Abstract
The novel benzoyl coenzyme A (benzoyl-CoA) biosynthesis pathway in “Streptomyces maritimus” was investigated through a series of target-directed mutations. Genes involved in benzoyl-CoA formation were disrupted through single-crossover homologous recombination, and the resulting mutants were analyzed for their ability to biosynthesize the benzoyl-CoA-primed polyketide antibiotic enterocin. Inactivation of the unique phenylalanine ammonia-lyase-encoding gene encP was previously shown to be absolutely required for benzoyl-CoA formation in “S. maritimus”. The fatty acid β-oxidation-related genes encH, -I, and -J, on the other hand, are necessary but not required. In each case, the yield of benzoyl-CoA-primed enterocin dropped below wild-type levels. We attribute the reduced benzoyl-CoA formation in these specific mutants to functional substitution and cross-talk between the products of genes encH, -I, and -J and the enzyme homologues of primary metabolism. Disruption of the benzoate-CoA ligase encN gene did not perturb enterocin production, however, demonstrating that encN is extraneous and that benzoic acid is not a pathway intermediate. EncN rather serves as a substitute pathway for utilizing exogenous benzoic acid. These experiments provide further support that benzoyl-CoA is formed in a novel bacterial pathway that resembles the eukaryotic assembly of benzoyl-CoA from phenylalanine via a β-oxidative path.
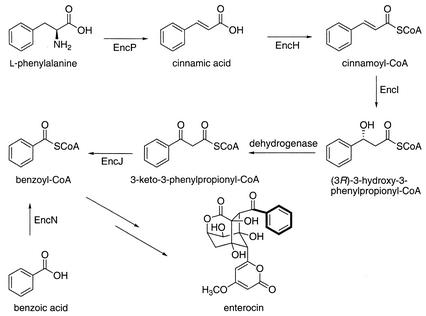
Benzoyl coenzyme A (benzoyl-CoA) is a rare bacterial metabolite that serves as a starter unit for the biosynthesis of the polyketides enterocin and soraphen (11) and as a central intermediate of anaerobic aromatic metabolism (3). Its biosynthesis involves at least two oxidative pathways from the amino acid phenylalanine. Feeding experiments with 2H- and 13C-labeled intermediates (5, 6) and sequence analysis of the enterocin biosynthetic gene cluster (enc) (12) revealed that benzoyl-CoA is biosynthesized in a plant-like manner in the sediment-derived bacterium “Streptomyces maritimus” (Fig. 1). In plants where benzoyl-CoA is a common metabolite and a component of numerous natural products, phenylalanine is nonoxidatively deaminated to cinnamic acid and then converted in a CoA-dependent manner to benzoyl-CoA via two proposed routes involving β-oxidation (5) and reverse aldol (1) reactions. The denitrifying bacterium Thauera aromatica, conversely, metabolizes phenylalanine to benzoyl-CoA, which is then reductively degraded (15). This anaerobic conversion involves the transamination of phenylalanine to phenylpyruvate, followed by two successive α-oxidative decarboxylations.
FIG. 1.
Proposed biosynthetic pathway for benzoyl-CoA, the starter unit of the polyketide enterocin. EncP, PAL; EncH, cinnamate-CoA ligase; EncI, cinnamoyl-CoA hydratase; EncJ, 3-keto-3-phenylpropionyl-CoA thiolase; EncN, benzoate-CoA ligase.
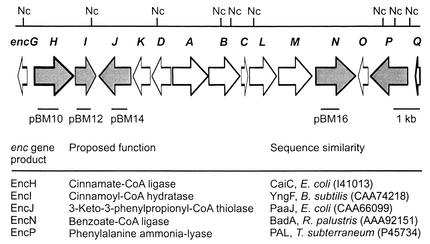
The biosynthesis of benzoyl-CoA in “S. maritimus,” which parallels common eukaryotic pathways in plants and fungi, is a novel bacterial pathway. Analysis of the 20-open-reading-frame (ORF) enc gene cluster revealed five ORFs (Fig. 2) putatively involved in the biosynthesis of the benzoyl-CoA starter unit of the bacteriostatic agent enterocin (12). These genes are arranged on four transcripts neighboring the enterocin polyketide synthase (PKS) genes encA, encB, and encC. We previously characterized the unique phenylalanine ammonia-lyase (PAL; EC 4.3.1.5) gene encP and showed that its inactivation resulted in the abolishment of de novo cinnamic acid and enterocin synthesis (20). Enterocin biosynthesis could be restored in the encP-inactivated mutant “S. maritimus” KP through supplementation with cinnamic or benzoic acid as well as complementation with plasmid-borne encP. Subsequent biosynthetic reactions are thought to involve the activation of cinnamic acid to its CoA thioester, presumably by the encH gene product cinnamate-CoA ligase, followed by one round of β-oxidation. Two β-oxidation homologous encoding genes, encI and encJ, which encode an enoyl-CoA hydratase and a β-oxoacyl-CoA thiolase, respectively, are clustered in the enc gene set. The third gene product necessary for a complete β-oxidation series, a hydroxyacyl-CoA dehydrogenase, was, however, not identified in the cluster. The fifth gene putatively involved in the formation of benzoyl-CoA in “S. maritimus” that was identified in the gene cluster is encN, a putative benzoate-CoA ligase.
FIG. 2.
Partial organization of the enterocin biosynthetic gene cluster (enc). The orientation of the arrows indicates the direction of each ORF, and those that are shaded are involved in benzoyl-CoA biosynthesis. Suicide plasmids pBM10, pBM12, pBM14, and pBM16 contain internal fragments of encH, encI, encJ, and encN, respectively, and were used for gene inactivation by single recombination. Nc, NcoI. B. subtilis, Bacillus subtilis; R. palustris, Rhodopseudomonas palustris; T. subterraneum, Trifolium subterraneum.
In the present study, we examined the genes encH, -I, -J, and -N in benzoyl-CoA formation. Each gene was inactivated, and the resulting mutants were biochemically examined. Our results describe for the first time the benzoyl-CoA biosynthesis genes in a bacterium whose products catalyze a plant-like β-oxidative pathway.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All strains and plasmids used in this work are listed in Table 1. “S. maritimus” strain BD26T (GenBank accession number AF233338) was grown as previously described (12). A1 medium was used for sporulation, and R2YE medium was used for the isolation of genomic DNA. Escherichia coli XL1-Blue was used for subcloning and grown on Luria-Bertani plates or in Luria-Bertani liquid medium. E. coli S17-1 was used as the host for E. coli-“S. maritimus” conjugation.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Properties or product | Source or reference |
|---|---|---|
| Strains | ||
| E. coli S17-1 | Contains RP4 integrated into the chromosome | 16 |
| “S. maritimus” BD-26T | Wild type, enterocin+ | 17 |
| “S. maritimus” KP | BD-26T exconjugant containing pBM4, enterocin−; Amr | 20 |
| “S. maritimus” KH | BD-26T exconjugant containing pBM10, enterocin+; Amr | This study |
| “S. maritimus” KI | BD-26T exconjugant containing pBM12, enterocin+; Amr | This study |
| “S. maritimus” KJ | BD-26T exconjugant containing pBM14, enterocin+; Amr | This study |
| “S. maritimus” KN | BD-26T exconjugant containing pBM16, enterocin+; Amr | This study |
| Plasmids | ||
| pCR2.1-TOPO | Vector for cloning PCR products | Invitrogen |
| pKC1139 | E. coli-streptomycete conjugal transfer vector; Amr | 2 |
| pJP15F11 | pOJ446 cosmid clone containing enc gene cluster | 13 |
| pBM9 | 0.8-kb amplified encH internal fragment cloned into pCR2.1-TOPO | This study |
| pBM10 | 0.8-kb EcoRI fragment from pBM9 cloned into pKC1139 | This study |
| pBM11 | 0.5-kb amplified encl internal fragment cloned into pCR2.1-TOPO | This study |
| pBM12 | 0.5-kb EcoRI fragment from pBM11 cloned into pKC1139 | This study |
| pBM13 | 0.7-kb amplified encJ internal fragment cloned into pCR2.1-TOPO | This study |
| pBM14 | 0.7-kb EcoRI fragment from pBM13 cloned into pKC1139 | This study |
| pBM15 | 0.7-kb amplified encN internal fragment cloned into pCR2.1-TOPO | This study |
| pBM16 | 0.7-kb EcoRI fragment from pBM15 cloned into pKC1139 | This study |
DNA manipulations.
“S. maritimus” total genomic DNA was isolated as described previously (12). Recombinant DNA procedures were performed by standard techniques (8, 14). Biotin-labeling and detection of chemiluminescent positives were performed with the DNADetector HPR Southern blotting kit (KPL, Inc.). Oligonucleotides were obtained from Sigma Genosys. PCR was carried out on a PTC-2000 thermal cycler (MJ Research) with Taq (GIBCO) DNA polymerase. DNA sequencing by a BigDye terminator cycle sequencing reaction with an ABI 377 sequencer was performed at the Laboratory of Molecular Systematics and Evolution at the University of Arizona.
Gene disruptions.
DNA fragments containing the targeted genes were PCR amplified (Table 2), cloned into the E. coli-streptomycete conjugal transfer vector pKC1139, and conjugated into “S. maritimus” as previously described for the construction of the related encP mutant “S. maritimus” KP (20). The single-crossover mutants were selected after propagating transconjugants on SGGP (21) plates at 37°C, and apramycin-resistant colonies were confirmed by Southern hybridization with biotinylated gene probes. Mutant strains were grown at 37°C on A1 plates containing 100 μg of apramycin/ml for approximately 24 to 30 h until sporulation.
TABLE 2.
Oligonucleotide primers used to amplify enc fragments for gene disruption experiments
| Targeted gene | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) | Product size (bp) |
|---|---|---|---|
| encH | CTGAAGGACTCTCAGGCCG | ATGGGCCTTCCCCGCACCAG | 789 |
| encI | TGACCGTCACGGTCGACAGCG | CTCGACGATGTCGTCGAGGAG | 529 |
| encJ | CGAGTGCCCTGGGCATGCGC | GCTGGGCTGGACCGTGCCGC | 699 |
| encN | CGCAGGGCCGGACGACGGC | GTGCCAGTACTGGGCCACG | 720 |
HPLC-MS analysis.
Cinnamic acid and polyketide production in “S. maritimus” mutants was analyzed by high-performance liquid chromatography (HPLC) and electrospray-mass spectrometry (MS) as previously described for the encP mutant “S. maritimus” KP (12). Feeding experiments involved overlaying ring-2H5-labeled intermediates (0.5 to 1 mg/plate).
Nucleotide sequence accession number.
The gene sequence of the enterocin biosynthetic gene cluster has been deposited at GenBank under accession number AF254925.
RESULTS
Disruption of the benzoyl-CoA formation genes encH-encJ and encN.
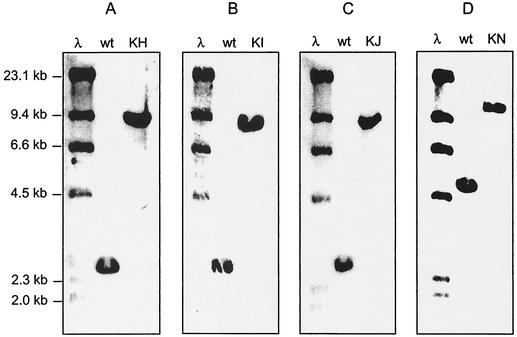
To examine which of the previously cloned and sequenced enc genes are involved in benzoyl-CoA biosynthesis in the thermotolerant marine bacterium “S. maritimus,” we developed a genetics system and disrupted several candidate genes by single-crossover homologous recombination. The PAL-encoding gene encP was previously inactivated (20) and verified to be involved in the initial reaction of the benzoyl-CoA pathway (Fig. 1). We employed a similar approach involving the E. coli-to-“S. maritimus” conjugal transfer of pKC1139-based temperature-sensitive plasmids carrying the internal 0.5- to 0.8-kb fragments of target enc genes. We constructed plasmids pBM10, pBM12, pBM14, and pBM16 to inactivate the genes encH, encI, encJ, and encN, respectively (Fig. 2). A single crossover between chromosomal and pKC1139-based enc sequences generated tandemly duplicated enc sequences with the vector containing an apramycin resistance gene between the sequences. In each plasmid, the enc genes were truncated at both termini. As a consequence, the mutant upstream- and downstream-targeted enc genes lacked the carboxyl and amino termini, respectively, thus resulting in nonfunctional enc gene products. In disrupting encH, the downstream gene encI may have been additionally inactivated due to polar effects. Transconjugants were grown under selective conditions, providing the “S. maritimus” mutant strains KH, KI, KJ, and KN in which encH, encI, encJ, and encN were respectively disrupted. Southern blot hybridization of genomic DNA from the wild-type and apramycin-resistant transconjugants with biotinylated enc gene fragment DNA probes verified all gene disruptions (Fig. 3). Predicted band shifts of approximately 7 kb were detected in NcoI digests of the total DNA. The four mutants did not exhibit any different phenotypes in comparison to the wild-type strain in A1 media when fermented at 37°C in the presence of apramycin.
FIG. 3.
Southern analysis of NcoI-digested genomic DNA from “S. maritimus” wild type (wt) and mutants KH, KI, KJ, and KN by using the following biotinylated DNA probes: 0.8-kb encH fragment from pBM9 (A); 0.5-kb encI fragment from pBM11 (B); 0.7-kb encJ fragment from pBM13 (C); 0.7-kb encN fragment from pBM15 (D). λ, bacteriophage λ DNA-HindIII digestion marker.
Analysis of the mutants and supplementation with deuterium-labeled intermediates.
To differentiate the roles of the two putative CoA ligases, the 535-amino-acid EncH and the 522-amino-acid EncN, cinnamic acid and enterocin production in the mutant strains KH and KN was compared. Although the encH mutant KH produced cinnamic acid and enterocin, HPLC analysis showed that their yields decreased to 15 to 20% of that of the wild-type strain (Table 3). Upon supplementation with d5-benzoate, enterocin production was fully recovered whereas cinnamic acid levels remained unchanged. MS analysis furthermore showed an 84% incorporation of the d5 label into enterocin. No significant difference, however, was observed by HPLC between the encN mutant KN and the wild-type strain, as wild-type levels of cinnamic acid and enterocin were detected. Supplementation with d5-benzoate provided no further increase in enterocin productivity and no notable incorporation of the deuterium label. Control feeding experiments with [ring-d5]phenylalanine gave similar incorporation percentages of approximately 9% in enterocin produced by the two enc CoA ligase mutants as well as the wild-type strain. These experiments support the DNA sequence analyses in assigning functional roles of the encH and encN gene products as cinnamate-CoA and benzoate-CoA ligases, respectively.
TABLE 3.
Characteristics of “S. maritimus” BD-26T-derived enc mutants
| Strain | Gene(s) inactivated | % Productiona
|
% Incorporation of ring-d5-labeled substrates in enterocin
|
|||
|---|---|---|---|---|---|---|
| Cinnamate | Enterocin | Phenylalanine | Benzoate | (3R)-3-Hydroxy-3-phenylpropionate | ||
| Wild type | none | 100 | 100 | 7 | 37 | 3 |
| “S. maritimus” KPb | encP, encO | 0 | 0 | 0 | 100 | Not tested |
| “S. maritimus” KH | encH, encI | 20 | 15 | 9 | 84 | 38 |
| “S. maritimus” KI | encI | 25 | 20 | 9 | 85 | 46c |
| “S. maritimus” KJ | encJ | 160 | 75 | 9 | 43 | 50c |
| “S. maritimus” KN | encN | 100 | 100 | 8 | 0 | 4 |
The relative production levels of cinnamic acid and enterocin were compared to those from the wild-type “S. maritimus” strain that were set to 100%. Each percentage is an average value from triplicate experiments with an error of 5 to 10%.
Data for mutant KP were previously reported (20) and are provided for comparison.
Previously reported in reference 5.
The functional roles of the two β-oxidation genes encI and encJ in benzoyl-CoA biosynthesis were similarly analyzed. The encI gene encodes a 258-amino-acid protein homologous to enoyl-CoA hydratases, and its initiation ATG codon overlaps the TGA stop codon of encH. The encI mutant KI behaved similarly to the encH mutant KH in that the production of cinnamic acid and enterocin significantly dropped 20 to 25% and that supplementation with d5-benzoic acid fully restored enterocin biosynthesis to wild-type levels (Table 3). The stop codon of the convergently transcribed encJ gene, which encodes a 400-amino-acid protein homologous to β-oxoacyl-CoA thiolase, is located 146 bases downstream of the encI gene. HPLC analysis of the encJ mutant KJ showed elevated levels of cinnamic acid but a 25% loss in enterocin yield. Supplementation with d5-benzoic acid restored enterocin biosynthesis to wild-type levels and resulted in a 43% incorporation of label.
It was previously demonstrated that β-hydroxy- and β-ketophenylpropionates were assimilated into the benzoate-derived unit of enterocin and concluded that their respective CoA thioester analogues were pathway intermediates (6). In order to determine to what degree the CoA ligases EncH and EncN may additionally serve in activating phenylpropanoid intermediates, we fed (3R)-[ring-2H5]-3-hydroxy-3-phenylpropionic acid (5) to the KH and KN mutants (Table 3). Similar incorporation levels of 3 to 4% were measured in enterocin produced by the wild type and mutant KN, suggesting that EncN does not appreciably activate the phenylpropanoid intermediate. Enterocin from the encH-encI mutant KH, on the other hand, was enriched at a much higher percentage of 38%. This mutant carries not only a disrupted cinnamate-CoA ligase encH but also an inactivated cinnamoyl-CoA hydratase encI gene due to polar effects. Thus, the enrichment from this experiment was compared not against wild-type levels but rather against the encI mutant KI in which a slightly higher 46% incorporation was previously documented (5). Consequently, 3-hydroxy-3-phenylpropionate may be activated in vivo by the action of more than one CoA ligase, including the encH gene product.
DISCUSSION
The biosynthesis of benzoyl-CoA in “S. maritimus” BD-26T was examined by mutating specific genes associated with the enterocin biosynthetic gene cluster. These experiments provide further support that benzoyl-CoA is formed in a novel bacterial pathway that resembles the eukaryotic assembly of benzoyl-CoA from phenylalanine via a β-oxidative route (5, 9). Inactivation of the unique PAL-encoding gene encP was previously shown to be absolutely required for benzoyl-CoA formation in “S. maritimus” (20). The fatty acid β-oxidation related genes encH, -I, and -J, on the other hand, were shown by target-directed mutagenesis in this study to be necessary but not required. In each case, the yield of benzoyl-CoA-primed enterocin significantly dropped below wild-type levels. We attribute the reduced benzoyl-CoA formation in these specific mutants to functional substitution and cross-talk between the products of genes encH, -I, and -J and their corresponding fatty acid β-oxidation homologues.
β-Oxidation of cinnamoyl-CoA and of fatty acids proceeds with the same absolute configuration of the 3-hydroxyacyl-CoA intermediate (5), thus supporting the evolutionary relationship between these pathways. Furthermore, there is considerable amino acid sequence similarity between the cinnamoyl-CoA hydratase EncI and fatty acid enoyl-CoA hydratases (12). In particular, the EncI amino acid residues Gly-116, Glu-119, and Glu-139, which are essential for catalytic activity in homologous-fatty-acid-type hydratases (4), suggest that the two enzyme systems operate by a common mechanism.
Inactivation of the encI gene in mutant KI resulted in the largest drop in cinnamic acid and enterocin productivity among the enc β-oxidation mutants, suggesting that EncI is the most pathway specific of the enterocin β-oxidation enzymes. While a similar loss was measured in the encH mutant KH, this mutant may effectively be an encI mutant and the overlapping encI gene may be translationally coupled. If this is indeed the case, the role of the encH CoA ligase gene product becomes less clear from these gene inactivation experiments. To this end, we have recently expressed and purified EncH as an octahistidyl-tagged recombinant protein and verified that this protein indeed catalyzes the in vitro CoA activation of trans-cinnamic acid (M. Izumikawa and B. S. Moore, unpublished observations).
Missing from the sequenced enc cluster is a homologue of β-hydroxyacyl-CoA dehydrogenase, the β-oxidation enzyme needed to catalyze the dehydrogenation of (3R)-3-hydroxy-3-phenylpropionoyl-CoA and complete the oxidative cycle. Homologues were found neither 2 kb upstream nor downstream of the 21.3-kb enc cluster (12). Heterologous expression of the enc-containing E. coli-streptomycete shuttle cosmid pJP15F11, nonetheless, was sufficient to produce enterocin (13), suggesting that either a dedicated dehydrogenase gene is located elsewhere on the cosmid clone or a dehydrogenase isoenzyme is provided by the streptomycete host. This second scenario is entirely plausible, as inactivation of the enterocin ketothiolase encJ gene in mutant KJ resulted in only a 25% loss in enterocin production, thereby suggesting strong complementation by the corresponding ketothiolase associated with fatty acid β-oxidation. The benzoyl-CoA pathway dehydrogenase may thus be entirely supplied through primary metabolism.
As products of ketothiolases are acyl-CoAs, we predicted that the homologous enc-encoded thiolase EncJ catalyzes the conversion of 3-keto-3-phenylpropionyl-CoA directly to benzoyl-CoA (Fig. 1). This conversion, however, bypasses the need to directly activate benzoic acid by a dedicated benzoate-CoA ligase such as EncN. Inactivation of the encN gene did not perturb the production of benzoate-primed enterocin, thereby confirming that encN is extraneous and that the product of the thiolase EncJ must be benzoyl-CoA and not free benzoic acid. Supplemental benzoic acid can efficiently enter the enterocin biosynthetic pathway in the wild-type strain but not in the corresponding encN mutant KN. This observation is consistent with our recent analysis of the encM mutant KM, in which the oxygenase encM as well as the coupled encN were disrupted (19). Although benzoate-primed polyketides were synthesized in this mutant strain, they were not enriched with administered d5-benzoic acid. These observations confirm that, although EncN is a functional benzoate-CoA ligase, EncN does not catalyze a reaction directly along the phenylalanine-to-benzoyl-CoA biosynthetic pathway. Rather, EncN serves as a substitute pathway for utilizing exogenous benzoic acid.
The redundancy of encN in the enterocin biosynthetic gene cluster may explain why benzoic acid is not accepted as a precursor in the biosynthesis of the macrolide soraphen A in the myxobacterium Sorangium cellulosum (7), even though the soraphen A PKS-loading domain harbors a benzoyl-CoA-specific acyltransferase (18). If benzoyl-CoA is synthesized in a similar manner in S. cellulosum, the bacterium may harbor homologues to just the encH-encJ and encP genes and not encN. Although the soraphen A biosynthetic gene cluster has been cloned and sequenced (10), genes encoding the benzoyl-CoA biosynthetic pathway are not clustered with the soraphen PKS genes and have not been reported to date.
In summary, we have assigned the enzymatic roles of the products of the novel benzoyl-CoA biosynthesis genes encH-encJ, encN, and encP through target-based mutagenesis. This pathway to benzoyl-CoA involves a plant-like conversion of phenylalanine to cinnamic acid, followed by a single round of β-oxidation, and is reported at the genetic level here for the first time in a bacterium.
Acknowledgments
This work was generously supported by grant AI47818 from the National Institutes of Health.
We thank Paul R. Shipley for assistance with MS experiments.
REFERENCES
- 1.Abd El-Mawla, A. M. A., and L. Beerhues. 2002. Benzoic acid biosynthesis in cell cultures of Hypericum androsaemum. Planta (Berl.) 214:727-733. [DOI] [PubMed] [Google Scholar]
- 2.Bierman, J., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 3.Harwood, C. S., G. Burchhardt, H. Herrmann, and G. Fuchs. 1998. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439-458. [Google Scholar]
- 4.He, X.-Y., and S.-Y. Yang. 1997. Glutamate-119 of the large α-subunit is the catalytic base in the hydration of 2-trans-enoyl-coenzyme A catalyzed by the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry 36:11044-11049. [DOI] [PubMed] [Google Scholar]
- 5.Hertweck, C., A. P. Jarvis, L. Xiang, B. S. Moore, and N. J. Oldham. 2001. A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid β-oxidation. Chembiochem 2:784-786. [DOI] [PubMed] [Google Scholar]
- 6.Hertweck, C., and B. S. Moore. 2000. A plant-like biosynthesis of benzoyl-CoA in the marine bacterium “Streptomyces maritimus.” Tetrahedron 56:9115-9120. [Google Scholar]
- 7.Höfle, G., and H. Reichenbach. 1995. The biosynthetic potential of the Myxobacteria, p. 61-78. In W. Kuhn and H.-P. Fiedler (ed.), Sekundärmetabolismus bei Mikroorganismen. Attempto Verlag, Tübingen, Germany.
- 8.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 9.Lapadatescu, C., C. Giniès, J.-L. Le Quéré, and P. Bonnarme. 2000. Novel scheme for biosynthesis of aryl metabolites from l-phenylalanine in the fungus Bjerkandera adusta. Appl. Environ. Microbiol. 66:1517-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligon, J. M., S. Hill, J. Beck, R. Zirkle, I. Molnár, J. Zawodny, S. Money, and T. Schupp. 2002. Characterization of the biosynthetic gene cluster for the antifungal polyketide soraphen A from Sorangium cellulosum So ce26. Gene 285:257-267. [DOI] [PubMed] [Google Scholar]
- 11.Moore, B. S., and C. Hertweck. 2002. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 19:70-99. [DOI] [PubMed] [Google Scholar]
- 12.Piel, J., C. Hertweck, P. R. Shipley, D. M. Hunt, M. S. Newman, and B. S. Moore. 2000. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate “Steptomyces maritimus”: evidence for the derailment of an aromatic polyketide synthase. Chem. Biol. 7:943-955. [DOI] [PubMed] [Google Scholar]
- 13.Piel, J., K. Hoang, and B. S. Moore. 2000. Metabolic diversity encoded by the enterocin biosynthesis gene cluster. J. Am. Chem. Soc. 122:5415-5416. [Google Scholar]
- 14.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Schneider, S., M. E. Mohamed, and G. Fuchs. 1997. Anaerobic metabolism of L-phenylalanine via benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 168:310-320. [DOI] [PubMed] [Google Scholar]
- 16.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 17.Sitachitta, N., M. Gadepalli, and B. S. Davidson. 1996. New α-pyrone-containing metabolites from a marine-derived Actinomycete. Tetrahedron 52:8073-8080. [Google Scholar]
- 18.Wilkinson, C. J., E. J. Frost, J. Staunton, and P. F. Leadlay. 2001. Chain initiation on the soraphen-producing modular polyketide synthase from Sorangium cellulosum. Chem. Biol. 8:1197-1208. [DOI] [PubMed] [Google Scholar]
- 19.Xiang, L., J. A. Kalaitzis, G. Nilsen, L. Chen, and B. S. Moore. 2002. Mutational analysis of the enterocin Favorskii biosynthetic rearrangement. Org. Lett. 4:957-960. [DOI] [PubMed] [Google Scholar]
- 20.Xiang, L., and B. S. Moore. 2002. Inactivation, complementation and heterologous expression of encP, a novel bacterial phenylalanine ammonia-lyase gene. J. Biol. Chem. 277:32505-32509. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto, H., K. H. Maurer, and C. R. Hutchinson. 1986. Transformation of Streptomyces erythraeus. J. Antibiot. 39:1304-1313. [DOI] [PubMed] [Google Scholar]