Abstract
The regulation by two transcriptional activators of flagellar expression (FlhD and FlhC) and the chemotaxis methyl-accepting protein Aer was studied with glass slide DNA microarrays. An flhD::Kan insertion and an aer deletion were independently introduced into two Escherichia coli K-12 strains, and the effects upon gene regulation were investigated. Altogether, the flhD::Kan insertion altered the expression of 29 operons of known function. Among them was Aer, which in turn regulated a subset of these operons, namely, the ones involved in anaerobic respiration and the Entner-Doudoroff pathway. In addition, FlhD/FlhC repressed enzymes involved in aerobic respiration and regulated many other metabolic enzymes and transporters in an Aer-independent manner. Expression of 12 genes of uncharacterized function was also affected. FlhD increased gltBD, gcvTHP, and ompT expression. The regulation of half of these genes was subsequently confirmed with reporter gene fusions, enzyme assays, and real-time PCR. Growth phenotypes of flhD and flhC mutants were determined with Phenotype MicroArrays and correlated with gene expression.
The Escherichia coli flagellar regulon consists of 14 operons expressed in a transcriptional hierarchy (for a review see reference 35). The flagellar master operon flhDC consists of flhD and flhC (4). The active complex is a heterotetramer (C2D2) that binds to the upstream sequences of class II promoters (24). FlhD/FlhC also has nonflagellar targets. Previous work with Panorama E. coli gene arrays (Sigma GenoSys, The Woodlands, Tex.) and lacZ fusions determined that FlhD/FlhC regulates the transporter for galactose (mglBAC), the rod-shape determination proteins (mreBCD), the tricarboxylic acid (TCA) cycle enzyme malate dehydrogenase (mdh), and five enzymes involved in anaerobic respiration (41).
Although FlhD alone does not bind to class II flagellar promoters, FlhD without the involvement of FlhC is involved in the regulation of the cell division rate (36). Mutants with mutations in flhD, but not those with mutations in flhC, continued to divide rapidly as they entered stationary phase. This was mediated by cadA (39), the second gene in the cadBA operon, encoding lysine decarboxylase (28, 29, 58).
In this study, glass slide genomic microarrays were used to study transcriptional regulation by FlhD and FlhC. Altogether, 29 operons that have a characterized function and 12 genes of unknown function were affected by FlhD or FlhD/FlhC. While the regulation of anaerobic respiration involved the oxygen sensor Aer, aerobic regulation appeared to be unaffected by this protein.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids are presented in Table 1. MC1000 was derived by crossing the araD139 Δ(araA-leu)7697 allele of D7091 Hfr into M182 (M. Casadaban). MC1000 flhD::Kan contains a kanamycin resistance gene inserted into flhD which exhibits a polar effect upon the expression of flhC (26). YK410 was used as a second parental strain (23). YK4131 (flhD A32D) and YK4136 (flhC N70T) contain point mutations (C. Barker and P. Matsumura, unpublished data). UU1245 [(aer)DE1 argD::Gen] contains a deletion in aer and an insertion of a gentamicin resistance element in the neighboring gene argD (6), and UU1246 contains only the argD::Gen allele. The lacZ fusion strains were described previously (41, 49, 57). Transcriptional fusions to Photorhabdus luminescens luxCDABE were from the LuxA library of E. coli chromosomal DNA segments in the moderate-copy-number plasmid pDEW201 (53-55) or the LuxZ library. The LuxZ library was constructed in the same manner as the LuxA library (53), except that E. coli chromosomal DNA fragments with an average size of approximately 0.7 kb were used. DNA sequence data from 4,608 plasmids were used for homology searching and mapping to the E. coli genome, as previously described (54), resulting in 1,799 genome-registered gene fusions.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| D7091 Hfr | araD139 Δ(araA-leu)7697 trp86 relA1 spoT1 metB1 rpoB mal | J. R. Beckwith |
| M182 | F− Δ(lacI-Y)74 galE15 galK16 λ−relA1 rpsL150 spoT1 e14− | S. Brenner |
| MC1000 | F−araD139 Δ(araA-leu)7697 Δ(lacI-Y)74 galE15 galK16 λ−relA1 rpsL150 spoT1 e14− | M. Casadaban |
| MC1000 flhD::Kan | flhD::Kan | 26 |
| YK410 | F−araD139 lacU169 strA thi pyrC46 nalA thyA his | 23 |
| YK4131 | YK410 flhD (A32D) | This study |
| YK4136 | YK410 flhC (N70T) | This study |
| UU1245 | (aer) DE1 argD::Gen | 6 |
| UU1246 | argD::Gen | J. S. Parkinson |
| PC25 | dmsA::lacZ | 49 |
| HW2 | napA::lacZ | 49 |
| HW1 | nrfA::lacZ | 57 |
| LK1 | frdA::lacZ | 49 |
| Plasmids | ||
| pBP18 | mglB::lacZ | 41 |
| pBP20 | glpA::lacZ | 41 |
| pDEW533 | aer::luxCDABE | T. K. Van Dyk |
| pDEW534 | serA::luxCDABE | T. K. Van Dyk |
| pDEW535 | edd::luxCDABE | T. K. Van Dyk |
| pDEW559 | gltBD::luxCDABE | T. K. Van Dyk |
| pDEW560 | gcvTHP::luxCDABE | T. K. Van Dyk |
| pDEW656 | narK::luxCDABE | T. K. Van Dyk |
Microarray fabrication.
E. coli genomic DNA from MG1655 was used as template for the PCR step. The E. coli annotation within the ERGO database (Integrated Genomics) identifies 4,485 open reading frames. Primers were designed such that the most distinctive 300-bp region of 4,442 of the open reading frames was amplified with consecutive rounds of PCR. PCR products were resuspended in 15 μl of spotting buffer (3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1.5 M betaine [14]) and printed in triplicate onto amino-alkylsilane slides (Sigma) with an OmniGrid arrayer (Gene Machines) by using Telechem SMP3 split pins. The DNA was cross-linked to the slides with UV light. Residual salt and unbound DNA were removed by rinsing the slides with 0.5% sodium dodecyl sulfate and water.
RNA isolation, cDNA probe synthesis, and hybridization conditions.
Overnight cultures of wild-type cells and the respective mutants were diluted 1:100 into 20 ml of Luria-Bertani broth (LB; 1% tryptone, 0.5% yeast extract, 1% NaCl) in a 250-ml Erlenmeyer flask. Cultures were grown aerobically at 34°C under constant shaking at 250 rpm. After reaching an optical density at 600 nm (OD600) of 0.5, growth of the culture and RNA degradation were inhibited by adding 2 ml of stop solution (5% phenol in ethanol). Bacterial pellets were flash frozen in liquid nitrogen and stored at −70°C. RNA was isolated with the hot phenol-sodium dodecyl sulfate method (9), with one phenol, one phenol-chloroform, and one chloroform extraction, followed by isopropanol precipitation. Final cleanup of the RNA was performed with an RNeasy mini column (Qiagen). Fluorescent cDNA probes were produced by aminoallyl reverse transcription and purified (http://cmgm.stanford.edu/pbrown/protocols/aadUTPCoupling Procedure.htm). Samples were dried and resuspended in 100 μl of Sigma Arrayhyb low-temperature hybridization solution (Sigma), supplemented with 1 μg each of salmon sperm DNA and yeast tRNA and subsequently combined with a complementary labeled cDNA. Hybridizations were performed in a GeneTAC HybStation instrument (Genomic Solutions).
Data analysis, quality control, and threshold determination.
Arrays were scanned on a GenePix 4000B array scanner (Axon Instruments) at 635 and 532 nm. Photomultiplier tube voltages were adjusted to give the maximum signal from each channel without bleaching any of the features within the confocal image. Images were analyzed with GenePix Pro 3.0 (Axon Instruments).
Median intensities were used to determine the log ratio of medians (log expression ratios), which are indicated as positive for induced genes and negative for repressed genes. Each experiment was performed four or five times, with RNA obtained from three or four independent bacterial cultures. The exact numbers of technical and biological replicates are indicated in the footnotes to Tables 2 and 3 . Averages of the means and standard deviations were determined for each gene.
TABLE 2.
E. coli nonflagellar genes regulated by FlhD/FlhC or Aera
| Gene | Log expression ratio
|
Function of the protein | ||
|---|---|---|---|---|
| MC/flhDCb | YK/flhDC | MC/aer | ||
| fdnG | 0.5 ± 0.1b | >1.3 | 1 ± 0.55 | Formate dehydrogenase (1.2.1.2) |
| csgG | NR | >1.3 | NR | Biosynthesis of curli |
| aer | 0.58 ± 0.15 | >1.3 | NR | Oxygen sensing |
| narK | 0.58 ± 0.15b | >1.3 | NR | Nitrate transport |
| yjiY | NR | >1.3 | NR | Hypothetical protein |
| tnaL | 0.3 ± 0.04 | 1.23 ± 0.58 | NR | Tryptophanase leader peptide |
| yfiD | NR | 1.2 ± 0.33 | 0.78 ± 0.39 | Pyruvate-formate lyase homolog |
| frdA | NR | 1.1 ± 0.4 | 0.73 ± 0.17 | Fumarate reductase (1.3.99.1) |
| tnaA | NR | 1.08 ± 0.04 | NR | Tryptophanase (4.1.99.1) |
| tnaB | NR | 1.08 ± 0.08 | NR | Tryptophan permease |
| yhjX | NR | 1.08 ± 0.04 | NR | Putative drug transport |
| napC | NR | 1.04 ± 0.33 | 0.85 ± 0.46 | Nitrate reductase, periplasmic (1.7.99.4) |
| ycgC | 0.49 ± 0.29 | 1.04 ± 0.11 | 0.82 ± 0.47 | Putative PTS system |
| fdnI | 0.32 ± 0.06b | 1 ± 0.25 | 0.78 ± 0.18 | Formate dehydrogenase |
| hybC | NR | 1 ± 0.4 | NR | Hydrogenase 2 |
| hybA | NR | 0.95 ± 0.34 | NR | Hydrogenase 2 |
| yjiX | NR | 0.95 ± 0.3 | NR | Hypothetical protein |
| ycdB | 0.65 ± 0.26 | 0.94 ± 0.22 | 0.8 ± 0.15 | Hypothetical protein |
| hybE | NR | 0.93 ± 0.29 | NR | Hydrogenase 2 |
| napF | NR | 0.89 ± 0.25 | 0.53 ± 0.17 | Nitrate reductase, periplasmic |
| napH | NR | 0.89 ± 0.19 | 0.73 ± 0.28 | Nitrate reductase, periplasmic |
| ybhB | 0.76 ± 0.26 | 0.85 ± 0.35 | NR | Hypothetical protein |
| gatZ | NR | 0.83 ± 0.17 | NR | Tagatose 6-phosphate kinase (2.7.1.144) |
| ydcD | >2 | 0.82 ± 0.38 | NR | Hypothetical protein |
| ybhA | >1.3 | 0.8 ± 0.42 | NR | Putative phosphatase |
| yjiA | NR | 0.8 ± 0.34 | NR | Putative carbon starvation protein |
| gatA | NR | 0.76 ± 0.14 | NR | PTS system, galactitol |
| dmsA | NR | 0.75 ± 0.36 | 1.2 ± 0.5 | DMSO reductase (1.8.99.−) |
| cadA | NR | 0.75 ± 0.26 | NR | Lysine decarboxylase (4.1.1.18) |
| carB | 0.66 ± 0.1 | 0.74 ± 0.15 | NR | Carbamoylphosphate synthetase (6.3.5.5) |
| dmsC | NR | 0.72 ± 0.1 | 0.56 ± 0.05 | DMSO reductase |
| cadB | NR | 0.72 ± 0.23 | NR | Lysine-cadaverine transport |
| hybD | NR | 0.72 ± 0.28 | NR | Hydrogenase 2 |
| hdeA | NR | 0.72 ± 0.47 | >1.3 | Hypothetical protein |
| frdB | NR | 0.7 ± 0.34 | 0.4 ± 0.1 | Fumarate reductase |
| csgF | 0.7 ± 0b | NR | NR | Biosynthesis of curli |
| hybF | NR | 0.68 ± 0.21 | NR | Hydrogenase 2 |
| nrfB | 0.6 ± 0.01b | 0.67 ± 0.1 | 0.52 ± 0.14 | Nitrite reductase (1.6.6.4) |
| fdnH | 0.3 ± 0.06b | 0.67 ± 0.14 | 0.43 ± 0.08 | Formate dehydrogenase |
| frdC | NR | 0.65 ± 0.33 | 0.4 ± 0.11 | Fumarate reductase |
| csgA | NR | 0.66 ± 0.3 | NR | Biosynthesis of curli |
| carA | 0.8 ± 0.4 | 0.64 ± 0.2 | NR | Carbamoylphosphate synthetase |
| dmsB | NR | 0.63 ± 0.06 | 0.56 ± 0.05 | DMSO reductase |
| napB | 0.3 ± 0.06b | 0.57 ± 0.03 | 0.32 ± 0.09 | Nitrate reductase, periplasmic |
| dppA | 0.56 ± 0.3 | NR | NR | Dipeptide permease |
| dadA | 0.56 ± 0.14 | NR | NR | d-Amino acid dehydrogenase (1.4.99.1) |
| frdD | 0.34 ± 0.1 | 0.54 ± 0.28 | 0.36 ± 0.08 | Fumarate reductase |
| dadX | 0.5 ± 0.03 | NR | NR | Alanine racemase (5.1.1.1) |
| gltB | 0.5 ± 0.2 | ND | NR | Glutamate synthase, large subunit (1.4.1.13) |
| napG | NR | 0.46 ± 0.08 | 0.75 ± 0.37 | Nitrate reductase, periplasmic |
| nrfA | NR | 0.46 ± 0.09 | 0.43 ± 0.06 | Nitrite reductase |
| serA | 0.46 ± 0.01b | NR | NR | d-3-Phosphoglycerate synthase (1.1.1.95) |
| napA | NR | 0.45 ± 0.06 | 0.45 ± 0.06 | Nitrate reductase, periplasmic |
| gltD | 0.34 ± 0.05 | 0.45 ± 0.03 | NR | Glutamate synthase, small subunit |
| malE | NR | 0.45 ± 0.12 | NR | Maltose transport |
| glpB | NR | 0.43 ± 0.11 | NR | Glycerol 3-phosphate dehydrogenase, anaerobic (1.1.99.5) |
| hdeB | NR | 0.41 ± 0.09 | >1.7 | Hypothetical protein |
| gatY | NR | 0.41 ± 0.09 | NR | d-Tagatose-1,6-bisphosphate aldolase (4.1.2.40) |
| dppB | 0.4 ± 0.1 | NR | NR | Dipeptide transport |
| dppC | 0.4 ± 0.1 | NR | NR | Dipeptide transport |
| rbsA | 1 ± 0.4 | 0.39 ± 0.15 | NR | Ribose transport |
| glpA | NR | 0.38 ± 0.06 | 1.1 ± 0.9 | Glycerol 3-phosphate dehydrogenase, anaerobic |
| edd | 0.34 ± 0.05 | NR | 0.72 ± 0.23 | 6-Phosphogluconate dehydratase (4.2.1.12) |
| gatB | NR | 0.32 ± 0.08 | NR | PTS system, galactitol |
| dppD | 0.3 ± 0.1 | NR | NR | Dipeptide transport |
| gcvP | −0.3 ± 0.58 | NR | NR | Glycine dehydrogenase (1.4.4.2) |
| sdhA | NR | −0.34 ± 0.06 | NR | Succinate dehydrogenase (1.3.99.1) |
| cyoD | NR | −0.4 ± 0.18 | NR | Cytochrome o ubiquinol oxidase |
| cyoE | NR | −0.41 ± 0.19 | NR | Cytochrome o ubiquinol oxidase |
| gcvH | −0.46 ± 0.08 | NR | NR | Glycine cleavage system H protein |
| gcvT | −0.49 ± 0.08 | NR | NR | Aminomethyltransferase (2.1.2.10) |
| sdhB | NR | −0.58 ± 0.19 | NR | Succinate dehydrogenase |
| gltA | −0.34 ± 0.06 | −0.6 ± 0.06 | NR | Citrate synthase (4.1.3.7) |
| mglC | NR | −0.62 ± 0.31 | NR | Galactose binding |
| cyoB | NR | −0.65 ± 0.32 | NR | Cytochrome o ubiquinol oxidase |
| sdhD | NR | −0.65 ± 0.09 | NR | Succinate dehydrogenase |
| cyoC | NR | −0.7 ± 0.4 | NR | Cytochrome o ubiquinol oxidase |
| oppC | −0.32 ± 0.09 | −0.74 ± 0.27 | NR | Oligopeptide transport |
| oppD | −0.32 ± 0.03 | −0.74 ± 0.31 | NR | Oligopeptide transport |
| sdhC | NR | −0.75 ± 0.19 | NR | Succinate dehydrogenase |
Cultures of MC1000 and YK410 and their respective flhD::Kan mutants were grown in LB at 34°C to an OD600 of 0.5 (columns 1 and 2). MC1000 was grown in triplicate culture, and the combined data from four data sets are presented. YK410 was grown in four cultures, and data from five experiments were analyzed. Cultures of MC1000 and its Δaer derivative were grown in LB at 34°C to an OD600 of 0.5 (column 3). Each culture was grown in triplicate, and data from five experiments were analyzed. A control experiment was performed with MC1000 and its argD::Gen derivative. Only genes that appeared unaffected by the mutation in argD are listed. Genes are ordered by decreasing log expression ratios (column 2). EC numbers are given in parentheses for enzymes at first appearance. >1.3 indicates an expression ratio between 20- and 50-fold, and >1.7 indicates an expression ratio between 50- and 100-fold. NR, not regulated. ND, not detected.
In the early stage of this study, a slide was printed that contained the E. coli library printed in duplicate on three individual slides. These slides yielded a smaller number of genes regulated by FlhD or FlhD/FlhC. However, nine genes appeared to be regulated in MC1000 that were not picked up by the high-density slide.
TABLE 3.
E. coli genes differentially regulated by FlhD and FlhCa
| Gene | Log expression ratio
|
|
|---|---|---|
| YK/flhD | YK/flhC | |
| gltD | >1.3 | NR |
| dadA | 1.04 ± 0.33 | NR |
| dadX | 0.97 ± 0.11 | NR |
| gcvT | 0.78 ± 0.27 | −0.32 ± 0.16 |
| gcvP | 0.73 ± 0.2 | NR |
| gltB | 0.62 ± 0.31 | NR |
| gcvH | 0.6 ± 0.1 | NR |
| ompT | 0.57 ± 0.08 | NR |
Cultures of YK410, YK4131, and YK4136 were grown in LB at 34°C to an OD600 of 0.5. Each culture was grown in triplicate, and data from five experiments were averaged. >1.3 indicates an expression ratio between 20- and 50-fold. NR, not regulated.
When differences between arrays occurred, a cross normalization was performed. Each block on the slide contained 12 control spots of human DNA in ratios from 1:30 to 30:1. Experimental ratios of these spots were determined and compared with the theoretical ratios. In cases with more than a 25% difference between slides, ratios were normalized to the differences for the control spots. In a few cases, the control spots did not yield an acceptable correlation between theoretical and experimental ratios and the slides were discarded.
To define a threshold level of significant difference, RNA from MC1000 was labeled with Cy3 and Cy5 and hybridized to a slide. Spot intensities of one sample were plotted against the spot intensities of the other sample (data not shown). Over 95% of the data fell within the twofold range, and about 98% were within threefold. An arbitrary significant expression ratio threshold was defined as twofold (log expression ratio higher than 0.3 or lower than −0.3).
The presence of about 50 flagellar genes was used as positive control. RNA from the isogenic strain couples MC1000-MC1000 flhD::Kan and YK410-YK410 flhD::Kan was labeled with Cy3 and Cy5 and hybridized to a slide. Spot intensities were compared (data not shown). Most flagellar genes were regulated in both backgrounds. However, some genes appeared to be regulated in only one strain. Likewise, not every gene within an operon always appeared to be induced. As a practical matter, we have focused on operons rather than individual genes throughout this work. An operon is considered regulated when the majority of the genes exhibit regulation, and a gene is considered regulated when it is regulated in at least one strain. In the Results, we demonstrate with reporter gene fusions and enzyme assays that the majority of these genes are indeed regulated in both genetic backgrounds even though regulation could not be detected by gene array.
Phenotype MicroArrays.
Phenotype MicroArrays were obtained from Biolog. These are 96-well microtiter plates with a different cell culture medium dried to the bottom of each well. These media are designed to test unique cellular phenotypes. Electrons produced during respiration are transferred to an indicator dye, resulting in a dark purple color (8).
Bacteria were grown overnight on R2A or Biolog universal growth agar, as recommended by the manufacturer (Biolog). Standardized cell suspensions were produced with the 85%T turbidity standard. Thymine, thiamine, histidine, and uracil were added at a concentration of 0.2%, and 100 μl was added to each well. Growth was monitored with an EL311 Microplate Autoreader (Bio-Tek Instruments) at 630 nm and analyzed after 24 h. The blank value was subtracted from each well, and the OD630 of the wild-type culture was divided by the OD630 of the mutants. Growth ratios above 1 indicate nutrients that provided better growth conditions for wild-type cultures. The growth of MC1000 was compared in two independent experiments. Of 95 data points, 70% were within 1 standard deviation and 94% were within 2 standard deviations from the average. A threshold level of a significant difference in growth was defined as a growth ratio above 2 or below 0.5. Only nutrients that allowed one of the cultures to grow to an OD630 of at least 0.4 were considered.
Determination of enzyme activities.
Triplicate cultures were inoculated 1:100 from overnight culture and grown at 34°C in LB. The activity of β-galactosidase from the flhD::lacZ strain was determined and expressed as Miller units (41). Light production from the edd::luxCDABE strain was measured over 2 s in a MiniLumat 9506 luminometer (EG & G Berthold) and expressed as specific light units (light intensity per milliliter of culture/OD600). The activity of succinate dehydrogenase was determined as described previously (25) and expressed as specific activity (nanokatals per milligram).
Real-time PCR.
Real-time PCR was performed with the same RNA samples used in the arrays. The PCR was performed with the SYBR Green kit from PE Biosystems. The reaction mixture contained 100 ng of cDNA, 1× SYBR Green buffer, 2.5 mM MgCl2, 0.25 mM (each) deoxynucleoside triphosphates, 0.05 μM (each) primer (gcvT, CGAAGCTCACCAAAAGCGGC and GTATTAAACAGTGTGGCAGC; gcvH, ATGAGCAACGTACCAGCAGAAC and TTACTGGTCTTCTAACAATGC; gcvP, CACCGCCTGCAGCTACCGC and GCCACGAAGAAGCGGTTGGC; dadA, TTACCCAATGATGAAACCGG and AAGAATGGTGGATACCGGCGC; dadX, GCCTGACCACCTGCGTACACAG and CGTGGCGCGGATAACCGTCGGC; gltB, CCCTCTCCTCTCTGCCACGC and GTGATGGTGTTGATTTCTCCG; gltD, GCTCACCTTCGGTATTCCGGCC and CCGGTTCGCCCATTTCGGTACGC; ompT, CTGGGAATAGTCCTGACAACCCC and CTCCTCAGAACTGTAGATATAGG; fliA, GCGTAACCGTTTAATAGCCTGGC and GGCAGCGTTATGTCCCGCTGG; flhD, GCAAGCGTGTTGAGAGCATGATGC and GGAATAATGCACACGGGGTGCGG), 0.01 U of AmpErase uracil N-glycosylase, and 1 U of Taq gold polymerase. The reaction was performed with 50 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C and monitored in an iCycler iQ real-time PCR detection system (Bio-Rad). A standard curve was derived from plasmid and used to convert threshold crossings to log copy numbers. Expression ratios were obtained by dividing copy numbers of wild-type cells by those of the mutants. The fliA gene, used as a positive control, yielded expression ratios of more than 100-fold. All PCR fragments yielded a single band on an agarose gel.
RESULTS
Gene array analysis.
RNA from parental strains and flhD::Kan derivatives of MC1000 and YK410 was labeled with Cy3 and Cy5, respectively, and log expression ratios were determined. Nonflagellar genes affected by the flhD::Kan mutation were ordered in four functional categories and are listed in Table 2. Many of these genes are regulated in response to changes in the oxygen concentration of the medium. A single oxygen-sensor-related gene, aer, was affected by flhD::Kan. Aer is a chemoreceptor and senses intracellular energy levels and couples them to the chemotaxis signaling pathways (6, 42). RNA from strain MC1000 and its aer(DE1) argD::Gen derivative was isolated and labeled with Cy3 and Cy5, respectively. A control experiment with a strain containing only the argD::Gen insertion distinguished the effects of the aer deletion and the argD insertion. Genes affected by aer(DE1) argD::Gen and unaffected by argD::Gen are included in Table 2. In two additional array experiments, the expression profile of YK410 was compared to that of its isogenic flhD (YK4131) and flhC (YK4136) mutants. Four operons appeared differentially regulated (Table 3). The results of all array experiments are summarized in Fig. 1 and discussed below.
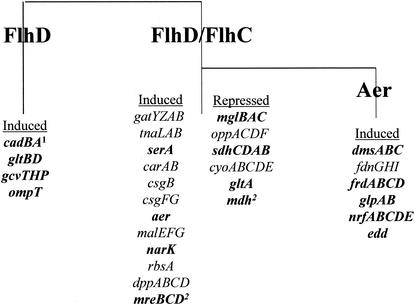
FIG. 1.
Pathways of FlhD regulation. The cartoon describes the operons that were regulated by FlhD, FlhD/FlhC, or Aer as determined by gene array. Operons that were confirmed by reporter gene fusion, enzyme assay, or real-time PCR are printed in boldface. Also included are targets that were previously identified (references 39 and 41, indicated by superscript numerals 1 and 2, respectively).
FlhD/FlhC regulates components of energy metabolism (functional group I).
Seven components of anaerobic respiration were induced by FlhD/FlhC (Table 2). In contrast, sdhCDAB (succinate dehydrogenase) and cyoBCDE (cytochrome o ubiquinol oxidase) of the aerobic respiratory chain were repressed by FlhD/FlhC. The edd gene encodes 6-phosphogluconate dehydratase and is part of the Entner-Doudoroff pathway. Regulation of genes involved in anaerobic respiration and the Entner-Doudoroff pathway involved the methyl-accepting chemotaxis protein and oxygen sensor Aer (Table 2).
To test the predictions derived from the array experiments, measurements of transcriptional activity were performed. Fourteen of 28 operons with known function that were affected by the flhD::Kan mutation in MC1000 or YK410 (Table 2) were tested with reporter gene fusions, enzyme assay, or real-time PCR. Cultures were grown for 10 h, and the maximal expression levels were compared between the parental strains and the mutants. All targets tested were confirmed in MC1000, YK410, or both (Table 4). Our initial hypothesis that regulation may take place in both backgrounds even when it can be detected by microarray in only one of the two parental strains was confirmed. For example, the reporter gene fusion was able to detect regulation of edd and serA in YK410. Overall, control experiments correlated well with the gene arrays. Regulation of dmsA, nrfA, frdA, napF, glpA, and edd by Aer was tested with lacZ and lux fusions (Table 4). As a sole exception, regulation of napF was not detectable with the lacZ fusion strain. Aer is confirmed as a regulator of enzymes involved in anaerobic respiration and the Entner-Doudoroff pathway and is, therefore, the first of the methyl-accepting chemotaxis proteins that has a demonstrated function in gene regulation.
TABLE 4.
Control experiments for all arraysa
| Gene | Log expression ratio
|
|||
|---|---|---|---|---|
| MC/flhDC | YK/flhD | YK/flhC | MC/aer | |
| lacZ fusions | ||||
| dmsA | 1.1 ± 0.14 | 0.3 ± 0.1 | 0.7 ± 0.05 | 1.4 ± 0.12 |
| napF | 1.25 ± 0.2 | 0.5 ± 0.1 | 0.5 ± 0.2 | NR |
| nrfA | 0.3 ± 0.02 | 0.4 ± 0.01 | 0.55 ± 0.03 | 0.4 ± 0.02 |
| frdA | 1.1 ± 0.2 | 0.3 ± 0.01 | NR | 1.2 ± 0.1 |
| glpA | NR | 0.35 ± 0.08 | 0.23 ± 0.04 | 0.3 ± 0.04 |
| mglB | −0.3 ± 0 | −0.45 ± 0.1 | −0.4 ± 0.19 | NR |
| lux fusions | ||||
| edd | 0.56 ± 0.1 | 0.5 ± 0.0.08 | 0.56 ± 0.06 | 0.43 ± 0.04 |
| serA | 1.23 ± 0.1 | 0.5 ± 0.03 | 0.53 ± 0.15 | NR |
| aer | >2 | 0.5 ± 0.08 | 0.45 ± 0.16 | NA |
| gltB | >1.7 | >2 | NR | NR |
| gcvT | −0.8 ± 0.04 | 0.7 ± 0.03 | −0.7 ± 0 | NR |
| narK | >3 | >2 | >3 | NR |
| Enzyme assays | ||||
| sdh | NR | −0.4 ± 0.15 | −0.4 ± 0.08 | NR |
| Real-time PCR | ||||
| gltA | 0.68 ± 0.26 | −0.7 ± 0.34 | −1 ± 0.9 | NR |
| ompT | ND | 0.8 ± 0.3 | NR | ND |
| dadA | ND | 0.8 ± 0.6 | NR | ND |
| dadX | ND | NR | NR | ND |
| gltB | ND | 1.2 ± 0.2 | NR | ND |
| gltD | ND | 1.1 ± 0.3 | −0.3 ± 0 | ND |
| gcvT | ND | 1.4 ± 0.6 | NR | ND |
| gcvH | ND | 1.4 ± 0.4 | NR | ND |
| gcvP | ND | 1.5 ± 0.5 | NR | ND |
The expression of dmsA, napF, nrfA, frdA, glpA, and mglB was determined as the activity of β-galactosidase from lacZ fusions. The expression of edd, serA, aer, gltB, gcvT, and narK was determined as the production of light from luxCDABE fusions. The enzyme activity of Sdh was determined. The expression of gltA, ompT, dadAX, gltBD, and gcvTHP was determined by real-time PCR. Maximal expression levels of wild-type cells were divided by those of the mutants and converted to log expression ratios (MC1000/MC1000 flhD::Kan [column 1], YK410/YK4131 [column 2], YK410/YK4136 [column 3], and MC1000/MC1000 Δaer [column 4]). The table shows the averages over three or more experiments; error bars indicate the standard deviations. NR, not regulated; ND, not determined; NA, not available.
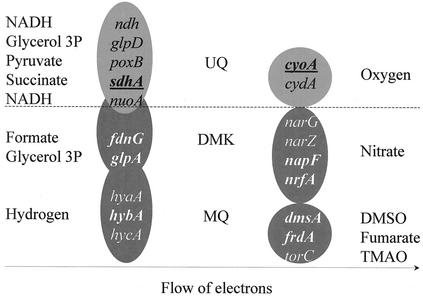
FlhD/FlhC is able to up- and down-regulate gene expression. While several complexes of the anaerobic respiration were up-regulated by FlhD/FlhC, some enzymes of the aerobic respiration were down-regulated. FlhD/FlhC may be an important factor in the switch from aerobic to anaerobic respiration as cultures deplete oxygen from the medium during prolonged growth in batch culture. These findings are summarized in Fig. 2.
FIG. 2.
Flow chart of respiration. The chart is drawn in a way that electrons flow from the left to the right. At the far left are electron donors, in the middle are quinones, and at the far right are electron acceptors. Genes encoding dehydrogenases are in the circles to the left, and genes encoding terminal reductases are in the circles to the right. Light gray circles above the dotted line indicate components of the aerobic respiratory chain, and dark gray circles below the dotted line indicate components of anaerobic respiration. All genes that are regulated by FlhD/FlhC, as determined by gene array, are printed in boldface. Genes that are repressed are underlined. All target operons except napF were confirmed with reporter gene fusions.
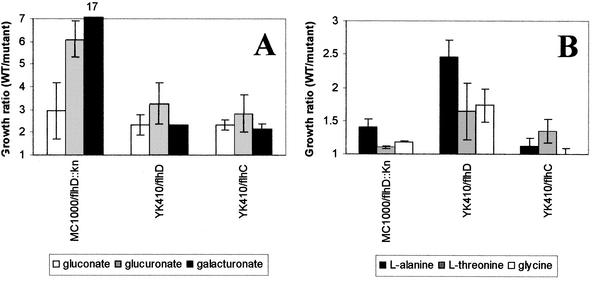
Since many of the nonflagellar targets of FlhD/FlhC are involved in metabolism, it is possible that flhD/flhC mutants may have multiple metabolic phenotypes. Phenotype MicroArray technology was applied to determine these phenotypes. Cell suspensions of strains MC1000, MC1000 flhD::Kan, YK410, YK4131, and YK4136 were inoculated onto PM1 plates, containing 95 single carbon sources. The only carbon sources that showed consistent differences in the two genetic backgrounds are shown in Fig. 3A, comparing the growth ratios for d-gluconate, d-glucuronate, and d-galacturonate in the strain pairings MC1000-MC1000 flhD::Kan, YK410-YK4131, and YK410-YK4136. Wild-type strains MC1000 and YK410 grew better on d-gluconate, d-glucuronate, and d-galacturonate than did their respective flhD, flhC, or flhD/flhC mutants. Mutants with mutations in aer did not grow on PM1 plates at all (data not shown).
FIG. 3.
Phenotype MicroArrays. Cultures of MC1000, MC1000 flhD::Kan, YK410, YK4131, and YK4136 were inoculated onto PM1 (carbon sources) (A) and PM3 (nitrogen sources) (B) plates. Growth was determined as the OD630 after 24 h. The values for the wild-type (WT) cultures were divided by the values for the mutants. The figure shows the growth ratios for the strain couples MC1000-MC1000 flhD::Kan, YK410-YK4131, and YK410-YK4136. Error bars indicate the standard deviations derived from two to five independent experiments.
The enzyme responsible for the degradation of d-gluconate, d-glucuronate, and d-galacturonate is 2-keto-3-deoxy-d-gluconate 6-phosphate aldolase (EC 4.2.1.14), encoded by eda and the key enzyme of the Entner-Doudoroff pathway (17). The eda gene is the second gene in the edd operon, which is transcribed as one transcript (15). The gene edd was regulated by FlhD/FlhC and Aer as shown by gene array (Table 2) and fusions of edd to luxCDABE (Table 4). The growth defect of the mutants on key metabolites of the Entner-Doudoroff pathway together with their reduced expression of edd indicates that FlhD/FlhC is a regulator of the Entner-Doudoroff pathway. This is the first demonstration of a correlation between phenotypes derived from Phenotype MicroArray technology with gene regulation and shows how Phenotype MicroArrays can be used to detect pathways that are regulated by a certain transcriptional regulator. Since d-gluconate, d-glucuronate, and d-galacturonate are the only carbon sources that yielded consistent results during three to five experiments, it is likely that the Entner-Doudoroff pathway is among the more important pathways that are regulated by FlhD/FlhC.
FlhD/FlhC and FlhD regulate other metabolic enzymes (functional group II).
Other metabolic enzymes that are regulated by FlhD/FlhC or FlhD are the products of cadBA (lysine-cadaverine transporter and lysine decarboxylase), dadAX (d-amino acid dehydrogenase and alanine racemase), gatYZAB (tagatose bisphosphate aldolase, tagatose 6-phosphate kinase, and phosphotransferase [PTS] system), tnaLAB (tryptophanase leader peptide, tryptophanase, and permease), gltBD (glutamate synthase), serA (d-3-phosphoglycerate dehydrogenase), carAB (carbamoylphosphate synthetase), and gcvTHP (glycine cleavage system). These genes are all involved in carbon and nitrogen metabolism (Table 2).
To distinguish between an effect of FlhD/FlhC and one of FlhD upon gene expression, the expression profile of YK410 was compared to that of the flhD (YK4131) and flhC (YK4136) mutants. Four operons appeared differentially regulated (Table 3). FlhD, but not FlhC, was an inducer of dadA, dadX, gcvT, gcvH, gcvP, gltB, gltD, and ompT expression. Even though regulation of ompT could not be detected with the FlhD/FlhC arrays (Table 3), we considered it a putative target for further analysis. The putative targets were tested with real-time PCR, with use of the same RNA sample as that for the gene array. With the exception of dadX, all genes were expressed more highly in the wild-type cells than in the flhD mutants. Occasionally, expression was even higher in the flhC mutant (Table 4).
Phenotypes related to these genes and caused by the mutation in flhD were determined with Phenotype MicroArrays. Cell suspensions of strains MC1000, MC1000 flhD::Kan, YK410, YK4131 (flhD), and YK4136 (flhC) were inoculated onto PM3 plates, containing 95 individual nitrogen sources. Figure 3B compares the growth ratios for the strain pairings MC1000-MC1000 flhD::Kan, YK410-YK4131, and YK410-YK4136 on l-alanine, l-threonine, and glycine, the only nitrogen sources that provided differential growth conditions consistently. The parental strain YK410 grew better on the three nitrogen sources than did its flhD isogen, whereas flhC mutants grew like the parental strain. No significant differences between wild-type cells and mutants were observed for MC1000.
The expression of the dadAX operon, encoding d-amino acid dehydrogenase and alanine racemase, is necessary for growth of E. coli on l- or d-alanine as a sole source of nitrogen (59, 60). As shown in Tables 3 and 4, the expression of dadA was affected by FlhD and not FlhC. Mutants in flhD exhibited reduced growth on l-alanine (Fig. 3B). This indicates that dadAX regulation is most likely the cause of the phenotype of the flhD mutant in the YK410 background. As a sole exception, regulation of dadX could be demonstrated only by gene array (Table 3) and not by real-time PCR (Table 4).
The gcvTHP operon encodes the glycine cleavage system, the major pathway of glycine and l-threonine degradation, and is required for growth on glycine and l-threonine as sole nitrogen sources (61, 62). As shown by gene array (Table 3) and real-time PCR (Table 4), expression of all three genes of this operon was 20- to 30-fold higher in wild-type cells than in flhD mutants. The mutation in flhC had no effect. Mutants with mutations in flhD exhibited reduced growth on glycine and l-threonine (Fig. 3B). In summary, FlhD regulates the expression of cadBA, dadAX, gcvTHP, gltBD, and ompT and improves growth on glycine, l-alanine, and l-threonine.
Functional group III contains membrane-associated complexes and transporters.
The csgA and csgFG genes are involved in biosynthesis of curli and were expressed more highly in wild-type cells than in the mutant (Table 2). The genes dppABCD (dipeptide transport), rbsA (ribose transport), narK (nitrate transport), and malE (maltose transport) were expressed more highly in the parental strains than in the mutant. The genes oppCDF (oligopeptide transport) and mglC (galactose transport) were repressed. The aer (oxygen sensor) gene was expressed more highly in the parental strains than in the mutants.
Functional group IV contains genes of uncharacterized function.
The group IV genes (Table 2) include hdeAB (putative acid stress chaperones), yfiD (pyruvate-formate lyase homolog), and ycgC (dihydroxyacetone kinase). While these genes seem interesting, this study focuses on the regulation of genes of known function.
DISCUSSION
Membrane arrays and glass slide arrays.
Membrane arrays identified eight operons as being regulated by FlhD/FlhC (41). The glass slide arrays used in this study identified 29 operons of characterized functions (Fig. 1) and 12 genes of uncharacterized functions as being regulated by FlhD/FlhC. This includes five of the targets identified with membrane arrays; the remaining three could not be confirmed with the glass slide array. The large number of operons that were affected by flhD::Kan as shown by glass slide arrays can in part be explained by the number of replicate slides, resulting in more data points and better statistical analysis. Overall, the glass slide arrays were more reproducible than the membrane arrays. In contrast, the membrane arrays were more sensitive, and operons like mreBCD (rod shape determination) and hydN (modulator of hydrogenases), which are expressed at very low levels, were detected. This shows that the use of different technologies is essential to identify as many targets as possible.
Gene arrays, reporter gene fusions, and real-time PCR.
Besides gene arrays, a number of other technologies have been described that enable us to study gene regulation on a large scale. The library of lux fusions that we have used in this study has been constructed and used by T. K. Van Dyk for the investigation of the responses to various stressful situations (53-55). Inconsistencies between array data and lux fusion assays have been described elsewhere (54). In this study, parts of Van Dyk's lux library together with several previously described lacZ fusions (49, 57) were used to confirm the data obtained by gene array. Of 14 targets that appeared regulated by array and were tested with reporter gene fusions and enzyme assays, all 14 were confirmed as being regulated by FlhD/FlhC. In addition, of six targets that appeared to be regulated by Aer during gene array assays, five were confirmed with lux and lacZ fusions. Only one gene among eight that were tested by real-time PCR proved wrong. False positives seem to be rare in our array. In contrast, false negatives were observed more frequently. Many genes appeared to be regulated in only one of the two parental strains that we used for this study. However, control experiments with reporter gene fusions and enzyme assays showed regulation in both genetic backgrounds. While we cannot explain this phenomenon, it indicates that the lack of regulation of some genes in one of the backgrounds is a technical problem with the array rather than a biological inconsistency between the strains.
Group I: components of energy metabolism.
Previous analysis with Sigma GenoSys membrane arrays and lacZ fusion strains (41) yielded a number of genes that were regulated by FlhD/FlhC and are involved in anaerobic respiration. These were napFAGHBC (periplasmic nitrate reductase), nrfABCDE (nitrite reductase), dmsABC (dimethyl sulfoxide [DMSO] reductase), and glpABC (glycerol 3-phosphate dehydrogenase). In this study, additional FlhD/FlhC-regulated genes involved in anaerobic respiration were identified. These are fdnGHI (formate dehydrogenase), frdABCDE (fumarate reductase), and hybACDEF (hydrogenase 2). Three components of the TCA cycle and aerobic respiration, sdhCDAB (succinate dehydrogenase), cyoABCDE (cytochrome o ubiquinol oxidase), and gltA (citrate synthase), were repressed in YK410. Apparently, FlhD/FlhC is able to repress aerobic pathways in addition to inducing anaerobic pathways (Fig. 2).
Regulation of gene expression in response to oxygen in E. coli is complex (for a review see reference 51), and many global regulators, such as flavin nucleotide reductase and ArcA/ArcB, are involved (11, 13, 27). It is possible that a global regulator links some of the metabolic targets of FlhD/FlhC. To find this common element, the gene list of the flhD::Kan array (Table 2) was searched for genes that are involved in the oxygen response. The only gene fulfilling these criteria was the one encoding the methyl-accepting chemotaxis protein Aer, and an array was performed with a strain containing an aer deletion (Table 2). Most of the genes involved in anaerobic respiration were regulated by Aer. Four of the operons were confirmed with lux and lacZ fusion strains. In contrast, expression of genes involved in aerobic respiration and the TCA cycle was unaffected by Aer. It seems that the pathway from FlhD/FlhC to its final targets splits at the level of aer expression. This is the first example of a methyl-accepting chemotaxis protein involved in gene regulation. Considering the large number of such proteins in some other bacteria (for example, Yersinia enterocolitica, Bacillus cereus, or Clostridium botulinum), this newly discovered mechanism may play a major role in bacterial gene regulation.
The mechanism by which Aer regulates anaerobic respiration remains unknown. Aer is known to sense intracellular energy levels and transduce oxygen signals (6, 42). The signal feeds into the CheA-CheY two-component system, and phospho-CheY binds to the flagellar switch, changing the direction of flagellar rotation. It is also possible that Aer is able to connect to a two-component system that is involved in gene regulation in response to oxygen. Candidates for such a role include ArcA/ArcB (20, 50) or NarQ/NarP (46, 47). Alternatively, Aer could regulate gene expression directly, without the involvement of a two-component system. Preliminary data with a cheY mutant indicate that this newly described regulation by Aer is independent from its function as a chemotaxis protein (data not shown).
In addition to anaerobic respiration, FlhD/FlhC also regulates the Entner-Doudoroff pathway through induction of Aer (Tables 2 and 4 and Fig. 3A). The occurrence of the Entner-Doudoroff pathway in all three phylogenetic domains together with the absence of the Embden-Meyerhof-Parnas pathway in saccharolytic archea (12, 38) indicates that the Entner-Doudoroff pathway is the older pathway for carbohydrate dissimilation (44). It is the major route for the degradation of sugar acids, such as d-gluconate, d-glucuronate, and d-galacturonate, all of which are present in high concentrations in the mouse large intestine (2). The Entner-Doudoroff pathway is also involved in the degradation of l-idonate (5) and in the response to several stress conditions, such as phosphate (52) and nitrogen (31) limitation. Mutants with mutations in Eda, the central enzyme of the Entner-Doudoroff pathway, have similar phenotypes as flhD/flhC mutants. They are unable to grow on d-gluconate, d-glucuronate, and d-galacturonate. Peekhaus and Conway (34) hypothesized that sugar acids are the daily bread of E. coli and that the Entner-Doudoroff pathway is an important aspect in the physiology of this bacterium.
Group II: other metabolic enzymes.
A number of genes were regulated that are related to carbon and nitrogen metabolism. Previous observations (37, 40) indicate that consumption of l-serine may lead to reduced acetyl phosphate levels and derepression of flhDC expression. Simultaneously, flagella are induced and the cell division rate is reduced (36). This study shows the regulation of additional metabolic targets by FlhD or FlhD/FlhC.
l-Serine can be directly converted to glycine, which is then degraded by the glycine cleavage system encoded by gcvTHP (61, 62). As l-serine is consumed, one might expect the glycine concentration to increase, increasing the demand for the Gcv system. On the other hand, l-serine is not only catabolized but is directly used in protein synthesis. Once l-serine is consumed, the demand for the biosynthetic pathway, the first gene of which is serA, is likely to increase (43). The gene products of gltBD (33) convert glutamine to glutamate. Consecutively, glutamate and 3-phosphohydroxypyruvate are converted to phosphoserine and α-ketoglutarate by the gene product of serC, the second gene in serine biosynthesis. Carbamoylphosphate synthetase, encoded by carAB, represents an alternative way of fixing ammonia (18). This reaction makes nitrogen available for pyrimidine nucleotide and arginine biosynthesis. The gatYZ gene products convert galactitol to glyceraldehyde 3-phosphate (30). It seems that FlhD and FlhD/FlhC play a role in balancing amino acid degradation and biosynthesis pathways.
Regulation of gcvTHP is complex. For YK410, gene array (Table 2), Phenotype MicroArray (Fig. 3B), and real-time PCR (Table 4) showed that gcv is induced by FlhD. Reporter gene fusions (Table 4) showed up-regulation by FlhD and down-regulation by FlhC. For MC1000, gene array (Table 2) and lux fusions (Table 4) showed repression by FlhD. On phenotype microarray plates, flhD mutants did not have a phenotype in MC1000 (Fig. 3B). While we cannot explain this phenomenon, it certainly underscores the complex regulation of many genes. The ratio of several regulatory factors may decide the final behavior of some operons.
One of the most interesting aspects of FlhD is its potential for combinatorial specificity. Induction of flagellar genes (24) and eight nonflagellar targets (41) required both FlhD and FlhC, acting as a heterotetramer. In contrast, reduction of the cell division rate (36) and the induction of cadBA (39) appeared to be regulated by FlhD alone. This is consistent with the hypothesis that FlhD may change its binding specificity by interacting with different partner proteins. In this study, three new targets of FlhD were identified. In summary, FlhD regulates cadBA, gcvTHP, gltBD, and ompT expression (Tables 3 and 4). Regulation of gcvTHP and dadAX leads to improved growth on glycine, l-alanine, and l-threonine (Fig. 3B).
Group III: membrane-associated complexes and transporters.
Curli are thin aggregative fibers, allowing E. coli to colonize inert surfaces (for a review see reference 19). Two operons are transcribed divergently, csgBA and csgDEFG. CsgF and CsgG may be involved in secretion and assembly of CsgA, the major fiber subunit. A mutation in OmpR significantly increased the expression of csgA (56). Since OmpR is a repressor of flhDC expression (45), it is possible that this effect was due to increased expression of flhDC in the OmpR mutant.
MalE is a maltose binding protein and interacts with the aspartate receptor Tar (64). RbsA and MglC are part of the high-affinity ribose and galactose transport systems RbsDACBKR (21, 32) and MglBAC (16). DppABCDE (1) and OppABCDE (48) constitute transport systems for dipeptides and oligopeptides. Aer is a signal transducer for aerotaxis (6, 42). All of these transporters feed into the chemotaxis signaling system.
Global regulation.
In recent years, the number of studies of global regulation has increased dramatically. Overlapping responses to a variety of stresses have been observed. For example, the effects of pH on gene expression are known to intersect with oxygenation (10, 29). This study describes the induction of several components of anaerobic respiration by FlhD/FlhC, constituting an intersection of FlhD/FlhC regulation and the response to oxygen. Several components of the acid response are regulated by FlhD/FlhC. Blankenhorn and coworkers (7) described the induction of tnaA and malE under high-pH conditions and the induction of yfiD and gatY under low-pH conditions. All of these were induced by FlhD/FlhC. Increased expression of hdeAB was observed upon treatment of bacteria with acetate (3) and in the presence of H-NS (63) and FlhD/FlhC (this study). Other acetate-regulated genes include oppA, tnaA, malE, mglB, and gatY (22), all of which are regulated by FlhD/FlhC. It seems that the response to many stresses is overlapping and that E. coli possesses sets of genes that are involved in multiple stress responses.
Acknowledgments
We thank R. P. Gunsalus (University of California, Los Angeles) and J. S. Parkinson (University of Utah, Salt Lake City) for providing strains; C. S. Barker (University of Illinois at Chicago, Chicago) for sequencing the mutations in YK4131 and YK4136; A. Radek and K. Luke (Integrated Genomics, Chicago, Ill.) and T. Hope, C. M. Steffens, and M. Melar (University of Illinois at Chicago) for technical help; B. Bochner (Biolog Inc., Hayward, Calif.) for helpful discussions; and P. O'Neill (Molecular Biology Consortium, Chicago, Ill.) and R. T. Fleming (University of Illinois at Chicago) for critically reading the manuscript.
This work was supported by grant GM59484 from the National Institutes of Health, DuPont Company Central Research and Development Department, and Integrated Genomics, Inc.
REFERENCES
- 1.Abouhamad, W. N., and M. D. Manson. 1994. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 14:1077-1092. [DOI] [PubMed] [Google Scholar]
- 2.Allen, A. 1984. The structure and function of gastrointestinal mucus, p. 3-11. In I. E. C. Boedeker (ed.), Attachment of organisms to the gut mucosa. CRC Press, Inc., Boca Raton, Fla.
- 3.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett, D. H., B. B. Frantz, and P. Matsumura. 1988. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5′ consensus sequences of operons under FlbB and FlaI control. J. Bacteriol. 170:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bausch, C., N. Peekhaus, C. Utz, T. Blais, E. Murray, E. Lowary, and T. Conwell. 1998. Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for l-idonic acid catabolism in Escherichia coli. J. Bacteriol. 180:3704-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochner, B., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang, S. E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, P. A., S. B. Melville, J. A. Albrecht, and R. P. Gunsalus. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of FNR and ArcA in repression and activation. Mol. Microbiol. 25:605-615. [DOI] [PubMed] [Google Scholar]
- 12.Danson, M. J. 1989. Central metabolism of the archaebacteria: an overview. Can. J. Microbiol. 35:58-64. [DOI] [PubMed] [Google Scholar]
- 13.Darwin, A. J., E. C. Ziegelhoffer, P. J. Kiley, and V. Stewart. 1998. FNR, NarP, and NarL regulation of Escherichia coli K-12 napF (periplasmic nitrate reductase) operon transcription in vitro. J. Bacteriol. 180:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diehl, F., S. Grahmann, M. Beier, and J. D. Hoheisel. 2001. Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res. 29:E38. [Online.] [DOI] [PMC free article] [PubMed]
- 15.Egan, S. E., R. Fliege, S. Tong, A. Shibata, R. E. Wolf, Jr., and T. Conway. 1992. Molecular characterization of the Entner-Doudoroff pathway in Escherichia coli: sequence analysis and localization of promoters for the edd-eda operon. J. Bacteriol. 174:4638-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Yaagoubi, A., M. Kohiyama, and G. Richarme. 1996. Defect in export and synthesis of the periplasmic galactose receptor MglB in dnaK mutants of Escherichia coli, and decreased stability of the mglB RNA. Microbiology 142:2595-2602. [DOI] [PubMed] [Google Scholar]
- 17.Entner, N., and M. Doudoroff. 1952. Glucose and gluconic acid oxidation of Pseudomonas saccharophila. J. Biol. Chem. 196:852-862. [PubMed] [Google Scholar]
- 18.Han, X., and C. L. Turnbough, Jr. 1998. Regulation of carAB expression in Escherichia coli occurs in part through UPT-sensitive reiterative transcription. J. Bacteriol. 180:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harel, J., and C. Martin. 1999. Virulence gene regulation in pathogenic Escherichia coli. Vet. Res. 30:131-155. [PubMed] [Google Scholar]
- 20.Iuchi, S., and E. C. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 85:1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, M. S., H. Oh, C. Park, and B. H. Oh. 2001. Crystallization and preliminary x-ray crystallographic analysis of Escherichia coli RbsD, a component of the ribose-transport system with unknown biochemical function. Acta Crystallogr. Sect. D 57:728-730. [DOI] [PubMed] [Google Scholar]
- 22.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komeda, Y., and T. Iino. 1979. Regulation of expression of the flagellin gene (hag) in Escherichia coli K-12: analysis of hag-lac fusions. J. Bacteriol. 139:721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maklashina, E., D. A. Berthold, and G. Cecchini. 1998. Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J. Bacteriol. 180:5989-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malakooti, J. 1989. Molecular analysis of the Escherichia coli motor components and regulation of internal control elements of the flaA operon. Ph.D. thesis. University of Illinois, Chicago.
- 27.Melville, S. B., and R. P. Gunsalus. 1996. Isolation of an oxygen-sensitive FNR protein of Escherichia coli: interaction at activator and repressor sites of FNR-controlled genes. Proc. Natl. Acad. Sci. USA 93:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng, S. Y., and G. N. Bennett. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol. 174:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng, S. Y., and G. N. Bennett. 1992. Regulation of the Escherichia coli cad operon: location of a site required for acid induction. J. Bacteriol. 174:2670-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobelman, B., and J. W. Lengeler. 1996. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J. Bacteriol. 178:6790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nystrom, T. 1994. The glucose-starvation stimulon of Escherichia coli: induced and repressed synthesis of enzymes of central metabolic pathways and role of acetyl phosphate in gene expression and starvation survival. Mol. Microbiol. 12:833-843. [DOI] [PubMed] [Google Scholar]
- 32.Park, Y., Y. J. Cho, T. Ahn, and C. Park. 1999. Molecular interactions in ribose transport: the binding protein module symmetrically associates with the homodimeric membrane transporter. EMBO J. 18:4149-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul, L., R. M. Blumenthal, and R. G. Matthews. 2001. Activation from a distance: roles of Lrp and integration host factor in transcriptional activation of gltBDF. J. Bacteriol. 183:3910-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peekhaus, N., and T. Conway. 1998. What's for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 180:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prüβ, B. M. 2000. FlhD, a transcriptional regulator in bacteria. Recent Res. Dev. Microbiol. 4:31-42. [Google Scholar]
- 36.Prüβ, B. M., and P. Matsumura. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prüβ, B. M., and A. J. Wolfe. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 12:973-984. [DOI] [PubMed] [Google Scholar]
- 38.Prüβ, B. M., H. E. Meyer, and A. W. Holldorf. 1993. Characterization of the glyceraldehyde 3-phosphate dehydrogenase from the extremely halophilic archaebacterium Haloarcula vallismortis. Arch. Microbiol. 160:5-11. [DOI] [PubMed] [Google Scholar]
- 39.Prüβ, B. M., D. Markovic, and P. Matsumura. 1997. The Escherichia coli flagellar transcriptional activator flhD regulates cell division through induction of the acid response gene cadA. J. Bacteriol. 179:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prüβ, B. M., J. M. Nelms, C. Park, and A. J. Wolfe. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 176:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prüβ, B. M., X. Liu, W. Hendrickson, and P. Matsumura. 2001. FlhD/FlhC regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol. Lett. 197:91-97. [DOI] [PubMed] [Google Scholar]
- 42.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rex, J. H., B. D. Aronson, and R. L. Somerville. 1991. The tdh and serA operons of Escherichia coli: mutational analysis of the regulatory elements of leucine-responsive genes. J. Bacteriol. 173:5944-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romano, A. H., and T. Conway. 1996. Evolution of carbohydrate metabolism. Res. Microbiol. 147:448-455. [DOI] [PubMed] [Google Scholar]
- 45.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart, V. 1993. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol. Microbiol. 9:425-434. [DOI] [PubMed] [Google Scholar]
- 47.Stewart, V., and R. S. Rabin. 1995. Dual sensors and dual response regulators interact to control nitrate- and nitrite-responsive gene expression in Escherichia coli, p. 233-252. In J. A. Hoch and T. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 48.Tame, J. R. H., G. N. Murshudov, E. J. Dodson, T. K. Neil, G. G. Dodson, C. F. Higgins, and A. J. Wilkinson. 1994. The structural basis of sequence-independent peptide binding by OppA protein. Science 264:1578-1581. [DOI] [PubMed] [Google Scholar]
- 49.Tseng, C.-P., J. Albrecht, and R. P. Gunsalus. 1996. Effect of microaerophilic cell growth conditions on aerobic (cyoABCDE, cydAB) and anaerobic (narGHJI, frdABCD, dmsABC) respiratory pathway gene expression in Escherichia coli. J. Bacteriol. 178:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unden, G., and J. R. Guest. 1985. Isolation and characterization of the FNR protein, the transcriptional regulator of anaerobic electron transport in Escherichia coli. Eur. J. Biochem. 146:193-199. [DOI] [PubMed] [Google Scholar]
- 51.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 52.Van Bogelen, R., E. R. Olson, B. L. Wanner, and F. C. Neidhardt. 1996. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J. Bacteriol. 178:4344-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Dyk, T. K., B. L. Ayers, R. W. Morgan, and R. A. LaRossa. 1998. Constricted flux through the branched-chain amino acid biosynthetic enzyme acetolactate synthase triggers elevated expression of genes regulated by rpoS and internal acidification. J. Bacteriol. 180:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Dyk, T. K., Y. Wei, M. K. Hanafey, M. Dolan, M. J. G. Reeve, J. A. Rafalski, L. B. Rothman-Denes, and R. A. LaRossa. 2001. A genomic approach to gene fusion technology. Proc. Natl. Acad. Sci. USA 98:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Dyk, T. K., E. J. DeRose, and G. E. Gonye. 2001. Lux array, a high-density, genomewide transcription analysis of Escherichia coli using bioluminescent reporter strains. J. Bacteriol. 183:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, H., C.-P. Tseng, and R. P. Gunsalus. 1999. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J. Bacteriol. 181:5303-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watson, N., D. S. Dunyak, E. L. Rosey, J. L. Slonczewski, and E. R. Olson. 1992. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J. Bacteriol. 174:530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wild, J., and T. Klopotowski. 1981. d-Amino acid dehydrogenase of Escherichia coli K12: positive selection of mutants defective in enzyme activity and localization of the structural gene. Mol. Gen. Genet. 181:373-378. [DOI] [PubMed] [Google Scholar]
- 60.Wild, J., J. Hennig, M. Lobocka, W. Walczak, and T. Klopotowski. 1985. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol. Gen. Genet. 198:315-322. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, R. L., L. T. Stauffer, and G. V. Stauffer. 1993. Roles of the GcvA and PurR proteins in negative regulation of the Escherichia coli glycine cleavage enzyme system. J. Bacteriol. 175:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wonderling, L. D., and G. V. Stauffer. 1999. The cyclic AMP receptor protein is dependent on gcvA for regulation of the gcv operon. J. Bacteriol. 181:1912-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida, T., C. Ueguchi, H. Yamada, and T. Mizuno. 1993. Function of the Escherichia coli nucleoid protein, H-NS: molecular analysis of a subset of proteins whose expression is enhanced in an hns deletion mutant. Mol. Gen. Genet. 237:113-122. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, Y., P. J. Gardina, A. S. Kuebler, H. S. Kang, J. A. Christopher, and M. D. Manson. 1999. Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc. Natl. Acad. Sci. USA 96:939-944. [DOI] [PMC free article] [PubMed] [Google Scholar]