Abstract
Borrelia burgdorferi, the causative agent of Lyme disease, is transmitted by the tick Ixodes scapularis. A 2.9-kb fragment containing a putative spoT gene was isolated from B. burgdorferi genomic DNA by PCR amplification and cloned into a pBAD24 vector. The cloned gene complemented Escherichia coli mutant strain CF1693, which contains deletions of both the relA and spoT genes. The spoT gene in E. coli encodes a bifunctional enzyme capable of synthesizing and degrading (p)ppGpp, which mediates the stringent response during carbon source starvation. B. burgdorferi has been reported to have a stress response to serum starvation. Thin-layer chromatography was used to detect (p)ppGpp extracted from H332PO4-labeled B. burgdorferi cells starved for serum in RPMI. B. burgdorferi spoT gene expression was characterized during fatty acid starvation. Northern analysis of spoT revealed detectable message at 2.5 min of starvation in RPMI. Expression of spoT during serum starvation increased ∼6-fold during the 30 min that starvation conditions were maintained. Further, expression of spoT decreased when serum was added to serum-starved cells. Reverse transcriptase PCR (RT-PCR) was used to detect spoT mRNA from ∼106 cells starved for serum in RPMI for 2.5 to 30 min or incubated in tick saliva for 15 min. Northern blot analysis suggests that spoT transcript was ∼900 nucleotides in length. RT-PCR amplification of the transcript using several sets of primers confirmed this finding. Additionally, a truncated clone containing only the first 950 bp of the 2,001-bp spoT open reading frame was able to complement E. coli CF1693. The data suggest that B. burgdorferi exhibits a stringent response to serum starvation and during incubation in tick saliva.
The spirochete Borrelia burgdorferi, the causative agent of Lyme disease (9, 50), is a tick-borne pathogen spread by members of the genus Ixodes. During a blood meal by the tick vector, B. burgdorferi cells migrate through the tick gut epithelium and pass into the hemolymph and then to various tissues, including the salivary glands. B. burgdorferi has been shown to enter the salivary glands of Ixodes scapularis during a blood meal (42, 58) and has been successfully isolated from the saliva of I. scapularis (17), indicating that saliva serves as the route of transmission. During its life cycle, B. burgdorferi must adapt to different host conditions. As a result, the pathogen survives a variety of stress conditions which include physiological and nutrient changes between hosts (28). Stress conditions encountered by B. burgdorferi within the tick include starvation and temperature shifts (1, 10). Heat shock occurs when cells encounter rapid increases in temperature (10, 46). Proteins synthesized in response to elevated temperatures are termed heat shock proteins. Heat shock proteins have been identified and characterized in B. burgdorferi (10). It is hypothesized that additional stress responses are used as mechanisms of survival by B. burgdorferi under adverse conditions. B. burgdorferi cells are probably subjected to periods of starvation within their tick hosts when the ticks molt and go months between blood meals. B. burgdorferi, like other bacteria, should respond to nutrient depletion by initiating the stringent response (11).
The stringent response has been characterized as a short-term stress response to amino acid and carbon starvation. This global response is brought about by the failure of tRNA aminoacylation to keep up with the demand of protein synthesis. It is mediated by the accumulation of guanosine 3′,5′-bispyrophosphate (ppGpp) (11, 30). In Escherichia coli and Vibrio angustum, two genes, relA and spoT, govern the stringent response (4, 19, 39). The relA gene encodes the enzyme (p)ppGpp synthase I (PSI). The enzyme is activated when a ribosome engaged in protein synthesis binds a nonaminoacylated cognate tRNA at the acceptor site (4, 11, 41). The study of relA null mutants led to the observation that SpoT is a bifunctional enzyme capable of both ppGpp synthesis and degradation (21, 22, 27, 45, 57). Mutational analysis of the spoT gene confirmed that there are two enzymatic sites contained within the amino acid sequence. One enzyme site is responsible for synthesis of (p)ppGpp, while the other site is for degradation of (p)ppGpp (22, 57). The accumulation of (p)ppGpp due to (p)ppGpp synthase II (PSII) activity is primarily due to carbon source depletion (21, 22). The function of (p)ppGpp is to act as an intracellular alarmone for nutritional deficiency (18). As a result of (p)ppGpp accumulation, bacteria shut down processes that consume amino acids and energy reserves, including stable RNA synthesis and the initiation of new rounds of DNA replication.
Increased ppGpp concentration affects many cellular processes, including the inhibition of tRNA synthesis and rRNA synthesis and the stimulation of biosynthetic and some catabolic pathways (26). In Myxococcus xanthus, the accumulation of ppGpp has been shown to be required for the initiation of fruiting body formation (25). During carbon starvation, Mycobacterium smegmatis cells exhibit a morphological change from elongated rods to cocci in response to elevated intracellular levels of ppGpp (38). Similarly, in the bacterium V. angustum the carbon starvation response (ultramicrocell formation) involves the loss of motility and a change in morphology to a spherical form, which is attributed to the expression of a number of genes (29). Thus, the stringent response, like other stress responses, acts as a global regulator of the bacterial cell.
B. burgdorferi responds to serum starvation by inducing a starvation response that results in changes in protein expression and eventually in the transformation of motile helical vegetative cells into nonmotile spherical starvation-stress forms (1). It has been recently reported that ppGpp affects the synthesis of cyclopropane fatty acids through σs (15) and also has a role during fatty acid starvation in E. coli (49). This is of significance because the genome of B. burgdorferi contains no fatty acid synthetic pathways (20). B. burgdorferi cells depend upon an external source of fatty acids. Serum is rich in lipids and is the source of fatty acids for B. burgdorferi cells growing in pure culture or in a host. To date, most bacterial genomes that have been fully sequenced encode the proteins RelA and/or SpoT, which are capable of synthesizing or hydrolyzing (p)ppGpp (51). B. burgdorferi possesses a spoT ortholog (BB0198) and an rpoS gene (BB0771), but no relA gene (20). Since SpoT has been shown to be a bifunctional enzyme in other bacteria, it is likely that the B. burgdorferi SpoT is bifunctional and responsible for the synthesis and degradation of ppGpp. Other bacteria with putative relA or spoT homologs have been cloned and or characterized (2, 19, 34-36, 39, 53-55, 57), suggesting that it is not uncommon for ppGpp synthesis and degradation to be controlled by a single gene. Therefore, if the spoT ortholog were functional in B. burgdorferi, the organism should have a stringent control mechanism. This mechanism could contribute to the ability of B. burgdorferi to adapt to and survive various host environments. In this study we show that the B. burgdorferi spoT ortholog is functional, able to complement an E. coli relA spoT mutant, and also is expressed in a manner similar to that in other bacteria that exhibit a stringent response.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
B. burgdorferi B31 (high passage) was grown in BSK H medium (Sigma, St. Louis, Mo.) supplemented with 6% rabbit serum (Life Technologies, Grand Island, N.Y.). For most experiments, cells were grown for 48 h at 33°C. Additional media used for B. burgdorferi was RPMI 1640 Select-Amine (RPMI) (Life Technologies) and saliva collected from I. scapularis (T. Mather, University of Rhode Island). RPMI is a defined medium containing glucose, vitamins, and all essential amino acids. I. scapularis saliva was collected as described by Ewing et al. (17). BSK II without albumin fraction V and lacking rabbit serum (termed BSK) was also used (5).
E. coli strains XL1 MRF′, CF1648 (wild-type MG1655; relA+ spoT+), CF1652 (ΔrelA251::Kn), CF1693 (ΔrelA251::Kn ΔspoT207::Cm) (a gift from Michael Cashel, National Institutes of Health) were routinely grown in Luria broth (LB) containing the appropriate antibiotics overnight in a shaking water bath at 37°C. E. coli was also grown in M-9 minimal medium to induce the stringent response (43, 45).
The plasmid pBAD24 (a gift of Jon Beckwith, Harvard University, Cambridge, Mass.) was used to clone the putative spoT. The plasmid contains an ampicillin resistance marker (23). The plasmid was introduced into E. coli CF1693 by electroporation.
DNA isolation and PCR conditions.
Genomic DNA was isolated from B. burgdorferi B31 cells by using a Qiagen DNeasy DNA isolation kit (Qiagen Inc., Valencia, Calif.) following the protocol of the manufacturer. The DNA was used to PCR amplify a digoxigenin (DIG)-labeled probe for Southern and Northern analysis. PCR was performed using either an Applied Biosystems GeneAmp 9600 or 9700 thermocycler (Applied Biosystems, Foster City, Calif.). Primers used to amplify the DIG-labeled probe were SpoT F1 and SpoT R1 (Table 1). The probe was synthesized according the protocol of the manufacturer (Roche Molecular Biochemicals, Indianapolis, Ind.), utilizing cycles of 94°C (5 min; denaturation), 35 cycles of 94°C (1 min), 51°C (2 min), 72°C (3 min), and final extension at 72°C (7 min).
TABLE 1.
Primers used in this study
| Primer | Primer sequence (5′ → 3′)a | Coordinates |
|---|---|---|
| SpoT F1 | TTT TTG TCG ACG AGC TC∗C GGT AGT GGA TGA GC | 195219-195233b |
| SpoT F2 | TT GCA CAT TTA ATC AAG ATA AAT G | 195713-195736b |
| SpoT R1 | CGG ATT AGC TGG CAC AT | 196044-196028b |
| SpoT R2 | TTT TTC TGC AG∗G GTT TTG TTC TAA AGA ATC ATC C | 198165-198142b |
| RT F −25 | GAA AAG AGA GCA AGG GCG CTG ATA | 195657-195680b |
| RT F −50 | ACA TTT AAA AAC AAT TTA CGA TCT T | 195632-195656b |
| RT R +800 | TAA GTT TGT TTG TTC TTG TTT GCA T | 196507-196483b |
| RT R +850 | TTT TGT TTT TTG CAA ATT ATT CTT A | 196556-196532b |
| RT R +950 | TTG ATA TTT ATT TTC TTT TGG GCT T | 196656-196632b |
| OspA F1 | GGG AAT AGG TCT AAT ATT AGC CTT | 9411-9434c |
| OspA R1 | CTG CTG ACC CCT CTA ATT TGG TGC C | 10171-10147c |
Bold letters indicate recognition sites for the restriction endonucleases used. SpoT F1 includes SacI and SalI sites. SpoT R2 contains a PstI site. ∗ indicates the start of the B. burgdorferi-specific sequence.
Coordinates from B. burgdorferi main chromosome.
Coordinates from B. burgdorferi linear plasmid 54 (lp54).
A 2.9-kb fragment containing spoT was amplified by PCR from B. burgdorferi genomic DNA using primers spoT F1 and spoT R2 (Table 1). The PCR amplification followed the protocol above with the exception of an annealing temperature of 61°C. The resulting amplicon was digested with the endonucleases PstI and SalI (Promega, Milwaukee, Wis.) according to the protocol of the manufacturer. The restricted amplicon was then ligated into the vector pBAD24 (23) using T4 ligase (Promega) according to protocols of the manufacturer. The recombinant plasmid was transformed into E. coli CF1693 by electroporation. Transformants were selected on LB plates containing ampicillin (200 μg/μl). Colonies resistant to ampicillin were replica plated onto M-9 minimal medium plates containing ampicillin (200 μg/ml), kanamycin (25 μg/ml), and chloramphenicol (30 μg/ml) to test for complementation of the spoT mutation (43).
Reverse transcriptase (RT) reactions were performed using either a Superscript II kit (Life Technologies) or a Qiagen RT kit (Qiagen Inc.) according to the protocols of the manufacturers. The primers used for the RT reactions were SpoT R1, SpoT RT R +800, SpoT R +850, RT R +850, and RT R +950 (Table 1). Prior to the RT reaction, total RNA was treated with RQ1 DNase (Promega) to eliminate any residual DNA contamination. Total RNA was quantified and RT reactions were performed on equal amounts of RNA (1 μg) from each sample. PCR amplification was performed on 2 μl of each RT reaction mix (22-μl total volume). Following the RT reaction, amplification was performed by PCR using forward primers SpoT F2, RT F −50, and RT F −25 and reverse primers SpoT R1, RT R +800, RT R + 850, and RT R +950 to yield amplification products. Sizes of the amplification products were determined using Kodak digital imaging software (Kodak 1D image analysis software, version 3.0.2; Eastman Kodak Co., Rochester, N.Y.) by comparison to a 1-kb DNA ladder (Promega).
Truncated and full-length spoT DNA sequences were amplified by PCR, and the amplicons were cloned into pSTBlue-1 using a Novagen Perfectly Blunt cloning kit (Novagen, Inc., Madison, Wis.). PCR was performed using pBAD24.spoT as the template. The forward primer for all reactions was SpoT F1 with the following reverse primers: SpoT R2, SpoT R1, and SpoT R + 950. PCR amplicons yielded full-length spoT (2.9 kb) and two truncated DNA sequences of ∼0.4 and ∼1.1 kbp, respectively. The clones were designated pMBC04, pMBC05, and pMBC06. The truncated clones were then transformed into E. coli CF1693 as previously described and grown on M-9 agar containing isopropyl-β-d-galactopyranoside to determine whether the clones could complement E. coli CF1693. Plasmids were purified from the appropriate E. coli strains grown overnight using a Qiagen spin miniprep kit (Qiagen) according to the protocol of the manufacturer.
32P labeling and detection of (p)ppGpp by thin-layer chromatography (TLC).
E. coli cells were grown in phosphate-limited medium and labeled with [32P]phosphate as described by Bochner and Ames (8, 37). Briefly, cells were grown for 16 h in LB at 37°C. Cells were harvested by centrifugation (8,000 × g, 8 min, 4°C) and then incubated in glucose-limited (0.04%) phosphate-limited medium for 16 h at 37°C. Supplementation with glucose differed in that for glucose-limited culture 0.04% glucose was used, and otherwise 0.4% was utilized. To label E. coli cells with H332PO4, cells were harvested by centrifugation (8,000 × g, 8 min, 4°C), washed, resuspended at 2 × 106 CFU/ml in phosphate-limited medium supplemented with H332PO4 (specific activity, 500 mCi/mmol; ICN Biomedical, Irvine, Calif.) at 150 μCi/ml, and allowed to incubate for 3 h (∼2 generations). For induction of the stringent response, the cells were transferred to M-9 broth and allowed to incubate for 15 min at 37°C and then extracted for (p)ppGpp as described below.
To label B. burgdorferi cells with H332PO4, cells were grown in BSK H as previously described, harvested by centrifugation (8,000 × g, 8 min, 4°C), washed in RPMI, and then resuspended (∼1 × 107 cells/ml) and labeled in RPMI medium without phosphate (Life Technologies) supplemented with 6% rabbit serum for 4 h. Cells were labeled with H332PO4 (specific activity, 500 mCi/mmol; ICN Biomedicals) at 150 μCi/ml. The 32P-labeled cells were then washed and resuspended in RPMI at 106 cells/ml and incubated for 15 min at 33°C. Samples of labeled cells (200 μl) taken before and after the stress were extracted with formic acid for analysis of (p)ppGpp (8). Briefly, 20 μl of cold 13 M formic acid was added to the samples, and samples were stored overnight at −75°C. Prior to separation and analysis by TLC, cell debris was removed by centrifugation (15,000 × g for 5 min). A 10-μl sample of the extracted supernatant was spotted onto a polyethyleneimine (PEI) cellulose TLC plate (Sigma) and separated in 1.5 M H3PO4 (pH 3.4) buffer for 2.5 h. Digital images of the TLC plate were captured using a Molecular Dynamics Storm 840 PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). TLC analyses were repeated three times.
RNA isolation.
B. burgdorferi B31 cells grown in BSK H (60 ml) with 6% rabbit serum at 33°C (∼1 × 107 cells/ml) were harvested by centrifugation (8,000 × g, 8 min, 4°C), washed in RPMI, and resuspended in the appropriate medium (108 cells/ml) for induction of the spoT gene (RPMI, RPMI with 6% rabbit serum, BSK, and a 1:1 dilution of tick saliva [in sterile nuclease-free H2O]). Samples (1 ml) were taken at various times (0, 2.5, 5, 10, 15, and 30 min) after incubation had begun, and total RNA was isolated using RNA Purescript (Gentra, Minneapolis, Minn.) according to the protocol of the manufacturer. Purified RNA samples were stored at −75°C until use. The RNA was quantified by absorption at 260 nm using an Ultrospec 4000 spectrophotometer (Amersham Pharmacia Biotech, Piscataway, N.J.) (43).
DNA sequencing.
DNA sequencing was performed by the HHMI Biopolymer/Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, Conn.). Sequence analysis was carried out on Applied Biosystems 377 and 3100 DNA analysis instruments. The sequencing reactions utilized fluorescently labeled dideoxynucleotides (Big Dye terminators) and Taq DNA polymerase (Applied Biosystems) in a thermal cycling protocol. Premixed samples of DNA template along with primers (Table 1) were sent to the facility, which then performed the sequencing reactions.
Southern and Northern analysis.
B. burgdorferi genomic DNA (2 μg) was treated with the endonucleases EcoRI and EcoRV according to the protocol of the manufacturer (Promega). The digested fragments were separated by electrophoresis in a 1% agarose gel run in Tris-acetate-EDTA (TAE) buffer (43). The gel was prepared for Southern blot analysis as described by Seldon and Ausubel (48). After the washes, the DNA was transferred from the agarose gel onto a nylon membrane (Magna Graph; Osmonics, Minnetonka, Minn.) using the Turboblotter system (Schleicher & Schuell, Inc., Keene, N.H.), employing a downward transfer with 20× SSC (1× SSC contains 0.03 M sodium citrate and 0.3 M sodium chloride) made in diethyl pyrocarbonate (DEPC) (Sigma)-treated sterile distilled water (dH2O). The DNA was bound to the membrane by UV cross-linking using a UV cross-linker (FB-UVXL-1000; Fisher Scientific). The blot was then developed according to the specifications of the manufacturer (Life Technologies). Sizes of hybridizable bands were determined by comparison with a DNA standard and using Kodak 1D digital imaging software.
For Northern blot analysis, RNA samples (5 to 15 μg) were treated by heating at 55°C for 15 min (47). The RNA samples were separated in a formaldehyde-agarose gel, which was made with DEPC-treated dH2O, 10× MOPS buffer solution (0.4 M 3-[N-morpholino] propanesulfonic acid, 0.2 M sodium acetate, and 10 mM EDTA, pH 7.0, in DEPC-treated dH20) and 37% formaldehyde and containing 0.634 nM ethidium bromide. Equal amounts of RNA were loaded into each gel lane for each experiment. The gel was run at 80 V for 1.5 h. Following electrophoresis, the 23S and 16S rRNA subunits were observed in the gels by UV light. Northern analysis was only performed if both subunits were observed and were present in equal amounts for each lane of the gel. The gel was prepared for transfer to the nylon membrane by washing with 10× SSC for 45 min (47). The RNA was transferred to a nylon membrane (Magna Graph; Osmonics) using a Turboblotter (Schleicher & Schuell, Inc.). The RNA was then bound to the nylon membrane by using a UV cross-linker.
The nylon blots for Southern and Northern analysis were hybridized at 55°C with a spoT-specific DIG-labeled probe, which targets both DNA and RNA, according to the protocol of the manufacturer (Roche Molecular Biochemicals). The detection of the hybridized DIG-labeled probe followed the protocol of the manufacturer. Briefly, the blots were probed for 18 h with the DIG-labeled probe; following hybridization, the membranes were washed, blocked, and visualized by chemiluminescence according to the protocol of the manufacturer of the DIG synthesis kit.
RESULTS
Cloning of B. burgdorferi spoT.
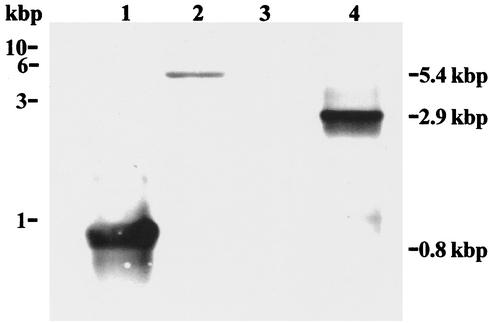
The putative B. burgdorferi spoT gene was PCR amplified from B. burgdorferi genomic DNA using the primers spoT F1 and spoT R2 (Table 1). This resulted in a 2.9-kb amplicon, which was treated with the endonucleases PstI and SalI and ligated into the plasmid vector pBAD24 to yield pBAD24.spoT. The recombinant plasmid was introduced into competent E. coli CF1693 cells by electroporation. Southern blot analysis was used to confirm the presence of the 2.9-kb PCR amplicon in pBAD24.spoT. B. burgdorferi genomic DNA treated with the endonucleases EcoRI and EcoRV and pBAD24.spoT DNA treated with PstI and SalI were separated by electrophoresis on a 1% agarose gel in TAE buffer and then examined by Southern blot analysis (47). The DNA was probed with a DIG-labeled spoT probe synthesized by PCR. Southern analysis (Fig. 1) revealed hybridization of the DIG-labeled probe with the PCR control (lane 1), as well as hybridization to a 5.4-kb fragment contained within the EcoRI- and EcoRV-digested B. burgdorferi genomic DNA (lane 2), and to a 2.9-kb PstI- and SalI-digested fragment of the cloned spoT gene (lane 4). No hybridization to the pBAD24 vector (lane 3) was detected. DNA fragment length was determined by comparison to a 1-kb molecular mass marker (Promega).
FIG. 1.
Southern analysis of the cloned B. burgdorferi spoT. A DIG-labeled spoT probe (see Materials and Methods) was used to probe a Southern blot containing the PCR product of spoT F1 and spoT R1 primers, which yielded a 0.8-kbp band (lane 1); B. burgdorferi genomic DNA digested with the restriction endonucleases EcoRI and EcoRV, yielding a 5.4-kbp band (lane 2); pBAD24 digested with the restriction endonucleases PstI and SalI (negative control) (lane 3); and pBAD24.spoT digested with PstI and SalI, yielding a 2.9-kbp band (lane 4). The positions of DNA size markers are shown on the left in kilobase pairs (kbp). The sizes of the detected DNA bands are shown on the right.
To verify that the cloned DNA contained the putative B. burgdorferi spoT gene (BB0198), the clone was sequenced. The recombinant plasmid pBAD24.spoT served as template PCR amplicons sequenced. The sequence data were compared to the B. burgdorferi genome and showed >99% sequence identity with the published spoT (BB0198) DNA from B. burgdorferi (20).
Complementation of E. coli CF1693 with B. burgdorferi spoT.
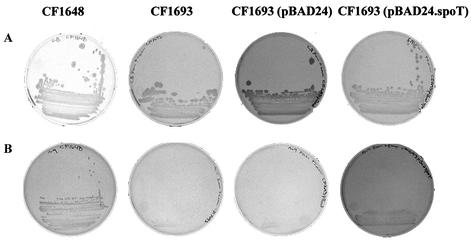
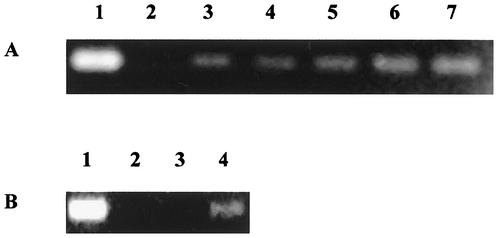
The cloned B. burgdorferi spoT gene was tested for function and ability to complement E. coli CF1693, which has deletions in both the relA and spoT genes, resulting in a phenotype unable to grow on M-9 agar (45). Figure 2 illustrates the growth of E. coli strains CF1648, CF1693, CF1693(pBAD24), and CF1693(pBAD24.spoT) on LB and M-9 media. As expected, the wild-type strain CF1648 grew on both LB and M-9 agar. In contrast, CF1693 and CF1693(pBAD24) grew on LB (Fig. 2A), but did not grow on M-9 agar (Fig. 2B). However, CF1693(pBAD24.spoT) did grow on both LB and M-9 agar plates (Fig. 2A and B), indicating that the B. burgdorferi spoT gene was able to complement the lack of functional relA and spoT genes in CF1693. The bacteria were also grown in LB and M-9 broth to determine the growth characteristics of the various strains. All strains grew at identical rates and to identical cell densities (∼3 × 109 CFU/ml) in LB. In contrast, while the wild-type E. coli CF1648 and E. coli CF1693(pBAD24.spoT) grew at identical rates in M-9 medium, E. coli CF1693 and E. coli CF1693(pBAD24) grew more slowly than either E. coli CF1648 or CF1693(pBAD24.spoT). After 9 h of growth in M-9 broth, the final cell density of E. coli CF1648 and CF1693(pBAD24.spoT) was ∼1 × 108 CFU/ml. The final cell density of E. coli CF1693 and CF1693(pBAD24) was ∼50-fold less, at ∼2 × 106 CFU/ml.
FIG. 2.
Complementation of E. coli CF1693 with the B. burgdorferi spoT gene cloned into the pBAD24 vector. E. coli CF1648, E. coli CF1693, E. coli CF1693(pBAD24), and E. coli CF1693(pBAD24.spoT) were streaked onto LB agar (A) and M-9 agar (B), incubated for 24 h at 37°C, and examined for growth.
Detection of the intracellular alarmone (p)ppGpp.
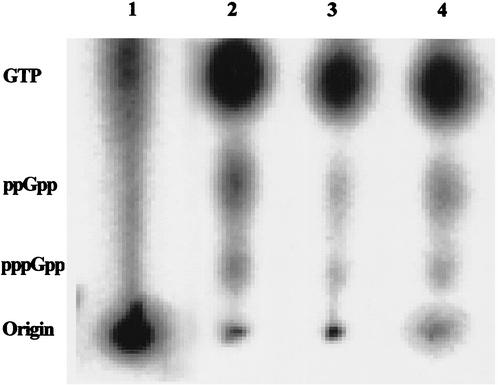
In order to detect the metabolic product of spoT, the alarmone (p)ppGpp, B. burgdorferi cells were grown and labeled with H332PO4 as described in Materials and Methods. Formic acid extracts from the 32P-labeled cells were prepared and separated by TLC and analyzed using a phosphorimager (see Materials and Methods). Intracellular nucleotide pools from B. burgdorferi cells in the presence and absence of serum were compared with those of E. coli CF1648 cells grown in LB or M-9. The accumulation of pppGpp and ppGpp was detected in B. burgdorferi cells starved for 15 min in RPMI and in E. coli cells grown in M-9 (Fig. 3). GTP was also detected in the cell extracts. A GTP (10 mmol) control was used to confirm the TLC separation and was viewed by UV light absorption (data not shown).
FIG. 3.
Detection of (p)ppGpp in formic acid extracts of H332PO4-labeled E. coli and B. burgdorferi cells by PEI TLC and phosphorimaging. Cells were labeled with H332PO4, starved, and then extracted for (p)ppGpp (see Materials and Methods). Lane 1, E. coli CF1648 incubated in LB; lane 2, E. coli CF1648 incubated in M-9 for 15 min; lane 3, B. burgdorferi cells labeled for 4 h and incubated in RPMI plus rabbit serum; lane 4, B. burgdorferi cells labeled for 4 h and then incubated in RPMI for 15 min.
Northern analysis of spoT expression.
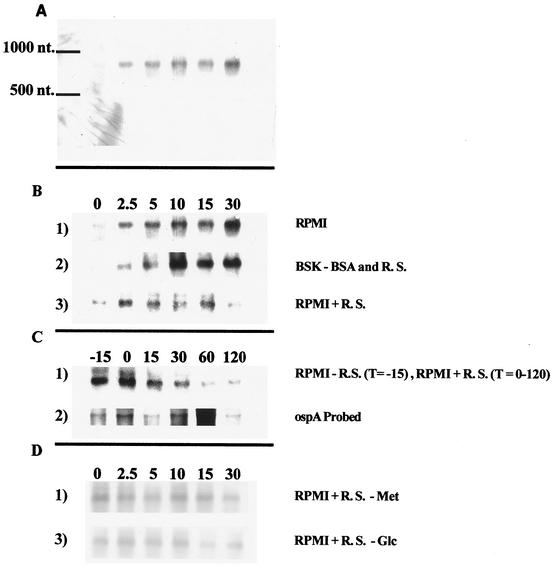
Expression of spoT was examined by Northern blot analysis. RNA extracted from B. burgdorferi cells incubated in either RPMI or BSK (without BSA or rabbit serum, sources of fatty acids) or RPMI plus rabbit serum was separated by agarose gel electrophoresis and examined by Northern blot analysis using a DIG-labeled probe (Fig. 4A, B, and C). A single transcript of ∼900 nucleotides (nt) in length was detected within 2.5 min of the initiation of serum starvation. Expression of spoT continued to increase (∼6-fold) during the 30 min that starvation conditions were maintained. In contrast, cells incubated in RPMI plus rabbit serum exhibited only a low basal level of spoT expression.
FIG. 4.
Northern blot analysis of spoT expression by B. burgdorferi cells in the absence and presence of rabbit serum. Cells samples were prepared and extracted for RNA as described in Materials and Methods. (A) Entire Northern blot of RNA from cells incubated in RPMI (T = 0, 2.5, 5, 10, 15, and 30 min), showing specificity of the DIG-labeled spoT probe, and approximate nucleotide marker. (B) Northern blot showing spoT expression in cells incubated in RPMI (row 1), BSK (row 2), and RPMI plus 6% rabbit serum (row 3) for 0, 2.5, 5, 10, 15, and 30 min. (C) Northern blot showing effect of addition of 6% rabbit serum on expression of spoT (row 1) and ospA (row 2). Cells were starved in RPMI for 15 min (T = −15), and then rabbit serum was added to the culture (T = 0) and samples were taken at 15, 30, 60, and 120 min. (D) Northern blot showing spoT expression in cells incubated in RPMI plus 6% rabbit serum minus methionine (row 1) and RPMI plus 6% rabbit serum minus glucose (row 2). Samples were taken and prepared from cells incubated for 0, 2.5, 5, 10, 15, and 30 min.
In order to determine whether spoT expression could be repressed by the addition of serum, cells were serum starved for 15 min to induce spoT expression and then rabbit serum (6% final concentration) was added to the culture (Fig. 4C, row 1). Samples were taken before and after the addition of rabbit serum extracted for RNA; serum-starved cells (T = −15) exhibited elevated levels of the spoT transcript. As expected, the amount of spoT mRNA remained high immediately after the addition of rabbit serum (T = 0). However, within 15 min after the addition serum to the starved cells, spoT expression exhibited a decline. Thirty minutes after the addition of serum, spoT expression returned to the basal level exhibited by cells not starved for serum (compare Fig. 4B, row 1, and C, row 1). In contrast, when the blot was stripped and probed with a DIG-labeled ospA probe, expression of the predicted transcript (∼1,000 nt) was observed (Fig. 4C, row 2). No decline in expression of ospA was observed, suggesting that the relief of serum starvation did not trigger a general mRNA degradation and that the effect was specific to the repression of spoT expression.
Conditions known to induce the stringent response in E. coli include starvation for amino acids and starvation for glucose (11). In order to determine whether these starvation conditions affect spoT expression in B. burgdorferi, the effects of starvation for amino acid (methionine) or for glucose were examined. Briefly, cells starved for methionine were incubated for up to 30 min in RPMI plus serum without methionine (Fig. 4D, row 1). Samples were collected at various times after the initiation of methionine starvation and extracted for RNA, and spoT expression was determined by Northern analysis (Fig. 4D, row 1). The amount of methionine contributed by the 6% serum was ∼1.3 μM. In a similar experiment, cells starved for glucose were incubated for up to 30 min in RPMI plus serum without glucose (Fig. 4D, row 2). Samples were collected at various times after the initiation of glucose starvation and extracted for RNA, and spoT expression was determined by Northern analysis (Fig. 4D, row 2). The amount of glucose contributed by the 6% rabbit serum was 0.26 μM, which was depleted during the experiment. Expression of spoT throughout the experiment remained at low basal levels and did not increase in response to the absence of methionine or glucose depletion.
Determination of spoT expression in I. scapularis saliva by RT-PCR.
It was of interest to determine whether incubation of B. burgdorferi cells in I. scapularis saliva would affect spoT expression. Increased expression of spoT would suggest that cells induce the stringent response during transmission to the mammalian host. Since only a very limited amount of tick (I. scapularis) saliva was available, RT-PCR was used to examine the expression of spoT. To determine whether RT-PCR amplification could detect spoT mRNA from ∼106 cells incubated under conditions known to induce spoT, RNA extracted from B. burgdorferi cells starved in RPMI (0 to 30 min) was used as a template for RT-PCR with the primers SpoT F2 and SpoT R1. Data presented in Fig. 5A demonstrate that spoT expression was detected within 2.5 min of the initiation of serum starvation and increased during the 30-min starvation. This result was in agreement with the results of Northern analysis of spoT expression (Fig. 4) and suggested that RT-PCR could be used to follow expression of spoT in small numbers of cells. The expected RT-PCR product was gel purified and sequenced to determine whether the RT-PCR product was due to spoT expression. The sequence obtained was 99% identical to the expected spoT product. Thus, RT-PCR could serve as a viable alternative to Northern analysis when determining spoT expression in low numbers of cells. When ∼106 B. burgdorferi cells were incubated in tick saliva, spoT expression increased (Fig. 5B), suggesting that incubation in tick vector saliva results in spoT expression consistent with the stringent response.
FIG. 5.
RT-PCR analysis of spoT expression in RNA isolated from B. burgdorferi cells incubated in RPMI (A) or I. scapularis saliva (B). The 371-bp amplicon was separated by gel electrophoresis on a 1% agarose gel using TAE buffer and stained with ethidium bromide. Equal volumes (10 μl) of the RT-PCR-amplified material were loaded onto each gel lane. (A) RT-PCR performed on RNA isolated from cells incubated in RPMI. Lane 1, PCR amplification from B. burgdorferi genomic DNA; lane 2, amplification using total RNA extracted at 0 min; lane 3, RNA extracted at 2.5 min; lane 4, RNA extracted at 5 min; lane 5, RNA extracted at 10 min; lane 6, RNA extracted at 15 min; lane 7, RNA extracted at 30 min. (B) RT-PCR performed on RNA extracted from B. burgdorferi cells incubated in the saliva of the tick vector I. scapularis (1:1 dilution in nuclease-free H2O). Lane 1, PCR amplification using B. burgdorferi genomic DNA as template; lane 2, negative control of nuclease-free H2O without DNA; lane 3, amplification using total RNA extracted from cells incubated for 0 min in tick saliva; lane 4, RNA extracted from cells incubated for 15 min in tick saliva.
Determination of start and end transcription sites for spoT.
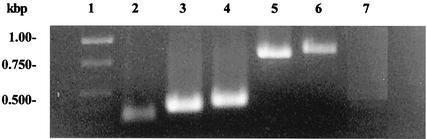
RT-PCR was used to determine the approximate start and end transcription sites for spoT. Since Northern analysis revealed that the spoT transcript was ∼900 nt long, several RT-PCR primer sets were selected to reverse transcribe and amplify possible spoT transcripts of this size and smaller. The RT primers selected started at positions +350, +800, +850, and +950 and the forward PCR primers selected started at positions +25, −25, and −50 from the start codon of spoT. RNA extracted from B. burgdorferi cells starved in RPMI for 15 min was used as the template for RT-PCR amplification. Examination for the resulting amplicons by gel electrophoresis revealed that RT-PCR successfully amplified products from all forward primers and from the +350, +800, and +850 primers (Fig. 6). No amplification was detected from the +950 RT reverse primer. The RT-PCR amplicons were sequenced to confirm their origin from spoT transcripts (data not shown) and were confirmed to be 99% identical to the expected spoT sequence; this experiment was repeated twice with identical results. As the spoT transcript is ∼900 nt long, these data suggest that spoT transcription begins at approximately −50 and extends to +850 to +875.
FIG. 6.
Determination of the approximate start and end transcriptional sites of spoT by RT-PCR. RNA was extracted and prepared for RT-PCR from B. burgdorferi cells starved in RPMI for 15 min. RT-PCR amplification was performed using several primer sets, and the reaction products were separated by gel electrophoresis and visualized using ethidium bromide and UV transillumination. Gel lanes: 1, DNA size markers; 2, amplification using primers SpoT F2 and SpoT R1; 3, amplification using primers SpoT F −25 and SpoT R1; 4, amplification using primers SpoT F −50 and SpoT R1; 5, amplification using primers SpoT F2 and SpoT R +800; 6, amplification using primers SpoT F2 and SpoT F2 and Spot R +850; 7, amplification using primers SpoT F2 and SpoT R +950.
Cloning of a truncated B. burgdorferi spoT gene.
In order to determine whether the truncated spoT mRNA observed by Northern blot analysis was capable of encoding a functional SpoT protein, three DNA sequences were amplified from pBAD24.spoT, using SpoT F1 as the forward primer with either SpoT R2 (to yield the full-length B. burgdorferi spoT gene), SpoT R1 (to yield the 300-bp 5′ portion of spoT), or SpoT +950 (to amplify the 950-bp 5′ portion of the spoT gene). The three amplicons were cloned into a Novagen Perfectly Blunt cloning vector. The clones pMBC04, pMBC05, and pMBC06 were transformed into E. coli CF1693 and the resulting transformants were tested for growth on M-9 minimal agar as previously described. As expected, the full-length spoT, E. coli CF1693(pMBC04), complemented the spoT mutation and restored growth on M-9 agar. The 300-bp truncated clone, E. coli CF1693(pMBC05), was unable to grow on the minimal medium. However, the 950-bp truncated clone, E. coli CF1693(pMBC06), was able to grow on M-9 agar, indicating that it encoded a protein capable of complementing the spoT mutation.
DISCUSSION
The spoT gene encodes a bifunctional enzyme capable of degrading and synthesizing (p)ppGpp (PSII). In E. coli, SpoT is constitutively expressed, resulting in low concentrations of (p)ppGpp during normal growth and resulting in changes to RNA polymerase promoter activity (27, 44). The SpoT/PSII system is thought to regulate growth rate by adjusting the rate of stable RNA synthesis (6, 7, 12). It is hypothesized that constitutive SpoT activity helps the cell monitor nutritional states with regard to carbon sources (11). The findings of Kvint et al. (31, 32) show that RpoS-dependent promoters require ppGpp for induction. It is proposed that RpoS-dependent promoters are positively affected by ppGpp due to the dissociation of σ70-programmed RNA polymerase from stringent promoters, increasing the availability of free RNA polymerase. Additionally, ppGpp reduces the stability of open complexes with RNA polymerase (6). The increased levels of ppGpp also can positively affect RpoS by increasing the half-lives of the RNA polymerase complexes. It is also thought that ppGpp may directly or indirectly aid in the ability of σs-dependent promoters to bind RNA polymerase holoenzyme.
Several relA or spoT homologs have been cloned and characterized (2, 19, 34, 35, 39, 53, 55, 57) These single genes appear to function like either the relA or spoT of E. coli or a combination of the two genes. For example, relA and spoT homologues of Bacillus stearothermophilus, Bacillus subtilis, Streptococcus pyogenes, and Streptococcus rattus are induced only under conditions of amino acid depletion. In contrast, the relA or spoT homologues of M. smegmatis, Mycobacterium tuberculosis, and Streptococcus equisimilis only respond to carbon starvation.
The stringent response is not only important in the regulation of short-term responses to nutritional depletion, it is also important in the regulation of long-term starvation-stress responses. For example, M. xanthus cells change from motile rods to nonmotile, spherical myxospores within fruiting bodies in response to starvation (25). The intracellular accumulation of (p)ppGpp is required for the initiation of fruiting body formation (25). In M. smegmatis, carbon starvation results in the activation RelA, increased ppGpp concentrations, and a morphological change from elongated rods to cocci (38). Thus, increased ppGpp concentration regulates FtsZ activity and, therefore, cell division. Increases in (p)ppGpp concentration have also been shown to control acid-stress tolerance in Lactococcus lactis (41).
B. burgdorferi undergoes several environmental transitions during transmission between the tick vector and a vertebrate host. These changes include temperature, nutrient availability, cell density, and pH (13, 16, 28). Some of these environmental transitions have been shown to affect gene expression in B. burgdorferi. Carreiro et al. (10) have demonstrated that rapid increases in temperature induce the heat shock response. More recently, Alban et al. (1) demonstrated that at least 20 proteins are induced in B. burgdorferi cells starved for serum. Additionally, motile helical cells convert over ∼48 h of starvation to round nonmotile starvation forms (or “cysts”). Since there are no fatty acid biosynthesis pathways in B. burgdorferi, these bacteria obtain fatty acids from their environment (20). Recently, Cox and Radolf (14) demonstrated that B. burgdorferi obtains fatty acids by diffusion through its outer membrane, which could result in the slow growth rate of the organism.
Experimental data presented in this communication demonstrate that the spoT gene of B. burgdorferi is functional and able to complement E. coli CF1693 (ΔrelA ΔspoT) to restore wild-type phenotypic growth in M-9 minimal medium (Fig. 2). Further, (p)ppGpp was detected in 32P-labeled cellular extracts of B. burgdorferi cells starved for serum (Fig. 3), suggesting that the spoT gene encodes PSII activity. Characterization of spoT expression was carried out by Northern blot analysis and RT-PCR.
Northern blot analysis of spoT expression in B. burgdorferi revealed that a 900-nt transcript was expressed within 2.5 min after the initiation of serum starvation (Fig. 4). Expression of spoT increased ∼6-fold over the first 30 min of starvation and returned to basal levels after 120 min of serum starvation (data not shown). Further, when rabbit serum was added to the serum-starved B. burgdorferi cells, the expression of spoT rapidly decreased to basal levels. The decrease in spoT expression was specific, since ospA expression remained nearly constant during the 120 min after the addition of rabbit serum. Thus, serum starvation induces spoT expression and serum feeding represses spoT expression. Additionally, neither glucose nor amino acid starvation (methionine) resulted in induction of spoT, indicating that in B. burgdorferi spoT expression and, therefore, the stringent response is induced only by serum starvation. In comparison, the E. coli stringent response is activated by starvation for amino acids, glucose, and fatty acids (11). While there are no data describing the transcriptional expression of either relA or spoT in E. coli or organisms other than B. burgdorferi, both TLC data for (p)ppGpp accumulation and Western blot analyses of RelA and SpoT expression have been used to characterize PSI and PSII activities and protein expression, respectively (2, 19, 34-36, 53, 55, 56). For example, Wendrich et al. (54) demonstrated by immunoblot analysis that the RelA/SpoT homologue of B. stearothermophilus is induced during amino acid starvation. Additionally, (p)ppGpp accumulation was detected within 3 min of the initiation of amino acid starvation. Our observations of spoT expression in B. burgdorferi are consistent with these previous reports.
A single ∼900-nt hybridizable transcript was always observed when mRNA preparations were examined using a spoT-specific DIG-labeled probe during Northern blot analysis. No other transcripts were observed when using the DIG-labeled spoT probe. The spoT transcript length was unexpected, because examination of B. burgdorferi genome data revealed a 2,001-bp open reading frame (ORF) (BB0198) identified as spoT (20). Further RT-PCR experiments suggest that the spoT transcript starts approximately 50 to 75 bp upstream of the start codon and ends approximately 850 to 875 bases downstream. The data strongly suggest that the spoT transcript is ∼900 nt long. Additionally, when a fragment of spoT containing the first 950 bp of the structural gene was cloned (pMBC06) it was able to complement E. coli CF1693 to allow growth on M-9 agar, indicating the restoration of the stringent response. We cannot distinguish between the possibility that the transcript is rapidly processed from a longer mRNA encoded from the entire ORF (40) or that the spoT ORF contains an internal transcriptional terminator. However, mutational analysis of the E. coli SpoT demonstrates that the first 203 amino acids of the 702-amino acid SpoT are necessary for (p)ppGpp phosphohydrolase activity and that amino acids 67 to 374 are necessary for PSII activity (22). The bacterium Streptomyces coelicolor has a functional relA/spoT homologue that has been characterized (34). Only 453 of the 874 amino acids of the protein are necessary for both (p)ppGpp phosphohydrolase (residues 93 to 397) and synthase (residues 267 to 453) activities (22, 35). These data support the idea that the entire spoT ORF is not needed to yield a functional SpoT protein.
When B. burgdorferi cells were incubated in the saliva of the tick vector I. scapularis, a 371-bp spoT-specific DNA fragment was amplified by RT-PCR. The RT-PCR product was sequenced and shown to be 99% identical to the expected spoT transcript. This result demonstrates that B. burgdorferi spoT is induced when the cells are incubated in the saliva of I. scapularis. This result is consistent with the hypothesis that the B. burgdorferi cells exhibit the stringent response during transmission from the tick vector to the mammalian host by tick saliva. Hammer and Swanson (24) demonstrated that when Legionella pneumophila is subjected to amino acid depletion, ppGpp accumulates and the cells convert from a replicative to a virulent state. More recently, Bachman and Swanson (3) showed that the accumulation of ppGpp activated an RpoS-dependent virulence pathway in L. pneumophila.
Finally, increases in intracellular concentration of (p)ppGpp have been shown to up regulate rpoS (σs), the main sigma factor involved in the expression of stationary-phase genes as well as the expression of genes involved in long-term starvation conditions (15, 33, 52). Recently, Elias et al. (16) showed that the B. burgdorferi genome contains a functional rpoS. It will be interesting to determine whether rpoS expression responds to serum starvation.
Acknowledgments
This work was supported by Public Health Service grant R01 AI 37230 from the National Institute of Allergy and Infectious Diseases.
We thank Michael Cashel for the gift of strains, Jon Beckwith for the gift of plasmids, and Thomas Mather for the gift of tick saliva.
REFERENCES
- 1.Alban, P. S., P. W. Johnson, and D. R. Nelson. 2000. Serum-starvation-induced changes in protein synthesis and morphology of Borrelia burgdorferi. Microbiology 146:119-127. [DOI] [PubMed] [Google Scholar]
- 2.Avarbock, D., J. Salem, L. S. Li, Z. M. Wang, and H. Rubin. 1999. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233:261-269. [DOI] [PubMed] [Google Scholar]
- 3.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 4.Baracchini, E., and H. Bremer. 1988. Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J. Biol. Chem. 263:2597-2602. [PubMed] [Google Scholar]
- 5.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 6.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. [DOI] [PubMed] [Google Scholar]
- 7.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 8.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257:9759-9769. [PubMed] [Google Scholar]
- 9.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 10.Carreiro, M. M., D. C. Laux, and D. R. Nelson. 1990. Characterization of the heat shock response and identification of heat shock protein antigens of Borrelia burgdorferi. Infect. Immun. 58:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss et al. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 12.Cashel, M., and J. Gallant. 1968. Control of RNA synthesis in Escherichia coli. I. Amino acid dependence of the synthesis of the substrates of RNA polymerase. J. Mol. Biol. 34:317-330. [DOI] [PubMed] [Google Scholar]
- 13.Cluss, R. G., and J. T. Boothby. 1990. Thermoregulation of protein synthesis in Borrelia burgdorferi. Infect. Immun. 58:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, D. L., and J. D. Radolf. 2001. Insertion of fluorescent fatty acid probes into the outer membranes of the pathogenic spirochaetes Treponema pallidum and Borrelia burgdorferi. Microbiology 147:1161-1169. [DOI] [PubMed] [Google Scholar]
- 15.Eichel, J., Y. Y. Chang, D. Riesenberg, and J. E. Cronan, Jr. 1999. Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (σs). J. Bacteriol. 181:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elias, A. F., J. L. Bono, J. A. Carroll, P. Stewart, K. Tilly, and P. Rosa. 2000. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J. Bacteriol. 182:2909-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing, C., A. Scorpio, D. R. Nelson, and T. N. Mather. 1994. Isolation of Borrelia burgdorferi from saliva of the tick vector, Ixodes scapularis. J. Clin. Microbiol. 32:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faxen, M., and L. A. Isaksson. 1994. Functional interactions between translation, transcription and ppGpp in growing Escherichia coli. Biochim. Biophys. Acta 1219:425-434. [DOI] [PubMed] [Google Scholar]
- 19.Flardh, K., T. Axberg, N. H. Albertson, and S. Kjelleberg. 1994. Stringent control during carbon starvation of marine Vibrio sp. strain S14: molecular cloning, nucleotide sequence, and deletion of the relA gene. J. Bacteriol. 176:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 21.Gentry, D. R., and M. Cashel. 1995. Cellular localization of the Escherichia coli SpoT protein. J. Bacteriol. 177:3890-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentry, D. R., and M. Cashel. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 19:1373-1384. [DOI] [PubMed] [Google Scholar]
- 23.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 25.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinemann, M., and R. Wagner. 1997. Guanosine 3′,5′-bis(diphosphate) (ppGpp)-dependent inhibition of transcription from stringently controlled Escherichia coli promoters can be explained by an altered initiation pathway that traps RNA polymerase. Eur. J. Biochem. 247:990-999. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez, V. J., and H. Bremer. 1991. Escherichia coli ppGpp synthetase II activity requires spoT. J. Biol. Chem. 266:5991-5999. [PubMed] [Google Scholar]
- 28.Indest, K. J., R. Ramamoorthy, M. Sole, R. D. Gilmore, B. J. Johnson, and M. T. Philipp. 1997. Cell-density-dependent expression of Borrelia burgdorferi lipoproteins in vitro. Infect. Immun. 65:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kjelleberg, S., M. Hermansson, P. Marden, and G. W. Jones. 1987. The transient phase between growth and nongrowth of heterotrophic bacteria, with emphasis on the marine environment. Annu. Rev. Microbiol. 41:25-49. [DOI] [PubMed] [Google Scholar]
- 30.Kuroda, A., H. Murphy, M. Cashel, and A. Kornberg. 1997. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272:21240-21243. [DOI] [PubMed] [Google Scholar]
- 31.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σs. J. Biol. Chem. 275:14795-14798. [DOI] [PubMed] [Google Scholar]
- 32.Kvint, K., C. Hosbond, A. Farewell, O. Nybroe, and T. Nystrom. 2000. Emergency derepression: stringency allows RNA polymerase to override negative control by an active repressor. Mol. Microbiol. 35:435-443. [DOI] [PubMed] [Google Scholar]
- 33.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Costa, O. H., M. A. Fernandez-Moreno, and F. Malpartida. 1998. The relA/spoT-homologous gene in Streptomyces coelicolor encodes both ribosome-dependent (p)ppGpp-synthesizing and -degrading activities. J. Bacteriol. 180:4123-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mechold, U., M. Cashel, K. Steiner, D. Gentry, and H. Malke. 1996. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J. Bacteriol. 178:1401-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mechold, U., and H. Malke. 1997. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J. Bacteriol. 179:2658-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ojha, A. K., T. K. Mukherjee, and D. Chatterji. 2000. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect. Immun. 68:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostling, J., L. Holmquist, and S. Kjelleberg. 1996. Global analysis of the carbon starvation response of a marine Vibrio species with disruptions in genes homologous to relA and spoT. J. Bacteriol. 178:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persson, M., E. Glatz, and B. Rutberg. 2000. Different processing of an mRNA species in Bacillus subtilis and Escherichia coli. J. Bacteriol. 182:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517-528. [DOI] [PubMed] [Google Scholar]
- 42.Ribeiro, J. M., T. N. Mather, J. Piesman, and A. Spielman. 1987. Dissemination and salivary delivery of Lyme disease spirochetes in vector ticks (Acari: Ixodidae). J. Med. Entomol. 24:201-205. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1987. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Sarubbi, E., K. E. Rudd, and M. Cashel. 1988. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol. Gen. Genet. 213:214-222. [DOI] [PubMed] [Google Scholar]
- 45.Sarubbi, E., K. E. Rudd, H. Xiao, K. Ikehara, M. Kalman, and M. Cashel. 1989. Characterization of the spoT gene of Escherichia coli. J. Biol. Chem. 264:15074-15082. [PubMed] [Google Scholar]
- 46.Scorpio, A., P. Johnson, A. Laquerre, and D. R. Nelson. 1994. Subcellular localization and chaperone activities of Borrelia burgdorferi Hsp60 and Hsp70. J. Bacteriol. 176:6449-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seldon, R., and F. M. Ausubel. 1987. Current protocols in molecular biology, p. 4.9.1-4.9.8. Wiley-Intersciences, New York, N.Y.
- 48.Seldon, R., and F. M. Ausubel. 1987. Current protocols in molecular biology, p. 2.8.1-2.9.15. Wiley-Intersciences, New York, N.Y.
- 49.Seyfzadeh, M., J. Keener, and M. Nomura. 1993. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:11004-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 51.Subramanian, G., E. V. Koonin, and L. Aravind. 2000. Comparative genome analysis of the pathogenic spirochetes Borrelia burgdorferi and Treponema pallidum. Infect. Immun. 68:1633-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teich, A., S. Meyer, H. Y. Lin, L. Andersson, S. Enfors, and P. Neubauer. 1999. Growth rate related concentration changes of the starvation response regulators σs and ppGpp in glucose-limited fed-batch and continuous cultures of Escherichia coli. Biotechnol. Prog. 15:123-129. [DOI] [PubMed] [Google Scholar]
- 53.van der Biezen, E. A., J. Sun, M. J. Coleman, M. J. Bibb, and J. D. Jones. 2000. Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl. Acad. Sci. USA 97:3747-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wendrich, T. M., C. L. Beckering, and M. A. Marahiel. 2000. Characterization of the relA/spoT gene from Bacillus stearothermophilus. FEMS Microbiol. Lett. 190:195-201. [DOI] [PubMed] [Google Scholar]
- 55.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26:65-79. [DOI] [PubMed] [Google Scholar]
- 56.Whitehead, K. E., G. M. Webber, and R. R. England. 1998. Accumulation of ppGpp in Streptococcus pyogenes and Streptococcus rattus following amino acid starvation. FEMS Microbiol. Lett. 159:21-26. [DOI] [PubMed] [Google Scholar]
- 57.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 58.Zung, J. L., S. Lewengrub, M. A. Raszinska, A. Speilman, S. R. Telford III, and J. Peisman. 1989. Fine structure evidence for the penetration of Lyme disease spirochete Borrelia burgdorferi through the gut and salivary tissue of Ixodes dammini. Can. J. Zool. 67:1737-1748. [Google Scholar]