Abstract
Cytokines secreted by cells of the immune system can alter the behavior and properties of immune or other cells. At a site of inflammation, sets of cytokines interact with immune cells, and their combined effect is often more important than the function of one isolated component. Conventional techniques, such as enzyme-linked immunosorbent assays, generally require large quantities of cells to characterize a complete cytokine profile of activated lymphocytes. The Bio-Plex system from Bio-Rad Laboratories combines the principle of a sandwich immunoassay with the Luminex fluorescent-bead-based technology. We developed a multiplex cytokine assay to detect different cytokines simultaneously in culture supernatant of human peripheral blood mononuclear cells stimulated with antigen and with mitogen. Fifteen human cytokines (interleukin 1α [IL-1α], IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-13, IL-15, IL-17, IL-18, gamma interferon, and tumor necrosis factor alpha) were validated with a panel of healthy individuals, rheumatoid arthritis patients, and juvenile idiopathic arthritis patients. Comparing the multiplex assay with a regular enzyme-linked immunosorbent assay technique with this donor panel resulted in correlation coefficients for all cytokines ranging from 0.75 to 0.99. Intra-assay variance proved to be less then 10%, whereas interassay variability ranged between 10 and 22%. This multiplex system proved to be a powerful tool in the quantitation of cytokines. It will provide a more complete picture in differences between activated lymphocyte cytokine profiles from healthy individuals and those from patients with chronic inflammatory diseases.
Cytokines are soluble proteins that are secreted by cells of the immune system. These proteins can alter the behavior and properties of different cell types. Although cytokine functions are complex, cytokine profiles are highly relevant parameters of an immune response. Different cytokines possess biological overlapping functions, and they have the ability to regulate production of other cytokines. Therefore, analysis of the function of the complete set of cytokines expressed within microenvironments (e.g., a site of inflammation) is often of more value than analysis of a single isolated cytokine (13).
Cytokines can be quantitated at various levels. mRNA can be detected by real-time PCR; intracellular proteins can be measured by fluorescence-activated cell sorter staining of permeabilized cells, and secreted cytokines can be quantified with bioassays, enzyme-linked immunosorbent assays (ELISAs), radioactive immunosorbent assays, and microarrays. Multiplex assays for detection of cytokines at the mRNA (6) and cellular levels (16, 18) are commonly used. However, these assays have one or more limitations, like the need for a large sample volume or detection of precursor proteins rather than native secreted proteins. In addition, these techniques are time-consuming and laborious.
Recent advances concerning applications for the simultaneous detection of proteins have resulted in different particle-based flow cytometric assays. These assays have proven to be very useful in the simultaneous detection of cytokines in body fluids. Unfortunately, at present, either the number of different microspheres or the availability of predefined kits limits these assays (1, 3). The Bio-Plex system employing the Luminex multianalyte profiling technology (Lab-MAP) allows individual and multiplex analysis of up to 100 different analytes in a single microtiter well (20).
Our laboratory focuses on immunoregulation and immunotherapy of children with autoimmune diseases—in particular, juvenile idiopathic arthritis (JIA). Sample volumes are relatively small due to our patient population. For a number of cytokines, ready-to-use beads are available, but not for the full spectrum. To overcome these limitations, we chose to develop and validate our own multiplex assays with the Bio-Plex system. With this assay, we were able to detect human cytokines in antigen-stimulated peripheral blood mononuclear cell (PBMC) culture supernatants from both autoimmune and healthy individuals. We showed that it is a reliable, fast, and reproducible technique with a sensitivity that is comparable to that of conventional ELISAs.
MATERIALS AND METHODS
Cell isolation and cultures.
Heparinized blood samples were collected from five patients with rheumatoid arthritis (RA) and five patients with JIA that were seen at our hospital, as well as from four healthy adult control donors. Informed consent was obtained either from parents or directly from the individuals who were older than 12 years.
PBMCs were isolated by Ficoll-Paque density gradient centrifugation (1.077 g/cm3; Amersham Pharmacia Biotech AB, Uppsala, Sweden). All cultures were performed in RPMI 1640 tissue culture medium supplemented with 100 U of penicillin-streptomycin per ml, 2 mM l-glutamine (all from Gibco BRL, Gaithersburg, Md.), and 10% heat-inactivated human AB serum containing no detectable levels of cytokine (<1 pg/ml; Sanquin Blood Bank, Utrecht, The Netherlands). Cells were cultured in round-bottomed microtiter plates (Greiner, Alphen aan de Rijn, The Netherlands) at 2 × 105 cells per well and incubated for 96 h at 37°C with either medium only, 2.5 μg of concanavalin A (ConA; Calbiochem, La Jolla Calif.) per ml, 1.5 μg of tetanus toxoid (TT; National Institute of Public Health and the Environment [RIVM], Bilthoven, The Netherlands) per ml, or 7 μg of phytohemagglutinin (PHA; Murex Biotech, Dartford, United Kingdom) per ml. At the end of the culture period, supernatants were collected and stored at −80°C until analysis.
Cytokine reagent Bio-Plex system.
All antibody pairs used were directed against different noncompeting epitopes of their respective cytokines and were purchased from different commercial sources (Table 1). If necessary, antibodies were reconstituted in phosphate-buffered saline (PBS), pH 7.4. Sodium azide (NaN3) was removed from the capture antibodies with a Vivaspin 500 concentrator with a 10,000-molecular-weight cutoff polyethersulfone membrane (Vivascience, Lincoln, United Kingdom), which was spun three times at 10,000 × g with PBS used as the wash matrix. The protein recovery was determined with a bicinchoninic acid protein assay (Pierce, Rockford, Ill.). All recombinant proteins except interleukin-1α (IL-1α), IL-8, and IL-13 were reconstituted in PBS, pH 7.4, containing 0.5% bovine serum albumin (BSA; Sigma-Aldrich, Zwijndrecht, The Netherlands) to a concentration of 10 μg/ml. IL-8 and IL-13 were reconstituted to a concentration of 1 μg/ml, and IL-1α was reconstituted to a concentration of 245 ng/ml. All proteins were aliquoted and stored at −80°C.
TABLE 1.
Reagents used for Bio-Plex immunoassaysa
| Cytokine | Recombinant protein
|
Capture Ab
|
Detection Ab
|
|||||
|---|---|---|---|---|---|---|---|---|
| Catalogue no. | Source | Catalogue no. | Clone | Source | Catalogue no. | Clone | Source | |
| IL-1α | SD901 | BS | AHC0912 | 624B 3F2 | BS | AHC0419 | 840C 20B8 | BS |
| IL-1β | M6058 | CLB | M9313 | CLB/IL1B-8 | CLB | M9313 | CLB/IL1B-4 | CLB |
| IL-2 | 554603 | BD | 555051 | 5344.111 | BD | 555040 | B33-2 | BD |
| IL-4 | 204-IL-005 | R&D | MAB604 | 3010.211 | R&D | BAF204 | Polyclonal | R&D |
| IL-5 | PHC0055 | BS | AHC0954 | JES1-39D10 | BS | AHC0859 | JES1-5A10 | BS |
| IL-6 | M6077 | CLB | M9316 | CLB/IL6-16 | CLB | M9316 | Polyclonal | CLB |
| IL-8 | 89/520 | NIBCS | M9318 | CLB/IL8-1 | CLB | M9318 | Polyclonal | CLB |
| IL-10 | M6086 | CLB | M9310 | B-N10 | CLB | M9310 | B-T10 | CLB |
| IL-12p70 | 219-IL-005 | R&D | MAB611 | 24945.11 | R&D | BAF219 | Polyclonal | R&D |
| IL-13 | 94/622 | NIBCS | M9313 | CLB/IL13-1 | CLB | M9313 | CLB/IL13-1 | CLB |
| IL-15 | 247-IL-005 | R&D | MAB647 | 34505.11 | R&D | BAM247 | 34593.11 | R&D |
| IL-17 | 317-IL-050 | R&D | MAB317 | 41809.111 | R&D | BAF317 | Polyclonal | R&D |
| IL-18 | B001-5 | MBL | D044-3 | 125-2H | MBL | D045-6 | 159-12B | MBL |
| TNF-α | M6145 | CLB | M9323 | CLB/TNF-7 | CLB | M9323 | CLB/TNF-5 | CLB |
| IFN-γ | 554616 | BD | 554548 | NIB42 | BD | 554550 | 4S.B3 | BD |
The reagents listed were obtained from the sources indicated by the following abbreviations: BS, Biosource; CLB, Sanquin; NIBSC, National Institute for Biological Standards and Controls (Potters Bar, United Kingdom); BD, BD Biosciences; R&D, R&D Systems; MBL, Medical & Biological Laboratories (Naka-ku Nagoya, Japan). All capture antibodies are mouse or rat monoclonal reagents. Detection antibodies for IL-4, IL-6, IL-8, IL-12, and IL-17 are either sheep or goat; others are monoclonal mouse or rat. Ab, antibodies.
Covalent coupling of antibodies to microspheres.
Carboxylated polystyrene microspheres, numbers 132, 133, 134, 136, 137, 138, 142, 143, 145, 147, 152, 156, 158, 160, and 162, each with a distinct emitting fluorescence pattern, were purchased from Luminex Corporation (Austin, Tex.). Covalent coupling of the capture antibodies to the microspheres was performed by following the procedures recommended by Luminex. In short, the microspheres' stock solutions were dispersed in a sonification bath (Sonicor Instrument Corporation, Copiaque, N.Y.) for 2 min. An aliquot of 2.5 × 106 microspheres was resuspended in microtiter tubes (Eppendorf, Hamburg, Germany) containing 0.1 M sodium phosphate buffer, pH 6.1 (phosphate buffer), to a final volume of 80 μl. This suspension was sonicated until a homogeneous distribution of the microspheres was observed. Solutions of N-hydroxy-sulfosuccinimide (Sulfo-NHS) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (Pierce), both at 50 mg/ml, were prepared in phosphate buffer, and 10 μl of each solution was sequentially added to stabilize the reaction and activate the microspheres. This suspension was incubated for 10 min at room temperature and then resuspended in 250 μl of PBS containing 50 μg of antibody. The mixture was incubated overnight in the dark with continuous shaking. Microspheres were then incubated with 250 μl of PBS-0.05% Tween 20 for 4 h. After aspiration, the beads were blocked with 1 ml of PBS-1% BSA-0.1% sodium azide. The microspheres were counted with a hemacytometer and stored at a final concentration of 106 microspheres per ml in the dark at 4°C.
Coupling efficiency of monoclonal antibodies was tested by staining 2,000 microspheres with either fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG; BD Biosciences, San Diego, Calif.) or FITC-conjugated goat anti-rat IgG (Zymed Laboratories, San Francisco, Calif.) antibodies. Functionality of the coupling was determined by incubating 500 microspheres with a biotinylated antibody directed to the source of the capture antibody and a standard of 2,500 pg of recombinant cytokine protein per ml in a final volume of 70 μl of high-performance ELISA buffer (HPE; Sanquin, Amsterdam, The Netherlands). After incubation with streptavidin R-phycoerythrin (SA-PE; BD Biosciences) and being washed with PBS-1% BSA-0.5% Tween 20, the microspheres were measured and analyzed with the Bio-Plex system (Bio-Rad Laboratories, Hercules, Calif.). This system distinguishes and classifies each microsphere by making use of the specific fluorescence dyes that are internalized in the microspheres. Furthermore, a fluorescence reporter signal which does not interfere with the classification signals is detected. Data analysis was done with Bio-Plex Manager software (Bio-Rad Laboratories) with five-parametric-curve fitting.
Multiplex cytokine assays.
Each cytokine was tested in a single bead assay to determine the optimal concentration of detection antibody. Next, the microspheres were multiplexed and optimized for incubation times and reporter signal. As a reporter signal, streptavidin Alexa-532 (Molecular Probes, Leiden, The Netherlands) and SA-PE were tested in different concentrations. All assays were performed with the same matrix as the culture supernatants.
Calibration curves from recombinant cytokine standards were prepared with threefold dilution steps in RPMI 1640 medium containing 10% AB serum. Samples were measured twice, and blank values were subtracted from all readings. All assays were carried out directly in a 96-well round-bottomed microtiter plate (Muller Ratiolab, Dreieich, Germany) at room temperature and protected from light. A mixture containing 500 microspheres per cytokine (total volume, 10 μl/well) was incubated together with a cocktail of biotinylated antibodies (10 μl/well) and a standard, sample, or blank in a final volume of 70 μl for 20 min under continuous shaking. Beads were washed twice with PBS-1% BSA-0.5% Tween 20 in order to remove any residual biotin present from the culture medium as well as any unbound antibody. After 10 min of incubation with SA-PE (50 ng/well) and washing with PBS-1% BSA-0.5% Tween 20, the fluorescence intensity of the beads was measured in a final volume of 100 μl of HPE buffer.
Measurements and data analysis of all assays were performed with the Bio-Plex system in combination with the Bio-Plex Manager software.
Cytokine ELISA.
ELISA kits for IL-2 (R&D Systems, Abingdon, United Kingdom), IL-4 (Biosource, Nivelles, Belgium), IL-12p70 (BD Biosciences), and IL-18 (Diaclone, Besançon, France) contained precoated ELISA plates, and the assays were performed as described by the manufacturers. ELISA kits for IL-1α and IL-5 were obtained from Biosource, and IL-1β, IL-6, IL-8, IL-10, IL-13, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) ELISA kits were obtained from Sanquin (Amsterdam, The Netherlands). ELISA was performed by coating 96-well polystyrene microtiter plates (Nalge Nunc International, Roskilde, Denmark) with a specific monoclonal antibody. Standards and samples were added after the plates were blocked, and the plates were incubated overnight at 4°C. A specific biotinylated antibody was added to all wells after they were washed, and they were incubated for 1 h at room temperature. The plates were washed and incubated for 30 min with horseradish peroxidase-conjugated streptavidin. After removal of nonbound horseradish peroxidase conjugate by washing, 3-3′,5,5′-tetramethylbenzidine substrate reagent solution (ICN Biomedicals Inc., Aurora, Ohio) was added to the wells. The reaction was stopped by the addition of 1.8 M H2SO4. The absorbency of all ELISAs was read at 450 nm with a Milenia microtiter plate reader (Diagnostic Products Corporation Nederland BV, Breda, The Netherlands). Standard curves for the various cytokines ranging from 0.25 to 5 up to 300 to 1,000 pg/ml were constructed by a four-parameter regression formula and plotted as a linear curve (log-log). Cytokine concentrations of experimental samples were calculated with Elisa plus software version 3.01 (Meddata Inc., New York, N.Y.).
RESULTS
Specific antibody pairs were used to develop and validate a 15-plex human cytokine assay. Critical parameters of this assay are the choice of (primary) antibodies, the concentration of the biotinylated detection antibodies, the amount and choice of conjugated fluorochrome, and the incubation times of samples and the conjugated fluorochrome. Several commercial cytokine antibody pairs suitable for use in ELISA did not perform well in our multiplex system. Either the coupling to the beads was ineffective (based on negative signal with FITC-labeled IgG) or the beads failed to report a signal from recombinant cytokine.
Biotin-conjugated antibodies were optimized in a single bead assay before they were added to the multiplex profile. The amount of the conjugated antibody used varied per cytokine. Optimal concentrations for this assay ranged between 8 and 16 ng of conjugated detection antibody per well.
As a reporter signal, streptavidin Alexa-532 and SA-PE were tested in different concentrations ranging from 10 to 500 ng/well for Alexa-532 and 10 to 100 ng/well for SA-PE. Alexa-532 turned out to be less suitable for several of our antibodies because we were unable to pick up a signal at a different concentration of fluorochrome, whereas SA-PE did result in a signal. Therefore, we used SA-PE for further experiments. The optimal amount of SA-PE was 50 ng per well. Lower concentrations resulted in a decrease of the fluorescence intensity, while higher concentrations did not further increase the signal.
Prolonging incubation times for either the sample or SA-PE did not improve our ability to detect any cytokine. Overall median fluorescence intensity (MFI) signals did increase with longer incubation times, but sensitivity was lost due to a proportionally higher increase in the background signal. Washing the plates after the SA-PE incubation increased the sensitivity when short incubation times were used but had no effect with longer incubation times because it reduced both the background and overall MFI readings equally.
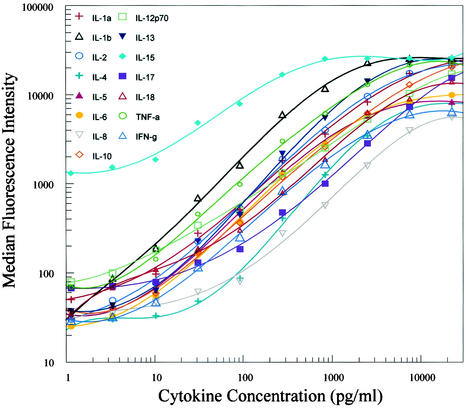
By combining threefold dilutions (22,500 to 1.1 pg/ml) of recombinant cytokines, standard curves for 15 cytokines were generated over a 4-log range. Data were obtained by measuring the MFI of each standard concentration with the Bio-Plex system. Using the Bio-Plex Manager software, we calculated curves with a five-parameter regression formula and the data were plotted as a log-log curve (Fig. 1). Large dynamic ranges were used to generate the curves so that, eventually, samples would not have to be concentrated or diluted. However, dynamic ranges for the standard curves differ. Most standard curves range between 5 and 5,000 pg/ml (Table 2). Some cytokines have a different dynamic range that can be accurately derived from this system. IL-5, IL-15, and IL-17 can be detected at a range from 10 to 5,000 pg/ml, whereas IL-4 and IL-8 can be detected accurately from approximately 30 pg/ml. At higher and lower concentrations, standard curves flattened out and lost linearity (Fig. 1).
FIG. 1.
Standard curves for recombinant cytokines. Data were generated by combining a threefold dilution of the standards starting at 22,500 pg/ml, coupled beads, and biotinylated antibodies in a 70-μl matrix solution incubated for 20 min. SA-PE was added after two washes and incubated for 10 min. After an additional wash, the median fluorescence intensity was measured with the Bio-Plex system. Standard curves were calculated with Bio-Plex Manager software by a five-parameter regression formula.
TABLE 2.
Dynamic concentration ranges of ELISA and the Bio-Plex systema
| Cytokine | Dynamic concn range (pg/ml)
|
|
|---|---|---|
| ELISA | Bio-Plex | |
| IL-1α | 3.3-800 | 2.4-5,000 |
| IL-1β | 3.6-300 | 2.4-5,000 |
| IL-2 | 31.2-2,000 | 4.8-5,000 |
| IL-4 | 0.8-50 | 39-2,500 |
| IL-5 | 7.8-1,000 | 9.8-5,000 |
| IL-6 | 2.0-450 | 4.8-5,000 |
| IL-8 | 6.1-480 | 30-10,000 |
| IL-10 | 3.1-600 | 4.8-10,000 |
| IL-12p70 | 7.8-500 | 4.8-10,000 |
| IL-13 | 1.3-250 | 4.8-5,000 |
| IL-15 | ND | 9.8-1,000 |
| IL-17 | ND | 9.8-10,000 |
| IL-18 | 15.6-1,000 | 2.4-5,000 |
| TNF-α | 4.1-1,000 | 2.4-5,000 |
| IFN-γ | 5.1-500 | 4.8-5,000 |
Dynamic concentration ranges of the linear part of the standard curve of each cytokine were measured with ELISA or in a multiplex assay using the Bio-Plex system. ND, not done.
Cross-reactivity between components during the assay had to be carefully assessed to monitor false-positive findings. To detect whether cross-reactivity occurred, a full-bead mixture with the detection antibodies for each bead was incubated three times in the presence of single cytokine standards at 2,500 pg/ml. Although the background varied with each bead, none of the specific antibody sets gave readings above background with nonrelevant cytokine standards, indicating that there was no detectable cross-reactivity. Positive readings were found for the microspheres labeled with the specific capture antibody (Fig. 2A).
FIG. 2.
Specificities of cytokine determination in a multiplex system. A complete set of 15 microspheres were mixed with all biotinylated antibodies and one single standard (2,500 pg/ml). (A) MFI was measured using the Bio-Plex system for IL-10 at 2,500 pg/ml; (B and C) comparison of Bio-Plex and ELISA results. Shown are regression lines for IL-1β (correlation coefficient, 0.96) (B) and IL-6 (correlation coefficient, 0.85) (C).
To determine the recovery of each cytokine, known amounts of recombinant cytokines (10,100 and 1,000 pg/ml) were spiked in a culture medium containing 10% human AB serum. These samples were treated as unknown samples and measured in a multiplex as duplicates in different assays. The recovery from the 100- and 1,000-pg/ml spiked samples averaged from 91 to 108% and from 98 to 107%, respectively (Table 3). There was a greater variance with the 10-pg/ml spiked samples. In general, recoveries varied between 81 and 121%, with the exception of IL-4, IL-8, and IL-17. Tests of IL-4 resulted in a recovery of 17 pg/ml, which is 170% of the spiked target, whereas tests of IL-8 and IL-17 resulted in a recovery that exceeded more than 250% (e.g., IL-8, 25.2 pg/ml; IL-17, 26.7 pg/ml).
TABLE 3.
Recovery of spiked cytokines in a multiplex assaya
| Cytokine | Cytokine recovery (pg/ml) at expected concn indicatedb
|
||
|---|---|---|---|
| 10 pg/ml | 100 pg/ml | 1,000 pg/ml | |
| IL-1α | 10.5 ± 1.3 | 97 ± 6 | 1,017 ± 51 |
| IL-1β | 10.1 ± 1.3 | 102 ± 12 | 1,020 ± 47 |
| IL-2 | 9.6 ± 1.1 | 100 ± 8 | 1,003 ± 57 |
| IL-4 | 17.0 ± 5.6 | 116 ± 19 | 998 ± 76 |
| IL-5 | 12.1 ± 3.7 | 108 ± 7 | 991 ± 66 |
| IL-6 | 10.4 ± 2.2 | 102 ± 19 | 1,003 ± 58 |
| IL-8 | 25.2 ± 16.1 | 97 ± 10 | 1,048 ± 87 |
| IL-10 | 9.3 ± 2.2 | 105 ± 5 | 1,014 ± 54 |
| IL-12p70 | 11.9 ± 3.1 | 102 ± 11 | 978 ± 60 |
| IL-13 | 11.7 ± 1.5 | 105 ± 6 | 1,018 ± 31 |
| IL-15 | 9.2 ± 1.1 | 94 ± 11 | 1,007 ± 47 |
| IL-17 | 26.7 ± 15.8 | 108 ± 18 | 1,045 ± 100 |
| IL-18 | 8.7 ± 1.2 | 94 ± 3.4 | 1,007 ± 81 |
| TNF-α | 11.6 ± 2.3 | 103 ± 12 | 1,042 ± 70 |
| IFN-γ | 8.1 ± 3.2 | 91 ± 6 | 1,073 ± 120 |
Known amounts of cytokines were spiked into RPMI culture medium. Samples were treated as unknown samples and determined in a multiplex assay. The mean ± standard deviation recoveries (picograms per milliliter) for the different amounts of each cytokine are shown.
Six experiments were done for each spiked cytokine.
For these latter cytokines, the error of spiking with 10 (and to a lesser degree also 100) pg/ml clearly comes from the lower detection range (e.g., flattened section) of the standard curves (Fig. 1).
The results of Bio-Plex analysis were compared with those of ELISA for PBMC culture supernatants from four healthy individuals containing an unknown concentration of each cytokine. The PBMCs were cultured under three different conditions: unstimulated, stimulated with antigen (TT), or stimulated with mitogen (either ConA or PHA). These stimulations resulted in at least four different concentrations that were widely disparate at the linear part of the standard curves of the multiplex assay. Correlations between both techniques were determined by measuring the levels of all cytokines with the Bio-Plex system. The levels of these cytokines, with the exception of IL-15 and IL-17, were also measured with conventional ELISA. All samples measured with the Bio-Plex system were assayed twice and at different time points so that the interassay variance could be determined.
The concentrations of the majority of samples measured with ELISA did concur with the concentrations that were determined with the Bio-Plex system (Fig. 2B). High correlation coefficients (r2), ranging from 0.75 to 0.99, were found for all cytokines (Table 4). Slopes varied between 0.70 and 1.21, with a mean slope of 0.98.
TABLE 4.
Correlation between Bio-Plex and ELISA resultsa
| Cytokine | r2 (n = 15) | Slope | CV intraassay (%) (n = 28) | CV interassay (%) (n = 4) |
|---|---|---|---|---|
| IL-1α | 0.986 | 1.15 | 8.2 | 17.8 |
| IL-1β | 0.964 | 0.97 | 9.6 | 10.3 |
| IL-2 | 0.991 | 0.93 | 9.5 | 14.4 |
| IL-4 | 0.950 | 0.82 | 8.9 | 16.4 |
| IL-5 | 0.853 | 1.10 | 9.6 | 20.1 |
| IL-6 | 0.847 | 0.87 | 7.6 | 15.5 |
| IL-8 | 0.946 | 1.04 | 7.1 | 14.7 |
| IL-10 | 0.926 | 0.86 | 8.6 | 16.1 |
| IL-12p70 | 0.850 | 1.02 | 8.5 | 11.8 |
| IL-13 | 0.867 | 0.70 | 8.4 | 21.4 |
| IL-15 | ND | ND | 8.9 | 13.4 |
| IL-17 | ND | ND | 9.5 | 17.9 |
| IL-18 | 0.976 | 1.21 | 9.6 | 21.3 |
| TNF-α | 0.911 | 0.86 | 7.6 | 16.6 |
| IFN-γ | 0.750 | 1.18 | 9.1 | 20.3 |
Regression coefficients (r2) of the cytokine concentration express the correlation of 13 human cytokines detected in culture supernatants by ELISA and a 15-plex assay run on the Bio-Plex system. Intraassay variability, expressed as coefficient of variation (CV = standard deviation divided by the mean), was calculated based on the average of 14 patient samples that were either untreated or treated with different stimuli (TT, ConA, or PHA) measured twice and run at two different time points. Interassay variation was measured by determining quadruplicates of one standard (1,000 pg/ml) at four different time points. n, number of samples; ND, not done.
Intra-assay variability, expressed as a coefficient of variation, was calculated based on the average for 14 patient samples that were either not stimulated or were treated with different stimuli (TT, ConA, or PHA) and measured twice in a multiplex assay repeated at two different time points. The intra-assay variability within the replicates of the samples is comparable to that of ELISA (variability of <10%) with an average coefficient of variation of 8.7% (Table 4).
Interassay variability was evaluated by testing quadruplicates of one standard that was positioned at the upper linear part of the standard curve of all cytokines (1,000 pg/ml) in a multiplex assay at four different time points. The variabilities of these samples were between 10 and 22%, with an average of 16.5% (Table 4).
To gain insight into the differences between activated lymphocyte cytokine profiles from healthy individuals and profiles from patients with chronic inflammatory diseases, we stimulated the PBMCs of healthy donors, RA patients, and JIA patients in vitro with antigen (TT) and with mitogen (PHA). The multiplex data were digitized to create a cytokine portrait, enabling the complete spectrum of cytokines to be visualized (Fig. 3). Differences in cytokine profiles under different stimuli can be observed. Stimulation with PHA resulted in higher cytokine concentrations than those from TT stimulation. Figure 3 shows that IL-10 was increased following TT activation in healthy donors (82 ± 74 pg/ml [mean ± standard deviation]) compared to that in RA patients (16 ± 13 pg/ml). IL-17 production, both after culture with medium or TT, was higher in adult donors (nonstimulated, 145 ± 105 pg/ml; TT-stimulated, 177 ± 73 pg/ml) as was IL-17 production in juveniles (medium, 0.2 ± 0.3 pg/ml; TT, 25.3 ± 15.8 pg/ml). Furthermore, IL-1α, IL-4, IL-12, IL-15, and IL-18 were hardly induced by stimulation with TT. IL-8 production was induced without external stimulation in cultures of all patient samples as measured in vitro (4,466 ± 3,031 pg/ml). These data indicate that multiplex cytokine analysis can be used either as a quantitative or comparative tool for the study of the human in vitro cellular immune response.
FIG. 3.
Cytokine profiles of in vitro-stimulated human lymphocytes. PBMCs from healthy controls (HC), (RA), and JIA patients were stimulated for 96 h with medium only, TT, or PHA. Supernatants were collected and cytokine levels were measured by the Bio-Plex system. The mean cytokine concentration (picograms per milliliter) of the TT stimulation is plotted at two-thirds of the logarithmic scale. Yellow squares indicate that the corresponding cytokine is undetectable (lower detection limit of the assay); black squares indicate the upper detection limit of the cytokine (see Table 2); blank squares indicate that the cytokine was not tested.
DISCUSSION
Previous studies have validated the Lab-MAP technology for the detection of cytokines in serum or in supernatants of cultures of mitogen-stimulated PBMCs from humans and mice (2, 11, 15). Here we report a validation of a multiplex cytokine assay for the detection of human cytokines in antigen- and mitogen-stimulated PBMC cultures. The results show that our multiplex assay is comparable in sensitivity, accuracy, and reproducibility to the “gold standard” which is the ELISA.
The fluorescent-bead-based detection assay has a clear advantage above the conventional ELISA, i.e., the ability to detect large numbers of analytes simultaneously, and therefore provides a powerful tool for profiling multiple cytokines (4). Theoretically, this assay can detect up to 100 analytes in a single sample. The volume required to detect all analytes would be sufficient to test a single cytokine by ELISA. Obviously, this is a major advantage when working with small sample volumes and will increase with each additional cytokine measured in a multiplex assay.
Moreover, this multiplex assay is sensitive and accurate since each fluorescence signal is the mean of 100 measurements of a single microsphere, and each microsphere represents an assay by itself. Although higher numbers of events are commonly used in particle-based flow cytometry, using as few as 100 events will not affect the precision of the assay (2).
Each standard curve can be adjusted to a biological range in which a sample can be expected depending on the origin of the specimen. With this wide dynamic range of standard curves, samples do not have to be diluted or concentrated. Nevertheless, for some applications, the lower limits of the standard curves will be the limiting factor of this assay. For example, we present a standard curve for IL-4 where the lowest concentration that can be detected in the linear part is approximately 40 pg/ml, whereas a sensitive ELISA for IL-4 will have a lower detection limit.
We have shown that this technique can be used to generate quantitative and comparative cytokine data in a given time period which is shorter than that required by a standard ELISA procedure. However, data acquisition becomes more complex when a large number of samples are measured with a multiplex assay. Therefore, we digitized our graphics for the analysis of cytokine profiles. With this kind of representation, this technique will improve the monitoring of cellular responses of individuals within a patient population (quantitative) or between different patient populations (comparative). Specific treatment of autoimmune patients can result in cytokine profile changes that are difficult to detect or monitor with existing techniques. For instance, nonresponders to a vaccination with recombinant hepatitis B will have a diminished Th1 and Th2 response compared to that of responders (10), whereas therapy of arthritic patients can increase the levels of the whole cytokine repertoire compared with those of control patients (8). Furthermore, immunotherapy of atopic patients can shift the allergen-specific response from a Th2 to a Th1 phenotype (17). These dynamic changes in cytokine profiles can be easily detected or monitored with only a small sample volume and can be used as a prognostic factor before, during, or after immunomodulatory therapy.
This multiplex cytokine assay involves mixing a large number of different antibodies in a single reaction. These antibodies could bind nonspecifically to any other component within the assay and serve as antigens for other immunoglobulins, causing false-positive values or blocking the readout (7). Thus, when sera are used as the matrix, the matrix has to be carefully monitored for blocking substances like heterophilic antibodies and for the presence of cytokines, even when used in high dilutions. We have encountered this phenomenon with the IL-13 standard curve that was blocked completely by both ELISA and the Bio-Plex system when a cell culture matrix was used. HPE buffer resulted in a good readout. This effect was overcome by switching to another batch of human serum that was tested for such a blocking event and did not contain any detectable cytokines.
Since this multiplex assay has been set up as a sandwich immunoassay, it will enhance specificity and reduce the risk of cross-recognition with other proteins (7). Hence, specificity could be increased when additional antibodies are added to the multiplex assay. Within a liquid phase, cytokines will bind with high-affinity antibodies. Due to this liquid phase, the chance of interaction of proteins with their complementary antibodies will increase. This effect will lead to less nonspecific binding and thus to lower background signals.
The Lab-MAP technique is not limited to measurement of cytokine profiles. Any soluble factor can be quantified by this system. Current applications include detection of subclasses of immunoglobulins (5), virus-specific antibodies (9, 12), pneumococcal capsular polysaccharides (14), analysis of single-nucleotide polymorphisms, (19, 22), and gene expression (21).
From the immunological point of view, the limitation of this technique is the availability of matched antibody pairs that do not cross-react in this system. With its current versatility, multiplex cytokine analysis will allow direct and complete measurement of regulatory circuits of the human immune system.
Acknowledgments
We thank Huib de Jong (Department of Rheumatology and Clinical Immunology, University Medical Center Utrecht), who performed the in vitro cultures of all RA patients. Furthermore, we thank Peter van Kooten, Corlinda ten Brink, Suzanne Berlo (Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University), Danielle Wolvers, and Diny Knol (Unilever Health Institute, Vlaardingen, The Netherlands) for their support and valuable discussions.
W. de Jager and B. J. Prakken were supported by the Dutch Rheumatoid Arthritis Foundation (Nationaal Reumafonds).
REFERENCES
- 1.Camilla, C., L. Mély, A. Magnan, B. Casano, S. Prato, S. Debono, F. Montero, J. P. Defoort, M. Martin, and V. Fert. 2001. Flow cytometric microsphere-based immunoassay: analysis of secreted cytokines in whole-blood samples from asthmatics. Clin. Diagn. Lab. Immunol. 8:776-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carson, R. T., and D. A. Vignali. 1999. Simultaneous quantitation of 15 cytokines using a multiplexed flow cytometric assay. J. Immunol. Methods 227:41-52. [DOI] [PubMed] [Google Scholar]
- 3.Cook, E. B., J. L. Stahl, L. Lowe, R. Chen, E. Morgan, J. Wilson, R. Varro, A. Chan, F. M. Graziano, and N. P. Barney. 2001. Simultaneous measurement of six cytokines in a single sample of human tears using microparticle-based flow cytometry: allergics vs. non-allergics. J. Immunol. Methods 254:109-118. [DOI] [PubMed] [Google Scholar]
- 4.Fulton, R. J., R. L. McDade, P. L. Smith, L. J. Kienker, and J. R. Kettman, Jr. 1997. Advanced multiplexed analysis with the FlowMetrix system. Clin. Chem. 43:1749-1756. [PubMed] [Google Scholar]
- 5.Gordon, R. F., and R. L. McDade. 1997. Multiplexed quantification of human IgG, IgA, and IgM with the FlowMetrix system. Clin. Chem. 43:1799-1801. [PubMed] [Google Scholar]
- 6.Halminen, M., M. Sjoroos, M. J. Makela, M. Waris, E. Terho, T. Lovgren, and J. Ilonen. 1999. Simultaneous detection of IFN-gamma and IL-4 mRNAs using RT-PCR and time-resolved fluorometry. Cytokine 11:87-93. [DOI] [PubMed] [Google Scholar]
- 7.Hennig, C., L. Rink, U. Fagin, W. J. Jabs, and H. Kirchner. 2000. The influence of naturally occurring heterophilic anti-immunoglobulin antibodies on direct measurement of serum proteins using sandwich ELISAs. J. Immunol. Methods 235:71-80. [DOI] [PubMed] [Google Scholar]
- 8.Hidaka, T., K. Suzuki, M. Kawakami, M. Okada, K. Kataharada, T. Shinohara, M. Takamizawa-Matsumoto, and F. Ohsuzu. 2001. Dynamic changes in cytokine levels in serum and synovial fluid following filtration leukocytapheresis therapy in patients with rheumatoid arthritis. J. Clin. Apheresis 16:74-81. [DOI] [PubMed] [Google Scholar]
- 9.Jones, L. P., H. Q. Zheng, R. A. Karron, T. C. T. Peret, C. Tsou, and L. J. Anderson. 2002. Multiplex assay for detection of strain-specific antibodies against the two variable regions of the G protein of respiratory syncytial virus. Clin. Diagn. Lab. Immunol. 9:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kardar, G. A., M. Jeddi-Tehrani, and F. Shokri. 2002. Diminished Th1 and Th2 cytokine production in healthy adult nonresponders to recombinant hepatitis B vaccine. Scand. J. Immunol. 55:311-314. [DOI] [PubMed] [Google Scholar]
- 11.Kellar, K. L., R. R. Kalwar, K. A. Dubois, D. Crouse, W. D. Chafin, and B. E. Kane. 2001. Multiplexed fluorescent bead-based immunoassays for quantitation of human cytokines in serum and culture supernatants. Cytometry 45:27-36. [DOI] [PubMed] [Google Scholar]
- 12.Martins, T. B. 2002. Development of internal controls for the Luminex instrument as part of a multiplex seven-analyte viral respiratory antibody profile. Clin. Diagn. Lab. Immunol. 9:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Garra, A., and K. Murphy. 1994. Role of cytokines in determining T-lymphocyte function. Curr. Opin. Immunol. 6:458-466. [DOI] [PubMed] [Google Scholar]
- 14.Pickering, J. W., T. B. Martins, R. W. Greer, M. C. Schroder, M. E. Astill, C. M. Litwin, S. W. Hildreth, and H. R. Hill. 2002. A multiplexed fluorescent microsphere immunoassay for antibodies to pneumococcal capsular polysaccharides. Am. J. Clin. Pathol. 117:589-596. [DOI] [PubMed] [Google Scholar]
- 15.Prabhakar, U., E. Eirikis, and H. M. Davis. 2002. Simultaneous quantification of proinflammatory cytokines in human plasma using the LabMAP assay. J. Immunol. Methods 260:207-218. [DOI] [PubMed] [Google Scholar]
- 16.Prussin, C., and D. D. Metcalfe. 1995. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J. Immunol. Methods 188:117-128. [DOI] [PubMed] [Google Scholar]
- 17.Romagnani, S. 2000. The role of lymphocytes in allergic disease. J. Allergy Clin. Immunol. 105:399-408. [DOI] [PubMed] [Google Scholar]
- 18.Tam, S. W., R. Wiese, S. Lee, J. Gilmore, and K. D. Kumble. 2002. Simultaneous analysis of eight human Th1/Th2 cytokines using microarrays. J. Immunol. Methods 261:157-165. [DOI] [PubMed] [Google Scholar]
- 19.Taylor, J. D., D. Briley, Q. Nguyen, K. Long, M. A. Iannone, M. S. Li, F. Ye, A. Afshari, E. Lai, M. Wagner, J. Chen, and M. P. Weiner. 2001. Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. BioTechniques 30:661-669. [DOI] [PubMed] [Google Scholar]
- 20.Vignali, D. A. 2000. Multiplexed particle-based flow cytometric assays. J. Immunol. Methods 243:243-255. [DOI] [PubMed] [Google Scholar]
- 21.Yang, L., D. K. Tran, and X. Wang. 2001. BADGE, beads array for the detection of gene expression, a high-throughput diagnostic bioassay. Genome Res. 11:1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye, F., M. S. Li, J. D. Taylor, Q. Nguyen, H. M. Colton, W. M. Casey, M. Wagner, M. P. Weiner, and J. Chen. 2001. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Hum. Mutat. 17:305-316. [DOI] [PubMed] [Google Scholar]