Abstract
Perturbations in the T-cell receptor (TCR) Vβ repertoire were assessed in the CD4 and CD8 T lymphocytes of human immunodeficiency virus (HIV)-infected children who were receiving therapy during the chronic phase of infection by flow cytometry (FC) and PCR analysis. By FC, representation of 21 TCR Vβ subfamilies was assessed for an increased or decreased percentage in CD4 and CD8 T cells, and by PCR, 22 TCR Vβ subfamilies of CD4 and CD8 T cells were analyzed by CDR3 spectratyping for perturbations and reduction in the number of peaks, loss of Gaussian distribution, or clonal dominance. The majority of the TCR Vβ subfamilies were examined by both methods and assessed for deviation from the norm by comparison with cord blood samples. The CD8-T-lymphocyte population exhibited more perturbations than the CD4 subset, and clonal dominance was present exclusively in CD8 T cells. Of the 55 total CD8-TCR Vβ families classified with clonal dominance by CDR3 spectratyping, only 18 of these exhibited increased expression by FC. Patients with high numbers of CD8-TCR Vβ families with decreased percentages had reduced percentages of total CD4 T cells. Increases in the number of CD4-TCR Vβ families with increased percentages showed a positive correlation with skewing. Overall, changes from normal were often discordant between the two methods. This study suggests that the assessment of HIV-induced alterations in TCR Vβ families at cellular and molecular levels yields different information and that our understanding of the immune response to HIV is still evolving.
The majority of peripheral blood CD4 and CD8 T cells express the αβ T-cell receptor (TCR), with the β chain represented by variable segments which are grouped into families based on sequence homology (16). A complete T-cell repertoire is indicative of an intact T-cell population with the potential to recognize a wide range of immunogens. Numerous reports have documented changes in the TCR Vβ repertoire during human immunodeficiency virus (HIV) infection in relation to disease progression and the effect of therapy (4, 5, 8-10, 19-23). CDR3 length spectratyping (8, 19, 21) and flow cytometric (FC) analysis of TCR Vβ families labeled with specific monoclonal antibodies (MAbs) (4, 7, 26) are among the most frequently used assays for the analysis of TCR Vβ repertoire in HIV infection. The CDR3 spectratype is an indicator of the relative proportion of cells in each TCR Vβ family with CDR3 of particular lengths, while labeling of cells with TCR-Vβ-specific MAbs provides a quantitative assessment of the percentages of particular TCR Vβ families in T cells. Thus, evaluation of the TCR Vβ repertoire with MAbs, in combination with CDR3 spectratyping, is expected to provide complementary information. In the present study, we analyzed the TCR Vβ repertoire in the CD4 and CD8 T cells of 22 HIV-infected children by CDR3 length analysis and by FC.
MATERIALS AND METHODS
Patient population.
TCR Vβ repertoire analyses by PCR and FC were performed concurrently in 22 HIV-infected children with a median age of 8.5 years (25th to 75th percentile, 5.5 to 13.0 years). The study cohort was comprised of patients in different stages of the disease with a median CD4 count of 26% (25th to 75th percentile, 22 to 31%) and a median plasma HIV RNA virus load of 19,629 RNA copies/ml (25th to 75th percentile, 1,070 to 96,216 RNA copies/ml). All except 2 patients were receiving antiretroviral therapy: 1 patient was on a single drug and 5 were on two drugs, while the remaining 14 patients were receiving ≥3 drugs. Peripheral blood samples were collected during regularly scheduled visits for routine clinical testing following the obtaining of informed consent as per institutional review board-approved protocols. This research was performed in compliance with all relevant federal guidelines and institutional policies. Patient characteristics at the time of study are shown in Table 1.
TABLE 1.
Immunologic and virologic characteristics of the study patients
| Patient no. | Patient identification no. | Age (yr) | T-cell count
|
Virus load (RNA copies/ml) | Treatment history (no. of drugs/duration [months])a | Virologic/immunologic responseb | |||
|---|---|---|---|---|---|---|---|---|---|
| CD4
|
CD8
|
||||||||
| % | Absolute | % | Absolute | ||||||
| 1 | 6750001208 | 0.2 | 37 | 3,713 | 28 | 2,785 | 125,482 | None | NA |
| 2 | 6750000964 | 2.0 | 17 | 1,420 | 39 | 3,178 | <50 | 3/24 | +/− |
| 3 | 6750001154 | 2.6 | 19 | 443 | 25 | 590 | 141,213 | 2/1 | −/− |
| 4 | 6750001146 | 3.2 | 20 | 715 | 42 | 1,466 | 14,258 | 2/29 | −/− |
| 5 | 6750001085 | 4.9 | 29 | 1,067 | 39 | 1,559 | 41,000 | None | NA |
| 6 | 6700000121 | 5.8 | 23 | 480 | 44 | 938 | 35,174 | 3/20 | −/− |
| 7 | 6750001028 | 6.8 | 29 | 708 | 38 | 918 | 79,267 | 3/2 | −/+ |
| 8 | 6750000946 | 7.0 | 23 | 502 | 27 | 598 | <50 | 3/9 | +/− |
| 9 | 6750000649 | 7.0 | 27 | 1,534 | 62 | 3,559 | 25,000 | 2/12 | −/+ |
| 10 | 6750000939 | 8.0 | 33 | 600 | 41 | 749 | 5,028 | 3/12 | +/+ |
| 11 | 6750000935 | 9.0 | 22 | 813 | 44 | 1,514 | 96,216 | 3/14 | −/+ |
| 12 | 6700000116 | 9.0 | 29 | 1,312 | 45 | 2,061 | 13,391 | 3/16 | −/+ |
| 13 | 6750000717 | 9.0 | 32 | 494 | 34 | 526 | 1,953 | 1/24 | +/+ |
| 14 | 6750000860 | 9.5 | 34 | 1,134 | 39 | 1,234 | >750,000 | 3/19 | −/+ |
| 15 | 6700000087 | 10.6 | 19 | 277 | 23 | 343 | 39,979 | 3/6 | −/+ |
| 16 | 6700000113 | 11.8 | 26 | 979 | 46 | 1,713 | 972 | 3/12 | +/+ |
| 17 | 6700000061 | 13.1 | 23 | 964 | 45 | 1,928 | 600 | 4/3 | +/− |
| 18 | 6700000011 | 13.4 | 26 | 633 | 35 | 835 | 1,070 | 4/1 | +/+ |
| 19 | 6700000129 | 13.7 | 38 | 677 | 36 | 637 | 9,161 | 2/12 | −/+ |
| 20 | 6750000988 | 15.0 | 0 | 0 | 15 | 202 | 229,086 | 3/14 | −/− |
| 21 | 6750000626 | 15.5 | 38 | 1,064 | 49 | 1,389 | 128,628 | 2/7 | −/+ |
| 22 | 6700000123 | 16.5 | 23 | 535 | 37 | 85 | <50 | 4/22 | +/− |
Patients were on combinations of nucleoside reverse transcriptase inhibitors (NRTIs) (zidovudine [AZT], lamivudine [3TC], and didioxyinosine) or on NRTIs plus nonnucleoside reverse transcriptase inhibitors (Nevirapine or Efavirenz) or two NRTIs plus protease inhibitors (nelfinavir, ritonavir, or saquinavir) for two, three, or four drug combinations.
Virologic response was defined as a decrease in plasma HIV RNA of greater than 1.0 log or to undetectable levels or the continued state of an undetectable virus load. Immunologic response was defined as an increase in the percentage of CD4 greater than 10% or the continued maintainance of CD4 T cells at greater than 25%. NA, not applicable.
CDR3 length analysis using reverse transcription-PCR.
Peripheral blood mononuclear cells were isolated from heparinized venous blood by ficoll-metrizoate (Lymphoprep; Nyegard, Oslo, Norway) density gradient centrifugation. CD4 and CD8 T cells were positively selected by using magnetic beads coated with anti-CD4 and anti-CD8 MAbs according to the manufacturer's instructions (Dynal, Lake Success, N.Y.). The purity of recovered cells as assessed by FC was >98%. RNA was extracted directly from cells coated on beads with Ultraspec RNA solution (Biotecx, Houston, Tex.). RNA (1 to 5 μg) was reverse transcribed with Moloney murine leukemia virus reverse transcriptase enzyme (Gibco BRL, Grand Island, N.Y.) and TCR β-chain C region primer (Cβ14). Multiplex PCR was performed with forward Vβ primers for TCR Vβ family 1 (Vβ1), -2, -3, -4, -5.1, -5.2, -6, -7, -8, -9, -11, -12, -13.1, -13.2, -14, -15, -16, -17, -18, -20, -21, -22, -23, and -24 and 32P-labeled CβR reverse primer in the presence of AmpliTaq Gold DNA polymerase (PerkinElmer, Branchburg, N.J.) as described previously (12, 25). Vβ10 and -19 were not analyzed, as they are pseudogenes (3). PCR conditions were as follows: 12 min of incubation at 95°C to activate the AmpliTaq Gold enzyme, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. At the end of 35 cycles, a final extension was performed at 72°C for 10 min. The PCR products were resolved on a 6% polyacrylamide gel, followed by analysis on a Phosphorimager (Molecular Dynamics, Sunnyvale, Calif.). An immunoscope pattern was generated by using ImageQuant software (Molecular Dynamics), and the intensity of the signals from CDR3 segments in each TCR Vβ family was quantified. Standards for CDR3 lengths in an unperturbed TCR Vβ repertoire were established by examining purified CD4 and CD8 populations in 10 umbilical cord blood samples obtained from healthy women following uneventful deliveries. CDR3 length distribution for each Vβ family in cord blood CD4 and CD8 T cells showed a Gaussian distribution. The 5th and 95th percentiles of the numbers of peaks were calculated for each family. These were five to nine peaks, with the exception of Vβ23, which showed three to seven peaks (data not shown). In patient samples, TCR Vβ families that exhibited a Gaussian distribution of the CDR3 lengths were considered unperturbed. The term skewing was used for TCR Vβ families that did not show a Gaussian distribution or had a reduction in the number of peaks to <5 (28). The term clonal dominance was used for TCR Vβ families that contained a peak whose area was >50% of the total area for that family (6, 12, 25).
FC determination of Vβ family representation.
The percentage of each Vβ family was quantified by four-color FC with a TCR Vβ kit (kindly provided by Coulter-Immunotech, Miami, Fla.). This kit consists of premixed combinations of three anti-Vβ MAbs per vial for the quantification of TCR Vβ1, -2, -3, -5.1, -5.2, -5.3, -7, -8, -9, -11, -12, -13.1, -13.6, -14, -16, -17, -18, -20, -21.3, -22, and -23: one conjugated with fluorescein isothiocyanate, the second conjugated with phycoerythrin, and the third conjugated with both fluorochromes (14). These reagents were combined with MAbs directed against CD4 conjugated with phycoerythrin-cyanine 5 (Immunotech) and directed against CD8 conjugated to allophycocyanin (Caltag, Burlingame, Calif.). Samples were labeled within 6 h of phlebotomy, lysed with Immunolyse (Coulter), and fixed in 1% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, Pa.). Lymphocytes were gated on CD4bright or CD8bright for the determination of Vβ percentages. Normal values for the percent representation of individual Vβ families within both CD4- and CD8-T-cell subsets were determined by calculating the 5th and 95th percentiles for each Vβ family from 10 cord blood samples (data not shown). In patient samples, values in excess of (increase) or less than (decrease) the 5th and 95th percentiles established in cord blood T cells were considered perturbed.
Estimation of plasma viral load.
HIV RNA in plasma was quantified with the Amplicor HIV monitor kit (Roche Diagnostics, Somerville, N.J.). Sensitivity of the assay was <50 HIV RNA copies/ml of plasma.
Statistical analysis.
Statistical analysis was performed with Sigmastat Software (Jandel Scientific, San Rafael, Calif.). Distributions of data sets were checked for normality and compared by using Student's t test or the Mann-Whitney rank sum test as appropriate, with a P of <0.05 considered significant. Spearman's correlation coefficient was utilized to determine the relationships between examined parameters.
RESULTS
TCR Vβ repertoire perturbations within CD4- and CD8-T-lymphocyte populations.
The TCR Vβ repertoire of both the CD4- and CD8-T-cell subsets of HIV-infected children (Tables 2 and 3, respectively) was found to be significantly perturbed compared to the cord blood repertoire, which served as the standard for an unaltered repertoire.
TABLE 2.
TCR Vβ family analysis in CD4 T cells in HIV-infected children by CDR3 length and FC analysis
| Patient no.a | PCRb
|
FC
|
||
|---|---|---|---|---|
| Skewing | Clonal dominance | Expansion | Depletion | |
| 1 | None | None | None | 12 |
| 2 | 23, 24 | None | 3, 5.1, 14, 16, 17 | 9, 11, 12 |
| 3 | ND | ND | None | 5.2, 12, 13.1 |
| 4 | 11, 21, 23, 24 | None | 8, 11, 20 | 7 |
| 5 | 23 | None | 8 | 11, 12 |
| 6 | 4, 21, 23 | None | 14 | 12 |
| 7 | ND | ND | 3, 9, 14, 18 | 12 |
| 8 | 23 | None | 9, 16, 18 | 12 |
| 9 | 23 | None | 3 | 5.1, 12 |
| 10 | 1, 4, 16, 18, 23 | None | 9, 11, 14, 17 | 7, 12 |
| 11 | 4, 5.1, 8, 20, 23, 24 | None | 8, 11, 16, 17 | 12 |
| 12 | 4, 23 | None | 5.1, 8, 9, 16, 22 | None |
| 13 | 23 | None | 11 | None |
| 14 | ND | ND | 3, 7, 16 | 9, 11, 12 |
| 15 | 23 | None | 14 | 9, 12, 13.1 |
| 16 | 4, 21, 23, 24 | None | 16 | 5.1, 9, 12, 18 |
| 17 | 5.2, 9, 11 | None | 5.2, 9, 11, 13.1, 14, 17, 18 | 8, 12 |
| 18 | 21, 23, 24 | None | 3, 5.2, 11, 13.1, 14 | 7, 12 |
| 19 | None | None | 3, 14, 17 | 12, 20 |
| 21 | 4, 23, 24 | None | 8, 13.1, 14, 16 | 7, 9, 12 |
| 22 | 23, 24 | None | 8, 14, 16, 18 | 7, 12 |
| Total no. of TCR Vβ families altered (no. of patients) | 42 (18) | 0 (18) | 60 (21) | 40 (21) |
ND, not done.
Analysis not done for patient 20.
TABLE 3.
TCR Vβ family analysis in CD8 T cells in HIV-infected children by CDR3 length analysis and FC analysis
| Patient no. | PCR
|
FC
|
||
|---|---|---|---|---|
| Skewing | Clonal dominance | Expansion | Depletion | |
| 1 | 4, 5.1, 18, 20, 22, 23, 24 | None | 9, 22 | None |
| 2 | 2, 3, 4, 5.1, 5.2, 6, 7, 8, 13.1, 13.2, 14, 15, 16, 17, 18, 20, 21, 22, 23 | 2, 5.1, 5.2, 14, 16, 17, 18 | 2, 5.3, 7, 8, 13.6, 18 | 1, 13.1, 21.3 |
| 3 | 4, 8, 9, 11, 18 | 8, 11, 18 | 2, 8, 11, 18, 22, 23 | 13.1, 20 |
| 4 | 1, 2, 3, 4, 6, 8, 9, 11, 13.2, 16, 20, | None | 5.2, 5.3, 12, 21.3, 22 | 1, 2, 13.6, 14 |
| 5 | 8, 12, 16, 20, 21, 22, 23, 24 | 20 | 2, 9, 20, 22 | 1, 3, 7, 21.3 |
| 6 | 1, 3, 11, 14, 18, 20, 21, 22, 23, 24 | 1, 3, 11, 18, 20, 21, 24 | None | 5.1 |
| 7 | 3, 4, 5.1, 5.2, 6, 7, 12, 18, 23 | 5.1, 5.2, 12, 18 | 2, 3, 5.3, 9, 11, 18 | None |
| 8 | 4, 5.1, 22, 24 | 5.1, 22 | 5.1, 5.3, 9, 18, 22 | 3 |
| 9 | 5.1, 5.2, 8, 9, 15, 18, 23 | None | None | 5.1 |
| 10 | 1, 3, 4, 5.2, 9, 11, 13.2, 14, 15, 16, 17, 18, 21, 22, 24 | 11, 15 | 5.1, 5.3, 9, 13.1, 18 | 3, 7, 20 |
| 11 | 1, 3, 5.1, 11, 14, 16, 18, 21, 22, 23 | 11, 16 | 2, 5.3, 9, 11, 16, 18, 21.3, 22 | 7, 20 |
| 12 | 1, 4, 13.2, 21, 23, 24 | 21, 24 | 9, 13.6, 22 | 3 |
| 13 | 1, 12, 20, 21, 23, 24 | 12, 20, 21, 24 | 5.1, 12, 14, 22 | 1, 13.1 |
| 14 | 3, 4, 6, 5.1, 12, 13.2, 14, 17, 18, 20, 21, 22, 23, 24 | 3, 6, 13.2 | 5.1, 7, 12, 16, 23 | None |
| 15 | 4, 8, 20, 21 | None | 2, 22, 23 | 3 |
| 16 | 2, 4, 12, 15, 18, 20, 22, 24 | 4, 12, 15 | 16, 18 | 1, 3, 5.1, 7, 13.1, 14 |
| 17 | 1, 4, 8, 9, 11, 12, 13.1, 14, 15, 17, 18, 21, 22, 23 | 1, 9, 13.1 | 1, 5.3, 9, 11, 13.1, 18 | 5.1, 7, 8, 12, 20, 21.3 |
| 18 | 5.2, 6, 14, 15, 20 | 5.2, 15 | 5.2, 13.1, 16, 22 | 3 |
| 19 | 1, 2, 4, 6, 7, 12, 14, 13.2, 15, 16, 17, 18, 20, 21, 22, 23, 24 | None | 2, 14, 16, 17, 18, 22, 23 | None |
| 20 | 3, 14, 18, 21, 23 | 3, 14, 18 | 5.2, 5.3, 9, 11, 18, 22, 23 | 1, 13.1, 14, 20, 21.3 |
| 21 | 2, 3, 4, 6, 9, 12, 13.2, 15, 16, 17, 18, 20, 21, 22, 23, 24 | 3, 6, 13.2, 17, 20, 21, 24 | 2, 13.1, 16, 20, 23 | 3, 7 |
| 22 | 3 | None | 2, 5.2, 13.1, 22 | None |
| Total no. of TCR Vβ families altered (no. of patients) | 202 (22) | 55 (22) | 97 (22) | 45 (22) |
As shown in Table 2, CDR3 spectratyping of CD4 T cells revealed skewing in 9.7% of Vβ families based upon the loss of Gaussian distribution and reduction in the number of peaks in the CDR3 regions of particular Vβ families. There was no evidence of clonal dominance in any Vβ family in CD4 T cells. By FC, there was evidence for both an increase and a decrease in the percentage of CD4 T cells expressing particular Vβ families. Increased expression was detected in 13.6% (n = 60) of TCR Vβ families, and decreased expression was observed in 8.8% (n = 40) of TCR Vβ families of CD4 T cells. As indicated in Table 2, when data for CDR3 spectratyping and FC were evaluated together, of the 42 CD4 TCR Vβ families that exhibited skewing (by CDR3 spectratyping), percent TCR Vβ expression by FC was increased in only five individual TCR Vβ families and was not decreased in any.
In contrast to CD4 T cells, CD8 T cells exhibited profound alterations in the TCR Vβ repertoire of HIV-infected children by all measured parameters (Table 3). Abnormalities in CD8-TCR Vβ repertoire were most apparent in assays of CDR3 spectratyping, with 202 Vβ families (38.2%) exhibiting skewing; of these, 55 Vβ families (10.4%) exhibited clonal dominance. As seen for the CD4 T cells, FC revealed an abnormal expression of certain TCR Vβ families of CD8 T cells, only some of which were deemed skewed or dominant by CDR3 length analysis. Overall, in the CD8 T-cell repertoire of all the Vβ families tested in 22 HIV-infected children, 202 Vβ families showed skewing by CDR3 spectratyping; of these 202 families, the percentages of 43 families were increased as determined by FC and the percentages of 6 families were decreased. Of the 55 families that showed evidence of clonal dominance by CDR3 spectratyping, 18 exhibited increased expression by FC.
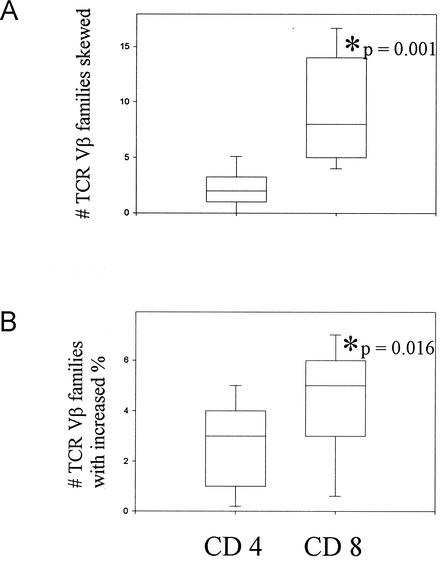
By both PCR-based CDR3 length spectratyping and FC, CD8 T cells exhibited more perturbations than the CD4 subset (Fig. 1). By CDR3 spectratyping, a median number of two TCR Vβ families for CD4 T cells and eight Vβ families for CD8 T cells (P < 0.001) were skewed in 18 subjects. Clonal dominance was observed exclusively in the CD8 T cells. By FC as well, increases in the percent expression of particular Vβ families in the CD8-T-cell compartment were greater than the increases of those within the CD4 subset (mean CD8 count 4.41 versus mean CD4 count = 2.86, P = 0.016) in 21 subjects. The absolute difference in percentages of each TCR Vβ family relative to that from the cord blood was significantly greater for CD8 T lymphocytes compared to that for CD4 T lymphocytes (mean CD8 count = 1.36 versus mean CD4 count = 0.92, P < 0.001).
FIG. 1.
Greater disruption of the CD8-TCR Vβ repertoire as compared to CD4-TCR Vβ repertoire in HIV-infected study patients. TCR Vβ repertoire in study patients was analyzed in relation to that for normal cord blood T cells, which represent an unperturbed repertoire. The CD8-T-lymphocyte population exhibited more changes by both PCR (A) and FC (B) than the CD4 population. Box plots indicate the median value as well as 5th, 25th, 75th, and 95th percentiles. The two groups were compared using the Mann-Whitney rank sum test with a P of ≤0.05 considered significant.
Relationship between TCR Vβ perturbations and clinical parameters.
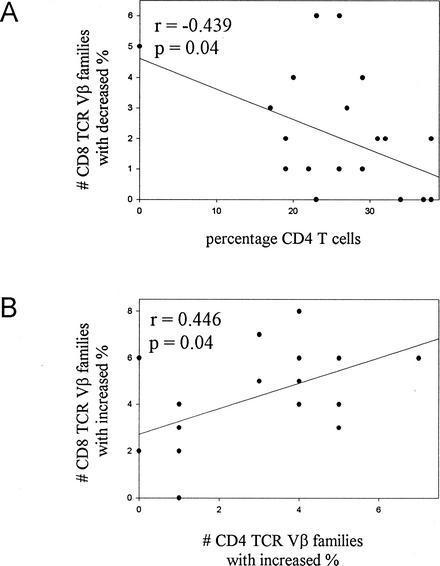
We investigated whether a relationship existed between the level of perturbation in the TCR Vβ repertoire and the variables of CD4-cell count and plasma viral load. Percent CD4 was negatively correlated with the number of CD8-TCR Vβ families showing decreased expression by FC (r = −0.439, P = 0.04) (Fig. 2A). No other correlations between repertoire deviation and CD4-cell count or plasma viral load were observed. In addition, no correlation was noted between perturbations detected by CDR3 spectratyping in the T-cell repertoire with the age of the patients, the nature of therapy, or plasma virus load.
FIG. 2.
Correlation of FC-based expression of TCR Vβ families with each other and with CD4 T cells. Decreases in the percent expression of CD8-TCR Vβ families correlate inversely with the percentage of CD4 T cells (A). Increases in the percent expression of CD8-TCR Vβ families correlate with increases in the percentages of CD4-TCR Vβ families (B).
Correlations between different assays used to determine TCR Vβ perturbations.
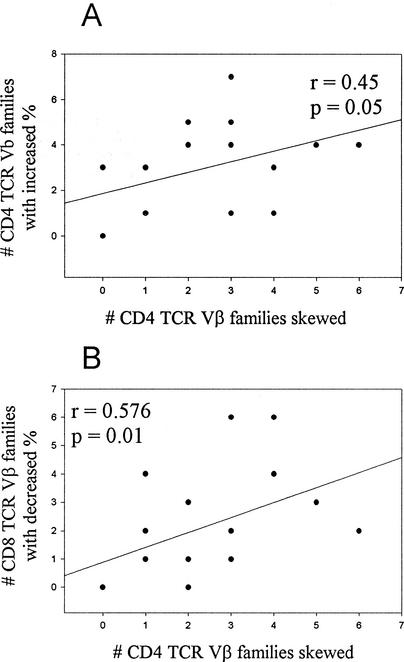
We sought to investigate if a relationship existed between the number of perturbations at the molecular level determined by CDR3 spectratyping (skewing or dominance) and the TCR Vβ usage at the cellular level as determined by FC (increase or decrease). The number of CD8-TCR Vβ families exhibiting increases in percent expression correlated with the number of CD4-TCR Vβ families that showed increases in percent expression (r = 0.446, P = 0.04) (Fig. 2B). The number of TCR Vβ families in CD4 T cells that exhibited skewing by CDR3 spectratyping correlated with the number of CD4 Vβ families that were overexpressed by FC (r = 0.45, P = 0.05) (Fig. 3A) and with the number of CD8-TCR Vβ families that were underexpressed by the FC assay (r = 0.576, P = 0.01) (Fig. 3B). Within the FC assay, overexpressed CD4-TCR Vβ families and CD8-TCR Vβ families correlated with each other. Within the CDR3 spectratyping assay, the number of CD8-TCR Vβ families showing skewing correlated with the number of Vβ families demonstrating clonal dominance (r = 0.43, P = 0.04). The individual TCR Vβ families exhibiting perturbations, however, were usually disparate and only rarely the same (Tables 2 and 3).
FIG. 3.
Relation of FC-based changes in TCR Vβ families with skewing observed by CDR3 length analyses. Increases in the percent expression of CD4-TCR Vβ families correlate with CD4-TCR Vβ skewing (A), and decreases in the percent expression of CD8-TCR Vβ families are correlated with CD4-TCR Vβ skewing (B).
DISCUSSION
Upon antigen encounter, the response of both CD4 and CD8 cells is deemed to be essential for mounting an efficient immune response. In the HIV-infected children studied, marked distortions were observed in TCR Vβ repertoire in CD4 and CD8 T cells, with more perturbations in CD8 compared to CD4 T cells, which is in concordance with previous reports (9, 13, 21, 22). While increases in the percentages of particular TCR Vβ families were detected by FC in CD4 and CD8 T cells, clonal dominance was exclusively present in the CD8-T-cell compartment (25), which has been previously shown to result from oligoclonal expansions by sequence analysis (12). A possible explanation lies in the robustness and kinetics of the response of each subpopulation subsequent to antigen encounter, as it has been demonstrated that the initial proliferation of CD8 cells is of greater magnitude than CD4 cells and of longer duration (13); a recent investigation of pediatric HIV infection found persistent high-level TCR Vβ-specific expansions only within the CD8 subpopulation (22). In the present study, increases in percent representation of TCR Vβ families in the CD4+ T cells were without clonotypic overrepresentation.
Our observation of an association between the number of CD4- and CD8-TCR Vβ families with increased percentages leads us to speculate that these CD4 populations may have specifically responded to HIV. If so, this would support the concept that helper T cells are imperative for CD8-T-cell function, e.g., cytotoxic lymphocyte activity. Indeed, patients with tremendously depleted CD4 pools no longer possess detectable HIV-specific CD8 cells (11) while those patients capable of controlling virus in the absence of therapy exhibit strong helper activity (27). HIV-infected children with preserved or restored CD4 compartments who are given therapy are perhaps more likely to exhibit virus-specific cytotoxic-T-lymphocyte expansions compared to patients in whom this population is depleted. While increased percentages of CD4 T cells in HIV-infected children occur transiently (22), their role in influencing the antigen-specific CD8-T-cell response remains to be elucidated. One possibility is that the CD4 help given to the CD8 lymphocytes is required only during the initial phase of activation. An alternative hypothesis is that, since CD4 T cells are the primary target of HIV infection, antigen-specific activity in these cells leads to their preferential infection and deletion and the remaining cells are dysregulated due to induction of anergy or apoptosis. A third possibility is that this may indicate an oligoclonal response but not with the dominance of particular clones.
How does the TCR Vβ response to HIV differ from that against other viral infections? For the response to Epstein-Barr virus, selective and massive expansion of a few dominant clones of CD8 T cells accounts for a significant proportion of the lymphocytosis observed (2). Similarly, only a limited number of clones appear to respond to measles virus (15). Importantly, neither infectious mononucleosis nor measles results in chronic persistent infection as does HIV and elicits only transient lymphocyte expansions. This is in contrast to HIV infection in which a much more heterogeneous response was observed. In an earlier study, it was found that HIV-specific lymphocytes displayed a heterogeneous TCR Vβ repertoire in certain patients (14). A key factor impacting the breadth of immune response may be the duration of disease; a pathogen-inducing chronic disease may, over time, involve more Vβ families than a transient infection. However, the normal mechanisms which result in focusing of the T-cell response may not be operative during HIV infection (1), which may be influenced by chronic immune activation, high mutation rate of the virus, or loss of the T-helper-cell population
It can be speculated that isolated TCR Vβ perturbations may result from the partial activation of lymphocytes by HIV or the response elicited could be against other antigens. Evidence from the B-cell compartment demonstrates a highly activated cell population that produces large amounts of antibody, very little of which is specific (18). A similar phenomenon could occur in the T-cell pool as well. Indeed, a mechanism of partial activation resulting from incomplete T-lymphocyte differentiation in response to viral antigens has recently been described (24). Experiments using major histocompatibility complex peptide tetrameric complexes in combination with functional assessment will serve to clarify this issue. Another important question is whether particular clones become susceptible to apoptosis. High levels of CD8-cell apoptosis in HIV-infected children have been demonstrated previously (17).
This study has demonstrated that the information gathered from qualitative PCR methods and from quantitative FC methods for the analysis of TCR Vβ repertoire is different. With many TCR Vβ families exhibiting discordance in the detection of deviation from the norm between the two methods, the significance of any single result remains unclear. Our study was conducted in chronically infected children receiving therapy; a clearer picture of the changes in TCR Vβ repertoire may be evident during the acute phase of response in the absence of treatment. Our understanding of the significance of assessment of the TCR Vβ repertoire during HIV infection in children is still evolving.
Acknowledgments
M.K. and T.W.M. contributed equally to this work.
This work was supported by Public Health Service grant NIH HD37345, AI48857, and AI28281.
We thank Maria Marecki, Herb Borrero, and Zhexiong Lian for assistance in sample preparation and Saroj Bakshi for assistance in obtaining samples.
REFERENCES
- 1.Blattman, J. N., D. J. D. Sourdive, K. Murali-Krishna, R. Ahmed, and J. D. Altman. 2000. Evolution of the T cell repertoire during primary, memory, and recall responses to viral infection. J. Immunol. 165:6081-6090. [DOI] [PubMed] [Google Scholar]
- 2.Callan, M. F. C., N. Steven, P. Krausa, J. D. K. Wilson, P. A. H. Moss, G. M. Gillespie, J. I. Bell, A. B. Rickinson, and A. J. McMichael. 1996. Large clonal expansions of CD8 T cells in acute infectious mononucleosis. Nat. Med. 2:906-911. [DOI] [PubMed] [Google Scholar]
- 3.Connors, M., J. A. Kovacs, S. Krevat, J. C. Gea-Banacloche, M. C. Sneller, M. Flanigan, J. A. Metcalf, R. E. Walker, J. Falloon, M. Baseler, R. Stevens, I. Feuerstein, H. Masur, and H. C. Lane. 1997. HIV induces changes in CD4+ T-cell phenotype and depletions within the CD4+ T-cell repertoire that are not immediately restored by antiviral or immune-based therapies. Nat. Med. 3:533-540. [DOI] [PubMed] [Google Scholar]
- 4.Cossarizza, A., C. Ortolani, C. Mussini, G. Guaraldi, N. Mongiardo, V. Borghi, D. Barbieri, E. Bellesia, M. G. Franseschini, B. D. Rienzo, and C. Franceschi. 1995. Lack of selective Vβ deletion in CD4 or CD8 T lymphocytes and functional integrity of TCR during acute HIV syndrome. AIDS 9:547-553. [DOI] [PubMed] [Google Scholar]
- 5.aDouek, D. C., J. M. Brenchley, M. R. Betts, D. R. Ambrozak, B. J. Hill, Y. Okamoto, J. P. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D. A. Price, M. Connors, and R. A. Koup. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95-98. [DOI] [PubMed] [Google Scholar]
- 6.Eiraku, N., R. Hingorani, S. Ijichi, K. Machigashira, P. K. Gregersen, J. Monteiro, K. Usuku, S. Yashiki, S. Sonoda, M. Osame, and W. W. Hall. 1998. Clonal expansion within CD4+ and CD8+ T cell subsets in human T lymphotrophic virus type I-infected individuals. J. Immunol. 161:6674-6680. [PubMed] [Google Scholar]
- 7.Faint, J. M., D. Pilling, A. N. Akbar, G. D. Kitas, G. D. Bacon, and M. Salmon. 1999. Quantitative flow cytometry for the analysis of T cell receptor Vβ chain expression. J. Immunol. Methods 225:53-60. [DOI] [PubMed] [Google Scholar]
- 8.Gorochov, G., A. U. Neumann, A. Kereveur, C. Parizot, T. Li, C. Katlama, M. Karmochkine, G. Raguin, B. Autran, and P. Debre. 1998. Perturbations of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat. Med. 4:215-221. [DOI] [PubMed] [Google Scholar]
- 9.Halapi, E., M. Jeddi-Tehrani, A. Blucher, R. Andersson, P. Rossi, H. Wigzell, and J. Grunewald. 1999. Diverse T-cell receptor CDR3 length patterns in human CD4+ and CD8+ T lymphocytes from newborns and adults. Scand. J. Immunol. 49:149-154. [DOI] [PubMed] [Google Scholar]
- 10.Hodara, V. L., M. Jeddi-Tehrani, J. Grunewald, R. Andersson, G. Scarlatti, S. Esin, V. Holmberg, O. Libonatti, and H. Wigzell. 1993. HIV infection leads to differential expression of T-cell receptor Vβ genes in CD4+ and CD8+ T cells. AIDS 7:633-638. [DOI] [PubMed] [Google Scholar]
- 11.Kalams, S. A., and B. D. Walker. 1998. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J. Exp. Med. 188:2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharbanda, M., S. Than, V. Chitnis, M. Sun, S. Chavan, S. Bakshi, and S. Pahwa. 2000. Patterns of CD8 T cell clonal dominance in response to change in antiretroviral therapy in HIV-infected children. AIDS 14:2229-2238. [DOI] [PubMed] [Google Scholar]
- 13.Maini, M. K., G. Casorati, P. Dellabona, A. Wack, and P. C. L. Beverley. 1999. T cell clonality in immune responses. Immunol. Today 20:262-266. [DOI] [PubMed] [Google Scholar]
- 14.McCloskey, T. W., V. Haridas, R. Pahwa, and S. Pahwa. 2002. T cell receptor Vβ repertoire of the antigen specific CD8 T lymphocyte subset of HIV infected children. AIDS 16:1459-1465. [DOI] [PubMed] [Google Scholar]
- 15.Mongkolsapaya, J., A. Jaye, M. F. C. Callan, A. F. Magnusen, A. J. McMichael, and H. C. Whittle. 1999. Antigen-specific expansion of cytotoxic T lymphocytes in acute measles virus infection. J. Virol. 73:67-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neisig, A., A. Vangsted, J. Zeuthen, and C. Geisler. 1993. Assembly of the T cell antigen receptor. J. Immunol. 151:870-879. [PubMed] [Google Scholar]
- 17.Niehues, T., T. W. McCloskey, J. Ndagijimana, G. Horneff, V. Wahn, and S. Pahwa. 2001. Apoptosis in T-lymphocyte subsets in human immunodeficiency virus-infected children measured immediately ex vivo and following in vitro activation. Clin. Diagn. Lab. Immunol. 8:74-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pahwa, S. 1988. HIV infection in children: nature of immunodeficiency, clinical spectrum, and management. Ped. Infect. Dis. J. 7:S61-S71. [PubMed] [Google Scholar]
- 19.Pantaleo, G., J. F. Demarest, H. Soudeyns, C. Graziosi, F. Denis, J. W. Adelsberger, P. Borrow, M. S. Saag, G. M. Shaw, R. P. Sekaly, and A. S. Fauci. 1994. Major expansions of CD8+ T cells with predominant Vβ usage during the primary immune response to HIV. Nature 370:463-467. [DOI] [PubMed] [Google Scholar]
- 20.Posnett, D. N., S. Kabak, A. S. Hodtsev, E. A. Goldberg, and A. Asch. 1993. T cell antigen receptor Vβ subsets are not preferentially deleted in AIDS. AIDS 7:625-631. [DOI] [PubMed] [Google Scholar]
- 21.Rebai, N., G. Pantaleo, J. F. Demarest, C. Ciurli, H. Soudeyns, J. W. Adelsberger, M. Vaccarezza, R. E. Walker, R. P. Sekaly, and A. S. Fauci. 1994. Analysis of the T-cell receptor β-chain variable region (Vβ) repertoire in monozygotic twins discordant for human immunodeficiency virus: evidence for perturbations of specific Vβ segments in CD4+ T cells of the virus-positive twins. Proc. Natl. Acad. Sci. USA 91:1529-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soudeyns, H., P. Champagne, C. L. Holloway, G. U. Silvestri, N. Ringuette, J. Samson, N. Lapointe, and R. P. Sekaly. 2000. Transient T cell receptor β-chain variable region-specific expansions of CD4+ and CD8+ T cells during the early phase of pediatric human immunodeficiency virus infection: characterization of expanded cell populations by T cell receptor phenotyping. J. Infect. Dis. 181:107-120. [DOI] [PubMed] [Google Scholar]
- 23.Soudeyns, H., N. Rebai, G. Pantaleo, C. Ciurli, T. Boghossian, R. P. Sekaly, and A. S. Fauci. 1993. The T cell receptor Vβ repertoire in HIV-1 infection and disease. Semin. Immunol. 5:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Spencer, J. V., and T. J. Braciale. 2000. Incomplete CD8(+) T lymphocyte differentiation as a mechanism for subdominant cytotoxic T lymphocyte responses to a viral antigen. J. Exp. Med. 191:1687-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Than, S., M. Kharbanda, V. Chitnis, S. Bakshi, P. K. Gregersen, and S. Pahwa. 1999. Clonal dominance pattern of CD8 T cells in relation to disease progression in HIV-infected children. J. Immunol. 162:3680-3686. [PubMed] [Google Scholar]
- 26.Wilson, J. K. D., G. S. Ogg, R. L. Allen, P. J. R. Goulder, A. Kelleher, A. K. Sewell, C. A. O'Callaghan, S. L. Rowland-Jones, M. F. C. Callan, and A. J. McMichael. 1998. Oligoclonal expansions of CD8+ T cells in chronic HIV infection are antigen specific. J. Exp. Med. 188:785-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zajac, A. J., J. N. Blattman, K. Murali-Krishna, et al. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng, W., S. Nakao, H. Takamatsu, A. Yachie, A. Takami, Y. Kondo, N. Sugimori, H. Yamazaki, Y. Miura, S. Shiobara, and T. Matsuda. 1999. Characterization of T-cell repertoire of the bone marrow in immune-mediated aplastic anemia: evidence for the involvement of antigen-driven T-cell response in cyclosporine-dependent aplastic anemia. Blood 93:3008-3016. [PubMed] [Google Scholar]