Abstract
Ebola virus consists of four genetically distinguishable subtypes. We developed monoclonal antibodies (MAbs) to the nucleoprotein (NP) of Ebola virus Zaire subtype and analyzed their cross-reactivities to the Reston and Sudan subtypes. We further determined the epitopes recognized by these MAbs. Three MAbs reacted with the three major subtypes and recognized a fragment containing 110 amino acids (aa) at the C-terminal extremity. They did not show specific reactivities to any 10-aa short peptides in Pepscan analyses, suggesting that these MAbs recognize conformational epitope(s) located within this region. Six MAbs recognized a fragment corresponding to aa 361 to 461 of the NP. Five of these six MAbs showed specific reactivities in Pepscan analyses, and the epitopes were identified in two regions, aa 424 to 430 and aa 451 to 455. Three MAbs that recognized the former epitope region cross-reacted with all three subtypes, and one that recognized the same epitope region was Zaire specific. One MAb, which recognized the latter epitope region, was reactive with Zaire and Sudan subtypes but not with the Reston subtype. These results suggest that Ebola virus NP has at least two linear epitope regions and that the recognition of the epitope by MAbs can vary even within the same epitope region. These MAbs showing different subtype specificities might be useful reagents for developing an immunological system to identify Ebola virus subtypes.
Ebola virus is a filamentous negative-strand RNA virus, which naturally infects humans and nonhuman primates. In these hosts, Ebola virus causes severe hemorrhagic fever with very high mortality (27, 28). Despite an extensive search, the natural reservoir of Ebola virus is not yet known (2, 10). Ebola virus consists of four genetically distinguishable subtypes: Zaire, Sudan, Côte d'Ivoire, and Reston (3). The former three cause severe hemorrhagic fever both in humans and nonhuman primates. Of them, the Zaire and Sudan subtypes have been the cause of major outbreaks (21, 26, 27, 29). The mortality in the infected patients varies depending on the subtype (1, 26, 29). Notably, the Reston subtype has infected humans on several occasions but with no associated clinical symptoms (12, 13, 17), whereas it caused severe hemorrhagic fever in nonhuman primates, especially in Macaca monkeys, like the other subtypes (4, 14). Therefore, it is important to distinguish between these subtypes and to elucidate the molecular basis for the differences.
Ebola virus contains seven structural proteins (16). Nucleoprotein (NP) is one of the most abundant structural proteins among the Ebola virus-encoded proteins (8) and consists of 739 (Zaire and Reston) or 738 (Sudan) amino acids (aa) (6, 20) (GenBank accession no. AF173836). The N-terminal half of the NP is highly hydrophobic and relatively conserved among subtypes, whereas the C-terminal half is variable. Cytotoxic T cells specific to aa 43 to 53 of NP have shown the potential to protect animals from experimental Ebola virus infection, although NP-specific antibodies failed to protect these animals (25).
Monoclonal antibodies (MAbs) specific to the glycoprotein (GP) of Ebola virus have been reported, and functional aspects of these MAbs were analyzed (7, 11, 23, 24). Several linear epitopes on GP molecules were also defined (24). However, considering its abundance and strong antigenicity, NP should be a better target for viral antigen detection. We have been developing MAbs to the NP of Ebola virus subtype Zaire for laboratory diagnostic purposes. We have already reported that an MAb that recognizes 26 aa near the C terminus is reactive with at least three subtypes of Ebola virus NP, and we applied this MAb to an antigen capture enzyme-linked immunosorbent assay (ELISA) (15). In the present study, we report MAbs that show different Ebola virus subtype specificities, and define linear epitopes on NP recognized by these MAbs.
MATERIALS AND METHODS
Cells.
The P3/Ag568 myeloma cell line and all hybridomas were maintained in RPMI 1640 (Life Technologies, Rockville, Md.) supplemented with 10% fetal bovine serum and antibiotics (100 U of penicillin and 100 μg of streptomycin/ml; Life Technologies). HAT supplement (100 μM sodium hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine; Life Technologies) was added as necessary. Tn5 cells were maintained in TC100 (Life Technologies) supplemented with 5% tryptose phosphate broth (Difco, Detroit, Mich.), 10% fetal bovine serum, and kanamycin (60 μg/ml; Meiji Seika, Tokyo, Japan).
Recombinant proteins.
The full-length recombinant NP (rNP) of Ebola virus Zaire subtype (Mayinga strain) was expressed in Tn5 cells with a histidine tag at the N-terminal end by using the recombinant baculovirus system and then purified as described previously (18). Partial peptides of Zaire NP were expressed with a glutathione S-transferase (GST) tag at the N-terminal end in bacteria by using a pGEX2T vector (Amersham Pharmacia, Little Chalfont, United Kingdom). The expressed fusion proteins were purified by using glutathione-agarose as described previously (18). NP-5, NP-6, NP-7, and NP-8 represent aa 361 to 461, aa 451 to 551, aa 541 to 640, and aa 630 to 739 (C-terminal end) of Zaire NP, respectively (18). For the comparison of reactivities among the subtypes, GST fusion peptides corresponding to aa 421 to 458 were expressed by pGEX2T in bacteria according to the amino acid sequences of Zaire (20), Reston (6), and Sudan (GenBank accession no. AF173836) subtypes, respectively.
Immunological assays.
ELISA was performed as reported previously (18) with purified rNP or partial NP peptides as the antigen. Linear epitopes on the NP were determined by using Pepscan (Chiron Technologies, Clayton, Australia) according to the manufacturer's instructions. Briefly, 194 peptides were prepared as 14-aa biotinylated peptides, including a 4-aa spacer sequence (SGSG) at the N-terminal end, according to the amino acid sequence of the Zaire subtype. The peptides were shifted by 1 aa, with a consecutive overlap of 9 aa to cover the entire NP-5 (aa 361 to 461) and NP-8 (aa 630 to 739) fragments. Immunoplates were coated with streptoavidin and blocked by sodium casein. The biotinylated peptides were added to each well, and the bound peptides were examined for their reactivity to MAbs with horseradish peroxidase-labeled anti-mouse immunoglobulin G (IgG; H+L) conjugate (Zymed, San Francisco, Calif.) and ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate (Roche Diagnostics, Mannheim, Germany). The average and standard deviation (SD) of each MAb were calculated based on the optical density (OD) values for the entire peptide set representing either NP-5 or NP-8. The peptides that showed ODs above the average + 2 SDs were considered positive. The peptide whose neighboring two peptides were both positive was also considered positive. To determine subtype specificities, 14-aa biotinylated-peptides, including a 4-aa spacer sequence (SGSG) at the N-terminal end, corresponding to aa 421 to 430 and aa 449 to 458 were synthesized according to the amino acid sequences of the Reston, Sudan, and Zaire subtypes, respectively (Sawady Technology, Tokyo, Japan).
Immunoblot analyses were performed with a semidry apparatus by a standard method with nylon membranes (Millipore, Bedford, Mass.). Reacted MAbs were detected with the horseradish peroxidase-labeled anti-mouse IgG (H+L) conjugate and a POD immunostain set (Wako Pure Chemicals, Osaka, Japan). Indirect immunofluorescence assay (IFA) was done as previously described with HeLa cells stably expressing rNP derived from either the Zaire or the Reston subtype as the antigen (19). IFA with the Sudan subtype was performed with an IFA antigen provided by J. B. McCormick (then at the Centers for Disease Control and Prevention, Atlanta, Ga.) in 1987. This antigen consisted of Maleo strain-infected Vero E6 cells fixed in acetone after gamma irradiation. The cDNA encoding the NP of the Reston subtype was isolated from infected monkey specimens as previously described (6) and stably expressed in HeLa cells (6a).
MAbs.
Hybridomas and MAbs were prepared as described previously (15). Briefly, BALB/c mice were immunized with purified rNP of the Zaire subtype that was expressed by using a baculovirus expression system. The spleen cells were fused with P3/Ag568 myeloma line cells, and hybridomas were established. The hybridomas were primarily screened by ELISA with purified rNP of the Zaire subtype. For IFA, either mouse ascitic fluids or culture supernatants of the hybridomas were used at a dilution of 1:100 or without dilution, respectively. For ELISA and immunoblotting, the ascitic fluids and culture supernatants were used at dilutions of 1:1,000 and 1:10, respectively. Isotypes of the MAbs were determined by using a mouse MAb isotyping kit (Life Technologies).
RESULTS
Subtype specificity of MAbs in IFA.
Ebola virus subtype specificity of 11 IgG MAbs was examined by IFA. Although these MAbs were initially selected on the basis of their reactivity to the rNP of the Zaire subtype in ELISA, all were reactive in IFA, as well with HeLa cells expressing rNP of Zaire (Table 1). Eight MAbs reacted with all three subtypes. MAb 1-7C reacted with the Zaire and Sudan subtypes but not with the Reston. 2-11G reacted with the Zaire and Reston subtypes but not with the Sudan. 3-9E reacted only with the Zaire subtype. These results that suggest these MAbs recognize different antigenic epitopes on the NP.
TABLE 1.
Summary of the MAb characteristics
| MAb | Iso- type | IFA resultd with subtype:
|
Recognition resultd
|
||||
|---|---|---|---|---|---|---|---|
| Zaire | Reston | Sudan | No. of fragmentsa | Peptide no.b | aac | ||
| 3-9E | G1 | + | − | − | 5 | 420-424 | D424DDIPF429 |
| 3-3A | G2b | + | + | + | 5 | 421-426 | D426IPFP430 |
| 1-5D | G1 | + | + | + | 5 | 421-426 | D426IPFP430 |
| 2-9D | G1 | + | + | + | 5 | 421-426 | D426IPFP430 |
| 1-7C | G1 | + | − | + | 5 and 6 | 446-451 | D451TTIP455 |
| 3-7D | G1 | + | + | + | 5 | ||
| 3-3D | G1 | + | + | + | 8 | ||
| 3-1E | G1 | + | + | + | 8 | ||
| 1-9E | G3 | + | + | + | 8 | ||
| 2-11G | G1 | + | + | − | 8 | ||
| 2-11C | G1 | + | + | + | |||
That is, the number of GST-fused partial fragments that showed reactivity with each MAb in an ELISA.
Peptide numbers reactive in the Pepscan analyses are shown as the first amino acid number of the representative amino acid sequence of the NP.
The minimum amino acid sequence recognized by each MAb was deduced from the results of the Pepscan analyses. The positions of the amino acids in the NP sequence are in subscript.
“+” indicates a positive reaction, and “−” indicates a negative reaction.
Definition of the epitopes recognized by the MAbs.
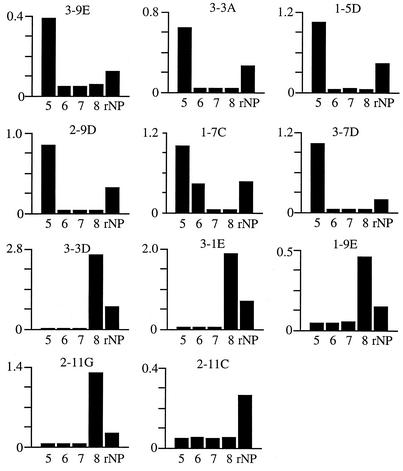
We attempted to elucidate the basis for the different patterns of specificity of these MAbs. The localization of epitopes was determined by ELISA with four partial overlapping fragments, NP-5 to NP-8, representing the C-terminal half of the NP (Fig. 1). Six clones recognized the NP-5 fragment, indicating that the epitopes of these MAbs were located within aa 361 to 461. Among them, MAb 1-7C was reactive with both NP-6 and NP-5. Four MAbs recognized NP-8, indicating that they recognize amino acid sequences within the sequence from aa 630 to 739. 2-11C did not show specific reactivity with any of the partial fragments and was not characterized further.
FIG. 1.
Localization of antigenic epitopes on the NP. The reactivity of each MAb with GST-fused partial polypeptides of the NP (Zaire) was examined in ELISA. NP-5, NP-6, NP-7, and NP-8 (columns 5, 6, 7 and 8, respectively) represent aa 361 to 461, aa 451 to 551, aa 541 to 640, and aa 630 to 739 of the NP, respectively. As a positive control, rNP (entire open reading frame) expressed with a histidine tag was included. Results are expressed as absorbance at OD at 405 nm (OD405).
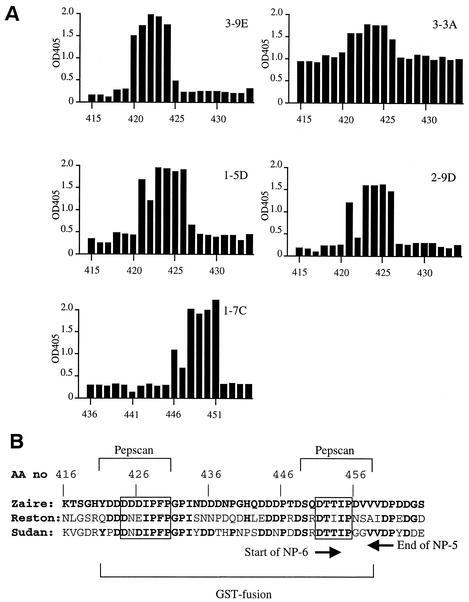
To further localize the epitopes recognized by these 10 MAbs, Pepscan analyses were employed. Selected results are shown in Fig. 2A. No MAb specific to NP-8 (3-3D, 3-1E, 1-9E, and 2-11G) was reactive to any oligopeptides (data not shown), indicating that these MAbs recognize conformational epitope(s). Five of six MAbs specific to NP-5 showed reactivities in the Pepscan analysis. 3-9E reacted with 10-aa oligopeptides starting from aa 420, 421, 423, and 424 of the NP, indicating the recognition of the amino acid sequence of D424DDIPF429 (Fig. 2B). 3-3A, 1-5D, and 2-9D reacted with peptides starting from aa 421 through 426, indicating the recognition of D426IPFP430. 3-3A inherently showed higher background in this assay; yet it showed a unique positive spike at these peptides. 1-7C reacted with peptides starting from aa 446 through 451, indicating the recognition of D451TTIP455, which is on both NP-5 and -6 (Fig. 2B). This agreed with the results that 1-7C was reactive to both NP-5 and -6 in ELISA. 3-7D failed to react with any of the 10-aa oligopeptides (data not shown).
FIG. 2.
(A) Pepscan analyses to determine the linear antigenic epitopes recognized by MAbs. Each overlapping peptide contains a 10-aa sequence of NP (Zaire) and is indicated by the position of the first amino acid. Results are expressed as absorbance at OD405. The average absorbance values (SDs) for each MAb were as follows 0.31 (0.36), 1.07 (0.17), 0.43 (0.38), 0.28 (0.30), and 0.33 (0.38) for 3-9E, 3-3A, 1-5D, 2-9D, and 1-7C, respectively. (B) Amino acid sequences, including the linear epitope regions on the NP of three Ebola virus subtypes. The sequence of Zaire and the identical amino acid in the Reston and Sudan sequences are shown in boldface. Amino acid positions in the entire NP (AA no) are indicated above the sequence. The last and the first amino acid for NP-5 and NP-6, respectively, are indicated by arrows below the sequences. The regions used in the immunoblot assay (GST fusion) and ELISA (Pepscan) for comparison among subtypes are also indicated below and above the sequences, respectively. Epitope regions identified by Pepscan are boxed.
Cross-reactivities of MAbs in immunoblotting and ELISA.
We next examined the subtype specificities of these five epitope-mapped MAbs by other immunological methods. Reactivities in immunoblot analyses were examined by using GST fusion peptides representing the epitope regions (aa 421 to 458) of the Zaire, Reston, and Sudan subtypes (Fig. 2B and 3). 3-3A, 1-5D, and 2-9D were reactive with all three subtypes in the immunoblotting, as in IFA. 3-9E and 1-7C also showed the same reaction pattern in the immunoblotting as in IFA; 3-9E reacted with only the Zaire subtype, whereas 1-7C reacted with the Zaire and Sudan but not with the Reston subtype.
FIG. 3.
(A) Reactivities of MAbs to Ebola virus subtypes in immunoblot assay. GST fusion polypeptides representing aa 421 to 458 of the NPs of Zaire, Reston, and Sudan subtypes were expressed in E. coli, and equal amounts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After samples were blotted to nylon membranes, the reactivity of the MAbs was examined. (B) Reactivities of MAbs to Ebola virus subtypes in ELISA. Oligopeptides containing the linear epitope regions (aa 421 to 430 and aa 449 to 458, respectively) of the NPs of Zaire (Z), Reston (R), and Sudan (S) subtypes were examined by ELISA. For 3-9E, 3-3A, 1-5D, and 2-9D, the oligopeptide representing aa 421 to 430 was used. For 1-7C, the oligopeptide representing aa 449 to 458 was used. The results are expressed as the absorbance at OD405.
The recognition patterns among the three subtypes were further examined by ELISA by using 14-mer oligopeptides representing the epitope regions (Fig. 2B and 3B). 3-3A, 1-5D, and 2-9D showed reactivity to all three subtypes, as in the other two assays. 3-9E reacted only with the Zaire, whereas 1-7C reacted with both the Zaire and Sudan subtypes but not with the Reston subtype, as in IFA and immunoblotting. The reactivity of 1-7C with the Sudan subtype was, however, weaker than that with the Zaire subtype. These results indicate that 3-9E is Zaire specific and that 1-7C is cross-reactive for Zaire and Sudan subtypes, respectively. 3-3A, 1-5D, and 2-9D were cross-reactive for all three subtypes.
DISCUSSION
We have established MAbs to the NP of Ebola virus subtype Zaire. With these MAbs, two linear epitopes that seem useful for the differentiation of Ebola virus subtypes were identified. In fact, the NPs of the Zaire, Sudan and Reston subtypes of Ebola virus were successfully segregated by using combinations of these MAbs. Unfortunately, since sequence information or antigenic materials of the NP of the Côte d'Ivoire subtype was not available to us, we could not examine whether these MAbs were reactive to this subtype. Comparison of the GP sequences among Ebola virus subtypes indicated that the Côte d'Ivoire subtype was closer to Zaire than to Reston (3, 22). Although only a single case of confirmed human infection caused by subtype Côte d'Ivoire has been reported (5, 9), it is necessary to examine whether these MAbs may react with the NP of this subtype to further determine the usefulness of this panel of MAbs for subtype identification.
We determined that 9 of 10 MAbs recognized the C-terminal half of NP. The highly hydrophobic nature of the N-terminal half of NP (16) may be one of the reasons why most of the MAbs recognized the C-terminal half. In the C-terminal half, only NP-5 and NP-8, which span aa 361 to 461 and aa 630 to 739, respectively, were reactive with these MAbs. We identified two linear antigenic epitope regions on NP-5 by Pepscan analyses. None of the MAbs specific to NP-8 showed reactivity in the Pepscan analyses, suggesting that these MAbs recognize conformational epitope(s) formed by more than 10 aa or discontinuous ones. We have previously reported that one of these MAbs, 3-3D, recognized a nonlinear epitope formed by 26-aa residues spanning aa 648 to 673 (15). Considering that 2-11G failed to cross-react with the Sudan subtype in IFA whereas the others were reactive with all three subtypes, we assume that at least two different recognition sites exist on this C-terminal region.
One linear epitope region on NP-5 was located between aa 420 and 426. Recognition of this epitope region by MAbs varied both in the minimum amino acid requirement and the critical amino acid residue within the epitope. This diversity resulted in differences in subtype specificities and cross-reactivities of the MAbs (Fig. 2B). Three MAbs—3-3A, 1-5D, and 2-9D—recognized amino acid sequence of D426IPFP430 and cross-reacted with Reston, in which D426 was changed to E. This means the amino acid replacement from D426 to E426 was not crucial for recognition by these three MAbs. 3-9E recognized the amino acid sequence of D424DDIPF429 and reacted only with the Zaire subtype. This specificity may be attributed to the amino acid difference at position 425 that is D for Zaire but N for both Reston and Sudan subtypes.
The other linear epitope recognized by 1-7C was identified as D451TTIP455. This amino acid sequence is conserved between the Zaire and Sudan subtypes, whereas aa 453 is I in Reston (Fig. 2B). MAb 1-7C was reactive with Zaire and Sudan subtypes as expected, but it failed to recognize Reston subtype in all three assays. In ELISA, 1-7C showed lower OD values with the Sudan subtype than with the Zaire subtype. It is not clear whether this is due to a partial interference by the surrounding amino acid on the binding of the MAb to the NP or to that on the binding of the biotinylated peptide to streptavidin coated on the plate.
1-7C recognized an amino acid sequence conserved between the Zaire and Sudan subtypes and was reactive with these two subtypes in all three immunological assays but not reactive with Reston. 3-9E showed a Zaire-specific reaction in all three assays. Thus, these two MAbs, as well as others reactive with all three subtypes, are useful for discriminating the subtypes of Ebola virus.
Acknowledgments
We gratefully acknowledge M. Ogata, Department of Virology 1, National Institute of Infectious Diseases.
This work was supported by grants from the Ministry of Health, Labor, and Welfare of Japan.
REFERENCES
- 1.Baron, C. B., J. B. McCormick, and O. A. Zubeir. 1983. Ebola virus disease in southern Sudan: hospital dissemination and intrafamilial spread. Bull. W. H. O. 61:997-1003. [PMC free article] [PubMed] [Google Scholar]
- 2.Breman, J. G., K. M. Johnson, G. van der Groen, C. B. Robbins, M. V. Szczeniowski, K. Ruti, P. A. Webb, F. Meier, and D. L. Heymann. 1999. A search for Ebola virus in animals in the Democratic Republic of the Congo and Cameroon: ecologic, virologic, and serologic surveys, 1979-1980. J. Infect. Dis. 179(Suppl. 1):S139-S147. [DOI] [PubMed] [Google Scholar]
- 3.Feldmann, H., and M. P. Kiley. 1999. Classification, structure, and replication of filovirus, p. 1-21. In H.-D. Klenk (ed.), Marburg and Ebola viruses. Springer-Verlag, Berlin, Germany.
- 4.Fisher-Hoch, S. P., T. L. Brammer, S. G. Trappier, L. C. Hutwagner, B. B. Farrar, S. L. Ruo, B. G. Brown, L. M. Hermann, G. I. Perez-Oronoz, C. S. Goldsmith, M. A. Hanes, and J. B. McCormick. 1992. Pathogenic potential of filoviruses: role of geographic origin of primate host and virus strain. J. Infect. Dis. 166:753-763. [DOI] [PubMed] [Google Scholar]
- 5.Formenty, P., C. Boesch, M. Wyers, C. Steiner, F. Donati, F. Dind, F. Walker, and B. Le Guenno. 1999. Ebola virus outbreak among wild chimpanzees living in a rain forest of Cote d'Ivoire. J. Infect. Dis. 179(Suppl. 1):S120-S126. [DOI] [PubMed] [Google Scholar]
- 6.Ikegami, T., A. B. Calaor, M. E. Miranda, M. Niikura, M. Saijo, I. Kurane, Y. Yoshikawa, and S. Morikawa. 2001. Genome structure of Ebola virus subtype Reston: differences among Ebola subtypes. Arch. Virol. 146:2021-2027. [DOI] [PubMed] [Google Scholar]
- 6a.Ikegani, T., M. Saijo, M. Niikura, M. E. Miranda, A. B. Calaor, M. Hernandez, D. L. Manalo, I. Kurane, Y. Yoshikawa, and S. Morikawa.2002. Development of an immunofluorescence method for the detection of antibodies to Ebola virus subtype Reston by the use of recombinant nucleoprotein-expressing HeLa cells. Microbiol. Immunol. 46:633-638. [DOI] [PubMed] [Google Scholar]
- 7.Ito, H., S. Watanabe, A. Takada, and Y. Kawaoka. 2001. Ebola virus glycoprotein: proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J. Virol. 75:1576-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiley, M. P., R. L. Regnery, and K. M. Johnson. 1980. Ebola virus: identification of virion structural proteins. J. Gen. Virol. 49:333-341. [DOI] [PubMed] [Google Scholar]
- 9.Le Guenno, B., P. Formentry, M. Wyers, P. Gounon, F. Walker, and C. Boesch. 1995. Isolation and partial characterisation of a new strain of Ebola virus. Lancet 345:1271-1274. [DOI] [PubMed] [Google Scholar]
- 10.Leirs, H., J. K. Mills, J. W. Krebs, J. E. Childs, D. Akaibe, N. Woollen, G. Ludwig, C. J. Peters, and T. G. Ksiazek. 1999. Search for the Ebola virus reservoir in Kikwit, Democratic Republic of the Congo: reflections on a vertebrate collection. J. Infect. Dis. 179(Suppl. 1):S155-S163. [DOI] [PubMed] [Google Scholar]
- 11.Maruyama, T., L. L. Rodriguez, P. B. Jahrling, A. Sanchez, A. S. Khan, S. T. Nichol, C. J. Peters, P. W. Parren, and D. W. Burton. 1999. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J. Virol. 73:6024-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, R. K., J. Y. Baumgardner, C. W. Armstrong, S. R. Jenkins, C. D. Woolard, G. B. Miller, Jr., L. D. Polk, D. R. Tavris, K. A. Hendricks, J. P. Taylor, D. M. Simpson, S. Schultz, L. Sturman, J. G. Debbie, D. L. Morse, P. E. Rollin, P. B. Jahrling, T. G. Ksiazek, and C. J. Peters. 1990. Filovirus infections among persons with occupational exposure to nonhuman primates. Morb. Mortal. Wkly. Rep. 39:266-273. [PubMed] [Google Scholar]
- 13.Miller, R. K., J. Y. Baumgardner, C. W. Armstrong, S. R. Jenkins, C. D. Woolard, G. B. Miller, Jr., P. E. Rollin, P. B. Jahrling, T. G. Ksiazek, and C. J. Peters. 1990. Filovirus infections in animal handlers. Morb. Mortal. Week. Rep. 39:221.
- 14.Miranda, M. E., T. G. Ksiazek, T. J. Retuya, A. S. Khan, A. Sanchez, C. F. Fulhorst, P. E. Rollin, A. B. Calaor, D. L. Manalo, M. C. Roces, M. M. Dayrit, and C. J. Peters. 1999. Epidemiology of Ebola (subtype Reston) virus in the Philippines, 1996. J. Infect. Dis. 179(Suppl. 1):S115-S119. [DOI] [PubMed] [Google Scholar]
- 15.Niikura, M., T. Ikegami, M. Saijo, I. Kurane, M. E. Miranda, and S. Morikawa. 2001. Ebola viral antigen-detection enzyme-linked immunosorbent assay using a novel monoclonal antibody to nucleoprotein. J. Clin. Microbiol. 39:3267-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters, C. J., A. Sanchez, P. E. Rollin, T. G. Ksiazek, and F. A. Murphy. 1996. Filoviridae: Marburg and Ebola viruses, p. 1161-1176. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Peters, C. J., and A. S. Khan. 1999. Filovirus diseases, p. 85-95. In H.-D. Klenk (ed.), Marburg and Ebola viruses. Springer-Verlag, Berlin, Germany.
- 18.Saijo, M., M. Niikura, S. Morikawa, T. G. Ksiazek, P. F. Meyer, C. J. Peters, and I. Kurane. 2001. Enzyme-linked immunosorbent assays for detection of antibodies to Ebola and Marburg viruses using recombinant nucleoproteins. J. Clin. Microbiol. 39:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saijo, M., M. Niikura, S. Morikawa, and I. Kurane. 2001. Immunofluorescence method for detection of Ebola virus immunoglobulin g, using HeLa cells which express recombinant nucleoprotein. J. Clin. Microbiol. 39:776-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez, A., M. P. Kiley, B. P. Holloway, J. B. McCormick, and D. D. Auperin. 1989. The nucleoprotein gene of Ebola virus: cloning, sequencing, and in vitro expression. Virology 170:81-91. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez, A., T. G. Ksiazek, P. E. Rollin, C. J. Peters, S. T. Nichol, A. S. Khan, and B. W. J. Mahy. 1995. Reemergence of Ebola virus in Africa. Emerg. Infect. Dis. 1:96-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez, A., S. G. Trappier, B. J. Mahy, C. J. Peters, and S. T. Nicole. 1996. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acd. Sci. USA 93:3602-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takada, A., S. Watanabe, K. Okazaki, H. Kida, and Y. Kawaoka. 2001. Infectivity-enhancing antibodies to Ebola virus glycoprotein. J. Virol. 75:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson, J. A., M. Hevey, R. Bakken, S. Guest, M. Bray, A. L. Schmaljohn, and M. K. Hart. 2000. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287:1664-1666. [DOI] [PubMed] [Google Scholar]
- 25.Wilson, J. A., and M. K. Hart. 2001. Protection from Ebola virus mediated by cytotoxic T lymphocytes specific for the viral nucleoprotein. J. Virol. 75:2660-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization/International Study Team. 1978. Ebola haemorrhagic fever in Sudan, 1976. Bull. W. H. O. 56:247-270. [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. 1978. Ebola haemorrhagic fever in Zaire, 1976. Bull. W. H. O. 56:271-293. [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. 1997. Part one: background information on the organisms and the disease, p. 1-3. In W.H.O. recommended guidelines for epidemic preparedness and response: Ebola haemorrhagic fever (EHF). World Health Organization, Geneva, Switzerland.
- 29.World Health Organization. 2001. Outbreak of Ebola haemorrhagic fever, Uganda, August 2000-January 2001. Wkly. Epidemiol. Rec. 76:41-46. [PubMed] [Google Scholar]