miriam Zolan for a cosmid clone containing the B43 genes. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council awarded to L.A.C. and A.J.B. L.A.C. is currently supported by an Emeritus Fellowship from the Leverhulme Trust.
References
- Binninger, D. M., C. Skryznia, P. J. Pukkila and L. A. Casselton, 1987. DNA mediated transformation of the basidiomycete Coprinus cinereus. EMBO J. 6 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A. J., and L. A. Casselton, 2001. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 17 393–400. [DOI] [PubMed] [Google Scholar]
- Burrows, D. M., 1991 Transformation studies in the basidiomycete fungi Coprinus cinereus and Coprinus bilanatus. Ph.D. Thesis, University of London, London.
- Caldwell, G. A., F. Naider and J. M. Becker, 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev. 59 406–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton, L. A., and A. de la Fuente-Herce, 1989. Heterologous gene expression in the basidiomycete Coprinus cinereus. Curr. Genet. 16 35–40. [DOI] [PubMed] [Google Scholar]
- Casselton, L. A., and N. S. Olesnicky, 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P., S. K. Sapperstein, J. D. Choi and S. Michaelis, 1997. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J. Cell Biol. 136 251–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S. L., J. Wood and J. B. Hicks, 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1 19–21. [Google Scholar]
- Felsenstein, J., 1993 PHYLIP (Phylogeny Inference Package), Version 3.5c. Department of Genetics, University of Washington, Seattle.
- Fowler, T. J., M. F. Mitton, L. J. Vaillancourt and C. A. Raper, 2001. Changes in mate recognition through alterations of pheromones and receptors in the multisexual mushroom fungus Schizophyllum commune. Genetics 158 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, T. J., M. F. Mitton, E. I. Rees and C. A. Raper, 2004. Crossing the boundary between the B alpha and B beta mating-type loci in Schizophyllum commune. Fungal Genet. Biol. 41 89–101. [DOI] [PubMed] [Google Scholar]
- Gola, S., and E. Kothe, 2003. The little difference: in vivo analysis of pheromone discrimination in Schizophyllum commune. Curr. Genet. 42 276–283. [DOI] [PubMed] [Google Scholar]
- Halsall, J. R., M. J. Milner and L. A. Casselton, 2000. Three subfamilies of pheromone and receptor genes generate multiple B mating specificities in the mushroom Coprinus cinereus. Genetics 154 1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, H. A., R. Kahmann and M. B Ölker, 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15 1632–1641. [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, M., H. Kishino and K. Yano, 1985. Dating of human ape splitting by molecular clock of mitochondrial DNA. J. Mol. Evol. 22 160–174. [DOI] [PubMed] [Google Scholar]
- Haylock, R. W., 1978 A study of functions controlled by the incompatibility factors of Coprinus lagopus with special reference to the B factor. Ph.D. Thesis, University of London.
- James, T. Y., S. R. Liou and R. Vilgalys, 2004. The genetic structure and diversity of the A and B mating-type genes from the tropical oyster mushroom, Pleurotus djamor. Fungal Genet. Biol. 41 813–825. [DOI] [PubMed] [Google Scholar]
- Johnson, A. D., 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5 552–558. [DOI] [PubMed] [Google Scholar]
- Jones, D. T., W. R. Taylor and J. M. Thornton, 1992. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8 275–282. [DOI] [PubMed] [Google Scholar]
- Koltin, Y., and J. R. Raper, 1967. The genetic structure of the incompatibility factor of Schizophyllum commune: three functionally distinct classes of B factors. Proc. Natl. Acad. Sci. USA 58 1220–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltin, Y., J. Stamberg and P. A. Lemke, 1972. Genetic structure and evolution of the incompatibility factors in higher fungi. Bacteriol. Rev. 36 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe, E., S. Gola and J. Wendland, 2003. Evolution of multispecific mating-type alleles for pheromone perception in the homobasidiomycete fungi. Curr. Genet. 42 268–275. [DOI] [PubMed] [Google Scholar]
- Kües, U., R. N. Asante-owusu, E. S. Mutasa, A. M. Tymon, E. H. Pardo et al., 1994. a Two classes of homeodomain proteins specify the multiple A mating types of the mushroom Coprinus cinereus. Plant Cell 6 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües, U., B. Göttgens, R. Stratmann, W. V. J. Richardson, S. F. O'Shea et al., 1994. b A chimeric homeodomain protein causes self compatibility and constitutive sexual development in the mushroom Coprinus cinereus. EMBO J. 13 4054–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan, J., 1993. The pheromone response pathway in Saccharomyces cerevisiae. Annu. Rev. Genet. 27 147–179. [DOI] [PubMed] [Google Scholar]
- Lewis, D., 1961. Genetical analysis of methionine suppressors in Coprinus. Genet. Res. 2 141–155. [Google Scholar]
- May, G., L. Lechevanton and P. J. Pukkila, 1991. Molecular analysis of the Coprinus cinereus mating type A factor demonstrates an unexpectedly complex structure. Genetics 128 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, G., F. Shaw, H. Badrane and X. Vekemans, 1999. The signature of balancing selection: fungal mating compatibility gene evolution. Proc. Natl. Acad. Sci. USA 96 9172–9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, M. J., 2000 Isolation of genes encoding HMG domain proteins from Coprinus cinereus and an analysis of their role in mating. Ph.D. Thesis, University of Oxford, Oxford.
- Müller, P., G. Weinzierl, A. Brachmann, M. Feldbrügge and R. Kahmann, 2003. Mating and pathogenic development of the smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky, N. S., A. J. Brown, S. J. Dowell and L. A. Casselton, 1999. A constitutively active G-protein-coupled receptor causes mating self-compatibility in the mushroom Coprinus. EMBO J. 18 2756–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky, N. S., A. J. Brown, Y. Honda, S. L. Dyos, S. J. Dowell et al., 2000. Self-compatible B mutants in Coprinus with altered pheromone-receptor specificities. Genetics 156 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea, S. F., P. T. Chaure, J. R. Halsall, N. S. Olesnicky, A. Leibbrandt et al., 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parag, Y., and Y. Koltin, 1971. The structure of the incompatibility factors of Schizophyllum commune: constitution of the three classes of B factors. Mol. Gen. Genet. 112 43–48. [Google Scholar]
- Rao, P. S., and D. J. Niederpruem, 1969. Carbohydrate metabolism during morphogenesis of Coprinus lagopus (sensu Buller). J. Bacteriol. 100 1222–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper, J. R., 1966 Genetics of Sexuality in Higher Fungi. Ronald Press, New York.
- Raper, J. R., M. G. Baxter and A. H. Ellingboe, 1960. The genetic structure of the incompatibility factors of Schizophyllum commune: the A factor. Proc. Natl. Acad. Sci. USA 46 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redhead, S. A., R. Vilgalys, J. M. Moncalvo, J. Johnson and J. S. Hopple, 2001. Coprinus Pers. and the disposition of Coprinus species sensu lato. Taxon 50 203–241. [Google Scholar]
- Rysavy, F. R., M. J. Bishop, G. P. Gibbs and G. W. Williams, 1992. The UK Human Genome Mapping Project online computing service. Comput. Appl. Biosci. 8 149–154. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmidt, H. A., K. Strimmer, M. Vingron and A. von Haeseler, 2000 Tree-Puzzle: Maximum Likelihood Analysis for Nucleotide, Amino Acid, and Two-State Data (http://www.tree.puzzle.de/). Software distributed by the authors.
- Shahriari, H., and L. A. Casselton, 1974. Suppression of methionine mutants in Coprinus I. Complementation and allele specificity as criteria of suppressor gene action. Mol. Gen. Genet. 134 85–92. [Google Scholar]
- Stamberg, J., and Y. Koltin, 1973. Genetic control of recombination in Schizophyllum commune: evidence for a new type of regulatory site. Genet. Res. 22 101–111. [DOI] [PubMed] [Google Scholar]
- Strimmer, K., and A. Von Haeseler, 1996. Quartet Puzzling: A Quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13 964–969. [Google Scholar]
- Swiezynski, K. M., and P. R. Day, 1960. Heterokaryon formation in Coprinus lagopus. Genet. Res. 1 114–128. [Google Scholar]
- Swofford, D. L., 2000 PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA.
- Taylor, P. L., 1996 Gene Jockey II Software. BIOSOFT, Cambridge, UK.
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., J. C. Thierry and O. Poch, 2003. RASCAL: rapid scanning and correction of multiple sequence alignments. Bioinformatics 19 1155–1161. [DOI] [PubMed] [Google Scholar]
- Urban, M., R. Kahmann and M. Bölker, 1996. Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251 31–37. [DOI] [PubMed] [Google Scholar]
- Vaillancourt, L. J., M. Raudaskoski, C. A. Specht and C. A. Raper, 1997. Multiple genes encoding pheromones and a pheromone receptor define the B β1 mating-type specificity in Schizophyllum commune. Genetics 146 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, J., L. J. Vaillancourt, J. Hegner, K. B. Lengeler, K. J. Laddison et al., 1995. The mating-type locus B-alpha-1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 14 5271–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]