Abstract
A semiquantitative PCR assay for the detection of BK virus in urine was developed using primers for BK virus that specifically amplified BK but not JC virus. DNA was extracted from urine through treatment with proteinase K followed by DNA precipitation with sodium acetate. Semiquantitation was achieved by amplifying serial dilutions (1:1, 1:10, 1:100, and 1:1,000) of the urine specimens. Each assay included both positive (stock BK virus and previously positive patient urine) and negative (no template) controls. A urine sample was interpreted as positive if any of the serial dilutions showed amplification of the DNA fragment of the expected size. For some patient-derived samples, amplification of the expected-size fragment was achieved with a dilute template whereas no amplification was achieved with a concentrated template. This was attributed to interfering substances in the urine. PCR results were compared with urine cytology and shown to be more sensitive. Validation studies were performed at the University of Nebraska Medical Center, utilizing a separate qualitative PCR assay that detects both BK and JC virus and distinguishes between them by restriction enzyme digestion patterns. Of 46 urine samples analyzed using both methods, 22 were positive by both assays, 18 were negative by both assays, 5 were positive only by the Nebraska method, and 1 was positive only by our method. In comparison with the Nebraska PCR, our PCR assay had a sensitivity of 81% and specificity of 95%. For twenty-one (43%) of 49 immunocompromised patients, tests were postive when specimens were submitted because of clinical suspicion of BK virus infection.
The human polyomavirus BK has been shown to be an important cause of cystitis in immunocompromised patients (13). It also produces interstitial nephritis in renal transplant patients that clinically resembles immune-mediated graft rejection (4, 8). Since graft rejection usually requires increasing the immunosuppressive regimen, whereas BK viral reactivation is often managed by decreasing immunosuppression, determining which of these entities is responsible for the pathology is critical. Several methods have been described for detecting BK virus, including renal biopsy, urine cytology, and urine PCR (1, 5, 14). PCR is the most sensitive of these assays and also may be quantitative. Because there was no commercially available test for BK viruria, we elected to develop one at the University of Minnesota and validate it with a procedure currently in use at the University of Nebraska Medical Center.
(This work was presented in part at the 17th Annual Meeting of the Pan American Society for Clinical Virology, Clearwater Beach, Fla., 30 April 2001, and at the 36th Annual Meeting of the Academy of Clinical Laboratory Physicians and Scientists, Seattle, Wash., 15 June 2001.)
MATERIALS AND METHODS
DNA extraction.
The urine specimen was inverted or swirled in a specimen cup a minimum of five times to create a homogenous suspension of cells. One milliliter was transferred into an Eppendorf tube and centrifuged for 10 min at 14,000 × g. The supernatant was removed, and the dry cell pellet was stored at −70°C if extraction was to be performed at a later time. To extract DNA, 500 μl of 10 mM Tris-1.2 mM EDTA-0.5% sodium dodecyl sulfate (pH 9.0) was added to the dry cell pellet, the pellet was resuspended by vortexing, 25 μl of 20-mg/ml proteinase K (Sigma, St. Louis, Mo.) was added, and the tube was incubated on a heating block at 56°C for 2 h. Next, 55 μl of 3 M sodium acetate and 0.5 ml of cold ethanol were added, mixed, and chilled at −70°C for 5 min, followed by centrifugation at 14,000 × g at 4°C for 20 min. The supernatant was decanted from the DNA pellet, which was then washed with 1 ml of 70% cold ethanol. The DNA pellet was resuspended in 50 μl of 10 mM Tris-1 mM EDTA (pH 7.6), and four serial 10-fold dilutions were made, from 1:1 to 1:1,000.
Semiquantitative PCR for BK virus (University of Minnesota assay).
For each patient's sample, seven tubes (four dilutions of the patient's sample, two positive controls, and one negative control) were prepared with a reaction mixture consisting of 5 μl of 10× Taq DNA polymerase buffer, 5 μl of PCR nucleotide mix (2 mM each), 2 μl of 5 μM BK forward primer, 2 μl of 5 μM BK reverse primer, 33 μl of sterile water, 2 μl of template DNA, and 1 μl of Taq DNA polymerase (5 U/μl; Promega, Madison, Wis.). The BK forward and reverse primers were specifically designed to amplify a portion of the gene encoding the large T antigen of BK virus without amplifying DNA derived from JC virus or simian virus 40 (SV40) virus (Fig. 1). PCR conditions were as follows: initial denaturation at 94°C for 5 min and then 40 amplification cycles (94°C for 1 min, 45°C for 1 min, 72°C for 1 min), and then extension at 72°C for 10 min. Twenty microliters of the PCR products were electrophoresed on 1.5% agarose gels, and the gels were stained with ethidium bromide and examined for a band of the appropriate size (Fig. 2). The negative control consisted of no template DNA, and the positive control consisted of BK virus from the American Type Culture Collection (ATCC) or a previously positive patient sample.
FIG. 1.
University of Minnesota BK primers show specificity for BK viral DNA. Primer sequences are highlighted by arrows. The multiple base mismatches between BK, JC, and SV40 viral DNA are shown in bold type. The PCR product is a 370-bp fragment that codes for part of the large T antigen.
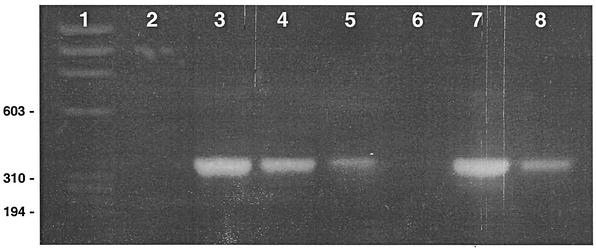
FIG. 2.
Agarose gel showing amplification of a 370-bp fragment of BK viral DNA from the urine of a renal transplant recipient. Lane 1 is the φx174 DNA/HaeIII marker. The template for lane 2 is a 1:1 dilution of DNA purified from the urine of a patient who was previously negative for BK virus by this assay (negative control). The templates for lanes 3 to 6 are serial dilutions (1:1, 1:10, 1:100, 1:1,000) of DNA purified from the urine of a renal transplant recipient. The template for lane 7 is BK virus stock from ATCC (positive control). The template for lane 8 is a 1:10 dilution of DNA purified from the urine of a patient who was previously positive for BK virus by this assay (positive control).
PCR amplification of JC virus.
JC virus was used to test the specificity of the BK primers. JC virus was obtained from ATCC. Two microliters of this concentrated virus as well as 2 μl of 10- and 100-fold dilutions of JC virus were used as a template in the University of Minnesota BK PCR assay as described above. In order to confirm the presence of JC virus, the same quantities of JC virus were used as a template in a JC-specific PCR assay using JC forward primer (5′-GAAGAACCCAAAAACTATTTGTTGAAA-3′) and JC reverse primer (5′-GCCTAACTGGAGACAATCTAGAATAATAGTC-3′). These primers were designed to amplify a portion of the JC VP2/VP3 genes without amplifying DNA derived from BK virus with the expected amplicon size of 133 bp. All other reaction conditions were the same as those described for the University of Minnesota BK PCR assay.
Validation of BK PCR.
The validation of the University of Minnesota assay was accomplished by sending duplicate samples to the University of Nebraska Medical Center, where a PCR was performed using primers as described by Arthur and Shah (2). DNA was extracted from the urine specimens using the Qiagen DNA purification kit. Reaction conditions were optimized for total reaction volumes of 50 μl containing 1 μg of sample DNA. The optimal concentrations of MgCl2 and deoxynucleoside triphosphates (dNTPs) were determined, as well as the optimal annealing temperature for the primers. Concentrations of Taq polymerase (1 U), 1× reaction buffer (Promega, Madison, Wis.), and primers (0.8 μM each) were kept constant. MgCl2 concentrations ranging from 1.0 to 3.0 mM (1.0, 1.25, 1.5, 1.75, 2.0, 2.25, 2.5, 2.75, and 3.0 mM) and dNTP concentrations of 20, 50, 100, and 400 μM were evaluated at annealing temperatures of 60 and 65°C. One hundred micromolar dNTPs, 1.5 mM MgCl2, and the 65°C temperature were determined to be optimal. 45 cycles of PCR were utilized: denaturation, 30 s at 94°C; annealing, 30 s at 65°C; extension, 30 s at 72°C. A 4-min denaturation step at 94°C was included in the first cycle, and a final extension of 72°C for 4 min followed the last cycle of PCR. Ten microliters of the PCR products were electrophoresed on 1.5% agarose gels, and the gels were stained with ethidium bromide and examined for a band of the appropriate size (172 bp). Since the primers have the potential to amplify JC virus as well as BK virus, restriction endonuclease digestion was performed. Each amplicon has a unique restriction site at its center: the JC virus amplicon has a BamHI site, and BK virus has a FokI site. Therefore, to determine if the amplicon was the result of BK virus or JC virus, 15 μl of the PCR products were digested with restriction endonucleases BamHI and FokI and electrophoresed on 1.5% agarose gels. The Nebraska laboratory personnel had no knowledge of the Minnesota PCR results at the time they performed their assays.
RESULTS
The University of Minnesota assay was performed on 60 samples from 49 immunocompromised patients who were thought to have BK virus-mediated renal dysfunction based on clinical signs and symptoms such as the presence of hematuria and/or an increase in serum creatinine. There were 25 bone marrow transplant patients (29 specimens), 10 renal transplant patients (13 specimens), 6 renal/pancreas transplant patients (9 specimens), 2 lung transplant patients (3 specimens), 1 heart transplant patient (1 specimen), and 5 patients with other diagnoses (5 specimens).
Results for a patient specimen were further analyzed only if there was no visible product in the negative control lane and there was a clearly visible 370-bp product in the positive control lane. A patient specimen was considered to be positive if any of the serial dilutions contained a visible band of the right size. Twenty-eight (47%) of the 60 urine samples tested positive for BK virus (Table 1). Twelve (43%) of these twenty-eight samples showed “partial inhibition,” meaning that at lower dilutions of template DNA there was no DNA amplification, whereas at higher dilutions of template DNA there was DNA amplification resulting in a band of the appropriate size (370 bp).
TABLE 1.
Results of PCR by highest dilution positive for 60 urine samples tested (University of Minnesota PCR)
| Positive endpoint | No. of samplesa | No. of samples with partial inhibitionb |
|---|---|---|
| 1:1,000 | 13 | 6 |
| 1:100 | 7 | 4 |
| 1:10 | 6 | 2 |
| 1:1 | 2 | 0 |
Thirty-two of the sixty samples tested were negative.
None of the 12 samples with partial inhibition was negative.
Patient-derived samples were submitted for BK viral testing on the basis of clinical signs and symptoms that suggested BK viral infection. Ancillary tests, such as urine cytology and renal biopsy, were performed concurrently at the discretion of the clinician but were not required for inclusion in our survey. Of the 60 urine specimens tested for BK virus infection by PCR, 28 were collected within 1 week of a urine cytology specimen. The results of these two tests were in agreement for 8 positive tests and 15 negative tests. In only one case was a sample positive by urine cytology but negative by PCR. Four urine specimens that were positive for BK virus by PCR were collected within 1 week of collection of urine specimens that did not show the nuclear changes characteristic of BK virus infection. This suggests that our PCR method is more sensitive than urine cytology for the detection of BK virus. Urine specimens were collected for BK PCR testing within 1 week of renal biopsy for only five patients. The results were in agreement between these two methods in all five cases (four negative and one positive for BK virus). Although the number of patients is not statistically significant, these results suggest a good correlation between these two tests.
Twenty-three of the 28 samples that were positive in the University of Minnesota assay were tested in the University of Nebraska assay. Of these 23 samples, all but 1 was positive by the University of Nebraska assay. In addition, of 32 samples that were negative in the University of Minnesota assay, 23 were tested in the University of Nebraska assay and 18 were negative in both assays. Therefore, when compared with the Nebraska assay, the sensitivity and specificity of the University of Minnesota assay were 81% (22 of 27 positive) and 95% (18 of 19 negative), respectively.
Of the six discrepant samples, two were collected within 1 week of a urine cytology specimen. In both cases, the urine cytology result and the University of Minnesota PCR results were in agreement (negative for BK virus), whereas the University of Nebraska PCR assay detected BK virus. None of the six discrepant samples was collected within 1 week of a renal biopsy.
The precision of our assay was tested by repeating the assay on five of the urine specimens, including two of the six specimens that were discrepant between our assay and the Nebraska assay. Of the five repeated assays, four reproduced the same result as the original assay (three positive, one negative) and one produced a positive result in the repeated assay, whereas it had produced a negative result in the original assay. (This specimen had tested positive for BK virus in the Nebraska assay.)
DISCUSSION
Our BK-specific PCR was developed for detection and semiquantitation of BK viruria in immunocompromised hosts. The method performed very well in comparison with an assay previously developed and in use at the University of Nebraska Medical Center with sensitivity and specificity of 81 and 95%, respectively. The University of Minnesota assay employs serial DNA dilutions in the PCR, which is important because if only undiluted DNA extracts had been tested, 12 of 28 positive specimens would have been false negatives. This partial inhibition effect is most likely due to inhibitory substances in the urine which are not completely removed by the DNA extraction procedure. Although the DNA extraction procedure could potentially be improved in order to reduce this inhibition, the high degree of correlation between the Minnesota and Nebraska assays demonstrates that the Minnesota assay in its present format is already both sensitive and specific. An improved DNA extraction procedure might reduce the number of dilutions of DNA that would need to be tested, but it may not significantly increase the sensitivity of the test and it would add a step that would increase the turnaround time and consume more reagents.
DNA from three members of the polyomavirus family (BK virus, JC virus, and SV40 virus) has previously been amplified from human tissues (17), although SV40 viral sequences have never been amplified from urine and SV40 is not thought to be a causative agent in immunosuppression-related cystitis (3, 16). Both JC and BK viral sequences have previously been amplified from human urine specimens (6, 11). However, whenever JC virus has been identified in the kidney of a patient with interstitial nephritis, BK virus has also been identified (3, 15). JC virus alone has not been shown to cause interstitial nephritis. Since the nucleotide sequence homologies of the BK virus with the JC virus and SV40 virus are 75 and 69%, respectively (18), the BK primers in this study were specifically designed to amplify BK while excluding amplification of JC and SV40 viruses (Fig. 1). In order to test the specificity of the Minnesota assay for BK virus, JC virus from ATCC was used as a template (Fig. 3). BK primers showed strong amplification of BK viral DNA, while JC viral DNA did not show significant amplification. The faint band observed when undiluted JC virus was amplified with BK primers (Fig. 3, lane 6) would not have been interpreted as a positive test result. The presence of JC virus was confirmed using JC-specific primers (Fig. 3, lanes 13 to 15).
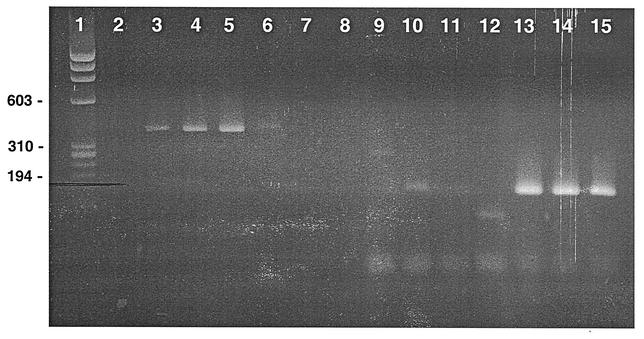
FIG. 3.
Agarose gel showing amplification of BK and JC viral DNA with BK-specific and JC-specific primers. Lane 1 is the φx174 DNA/HaeIII marker. Lanes 2 to 8 show results with BK-specific primers. Lane 2, no template; lanes 3 to 5, BK stock virus template at 1:1, 1:10, and 1:100 dilutions; lanes 6 to 8, JC stock virus template at 1:1, 1:10, and 1:100 dilutions. Lanes 9 to 15 show results with JC-specific primers. Lane 9, no template; lanes 10 to 12, BK stock virus template at 1:1, 1:10, and 1:100 dilutions; lanes 13 to 15, JC stock virus template at 1:1, 1:10, and 1:100 dilutions.
BK viruria was documented in 21 (43%) of the 49 patients whose symptoms suggested reactivation of BK infection. Although this implies a causal relationship, a prospective study will be required to define the incidence of posttransplant reactivation of BK virus and to establish the precise role of BK virus in renal allograft damage.
“Real-time” PCR using TaqMan technology has been demonstrated to be at least as sensitive as conventional PCR and in some reports up to 10 to 100 times more sensitive than conventional PCR (7, 9). We have begun TaqMan experiments using BK-specific primers. When these experiments are complete, we will be able to compare our standard method directly with real-time PCR for BK virus and choose the one that seems clinically more relevant for our planned prospective study of BK viruria. We also plan to test plasma samples from transplant patients for BK vire-mia, as it has been shown that BK viremia precedes the diagnosis of BK virus nephropathy in at least a subset of patients (10, 12).
Acknowledgments
We are grateful to Carrie James for her assistance in performing many of the BK PCR assays.
This work was supported by grants from the National Institutes of Health (AI-01411 and AI-27661), the Minnesota Medical Foundation, and the International Center for Antiviral Research and Epidemiology.
REFERENCES
- 1.Arthur, R. R., S. Dagostin, and K. V. Shah. 1989. Detection of BK virus and JC virus in urine and brain tissue by the polymerase chain reaction. J. Clin. Microbiol. 27:1174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, R. R., and K. V. Shah. 1989. Occurrence and significance of papovaviruses BK and JC in the urine. Prog. Med. Virol. 36:42-61. [PubMed] [Google Scholar]
- 3.Baksh, F. K., S. D. Finkelstein, P. A. Swalsky, G. L. Stoner, C. F. Ryschkewitsch, and P. Randhawa. 2001. Molecular genotyping of BK and JC viruses in human polyomavirus-associated interstitial nephritis after renal transplantation. Am. J. Kidney Dis. 38:354-365. [DOI] [PubMed] [Google Scholar]
- 4.Binet, I., V. Nickeleit, H. H. Hirsch, O. Prince, P. Dalquen, F. Gudat, and M. Mihatsch. 1999. Polyomavirus disease under new immunosuppressive drugs: a cause of renal graft dysfunction and graft loss. Transplantation 67:918-922. [DOI] [PubMed] [Google Scholar]
- 5.Corey, H. E., F. Alfonso, D. Hamele-Bena, S. M. Greenstein, R. Schechner, V. Tellis, P. Geva, and L. G. Koss. 1997. Urine cytology and the diagnosis of renal allograft rejection. II. Studies using immunostaining. Acta Cytol. 41:1742-1746. [DOI] [PubMed] [Google Scholar]
- 6.Flaegstad, T., A. Sundsfjord, R. R. Arthur, M. Pedersen, T. Traavik, and S. Subramani. 1991. Amplification and sequencing of the control regions of BK and JC virus from human urine by polymerase chain reaction. Virology 180:553-560. [DOI] [PubMed] [Google Scholar]
- 7.Hawrami, K., and J. Breuer. 1999. Development of a fluorogenic polymerase chain reaction assay (TaqMan) for the detection and quantitation of varicella-zoster virus. J. Virol. Methods 79:33-40. [DOI] [PubMed] [Google Scholar]
- 8.Howell, D. N., S. R. Smith, D. W. Butterly, P. S. Klassen, H. R. Krigman, J. L. Burchette, Jr., and S. E. Miller. 1999. Diagnosis and management of BK polyomavirus interstitial nephritis in renal transplant recipients. Transplantation 68:1279-1288. [DOI] [PubMed] [Google Scholar]
- 9.Lanciotti, R. S., A. J. Kerst, R. S. Nasci, M. S. Godsey, C. J. Mitchell, H. M. Savage, N. Komar, N. A. Panella, B. C. Allen, K. E. Volpe, B. S. Davis, and J. T. Roehrig. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 38:4066-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Limaye, A. P., K. R. Jerome, C. S. Kuhr, J. Ferrenberg, M.-L. Huang, C. L. Davis, L. Corey, and C. L. Marsh. 2001. Quantitation of BK virus load in serum for the diagnosis of BK virus-associated nephropathy in renal transplant recipients. J. Infect. Dis. 183:1669-1672. [DOI] [PubMed] [Google Scholar]
- 11.Marshall, W. F., A. Telenti, J. Proper, A. J. Aksamit, and T. F. Smith. 1991. Survey of urine from transplant recipients for polyomaviruses JC and BK using the polymerase chain reaction. Mol. Cell. Probes 5:125-128. [DOI] [PubMed] [Google Scholar]
- 12.Nickeleit, V., T. Klimkait, I. F. Binet, P. Dalquen, V. Del Zenero, G. Thiel, M. J. Mihatsch, and H. H. Hirsch. 2000. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. N. Engl. J. Med. 342:1309-1315. [DOI] [PubMed] [Google Scholar]
- 13.Peinemann, F., E. M. de Villiers, K. Dorries, O. Adams, T. A. Vogeli, and S. Burdach. 2000. Clinical course and treatment of haemorrhagic cystitis associated with BK type of human polyomavirus in nine paediatric recipients of allogeneic bone marrow transplants. Eur. J. Pediatr. 159:182-188. [DOI] [PubMed] [Google Scholar]
- 14.Purighalla, R., R. Shapiro, J. McCauley, and P. Randhawa. 1995. BK virus infection in a kidney allograft diagnosed by needle biopsy. Am. J. Kidney Dis. 26:671-673. [DOI] [PubMed] [Google Scholar]
- 15.Randhawa, P., F. Baksh, N. Aoki, D. Tschirhart, and S. Finkelstein. 2001. JC virus infection in allograft kidneys. Transplantation 71:1300-1303. [DOI] [PubMed] [Google Scholar]
- 16.Shah, K. V., R. W. Daniel, H. D. Strickler, and J. J. Goedert. 1997. Investigation of human urine for genomic sequences of the primate polymaviruses simian virus 40, BK virus, and JC virus. J. Infect. Dis. 176:1618-1621. [DOI] [PubMed] [Google Scholar]
- 17.Strizzi, L., G. Vianale, M. Giuliano, R. Sacco, F. Tassi, P. Chiodera, P. Casalini, and A. Procopio. 2000. SV40, JC, and BK expression in tissue, urine and blood samples from patients with malignant and nonmalignant pleural disease. Anticancer Res. 20:885-890. [PubMed] [Google Scholar]
- 18.Walker, D. L., and R. J. Frisque. 1986. The biology and molecular biology of JC virus, p. 327-377. In N. P. Salzman (ed.), The Papovaviridae, vol. 1. The polyomaviruses. Plenum Press, New York, N.Y.