Abstract
Microcystins represent an extraordinarily large family of cyclic heptapeptide toxins that are nonribosomally synthesized by various cyanobacteria. Microcystins specifically inhibit the eukaryotic protein phosphatases 1 and 2A. Their outstanding variability makes them particularly useful for studies on the evolution of structure-function relationships in peptide synthetases and their genes. Analyses of microcystin synthetase genes provide valuable clues for the potential and limits of combinatorial biosynthesis. We have sequenced and analyzed 55.6 kb of the potential microcystin synthetase gene (mcy) cluster from the filamentous cyanobacterium Planktothrix agardhii CYA 126. The cluster contains genes for peptide synthetases (mcyABC), polyketide synthases (PKSs; mcyD), chimeric enzymes composed of peptide synthetase and PKS modules (mcyEG), a putative thioesterase (mcyT), a putative ABC transporter (mcyH), and a putative peptide-modifying enzyme (mcyJ). The gene content and arrangement and the sequence of specific domains in the gene products differ from those of the mcy cluster in Microcystis, a unicellular cyanobacterium. The data suggest an evolution of mcy clusters from, rather than to, genes for nodularin (a related pentapeptide) biosynthesis. Our data do not support the idea of horizontal gene transfer of complete mcy gene clusters between the genera. We have established a protocol for stable genetic transformation of Planktothrix, a genus that is characterized by multicellular filaments exhibiting continuous motility. Targeted mutation of mcyJ revealed its function as a gene coding for a O-methyltransferase. The mutant cells produce a novel microcystin variant exhibiting reduced inhibitory activity toward protein phosphatases.
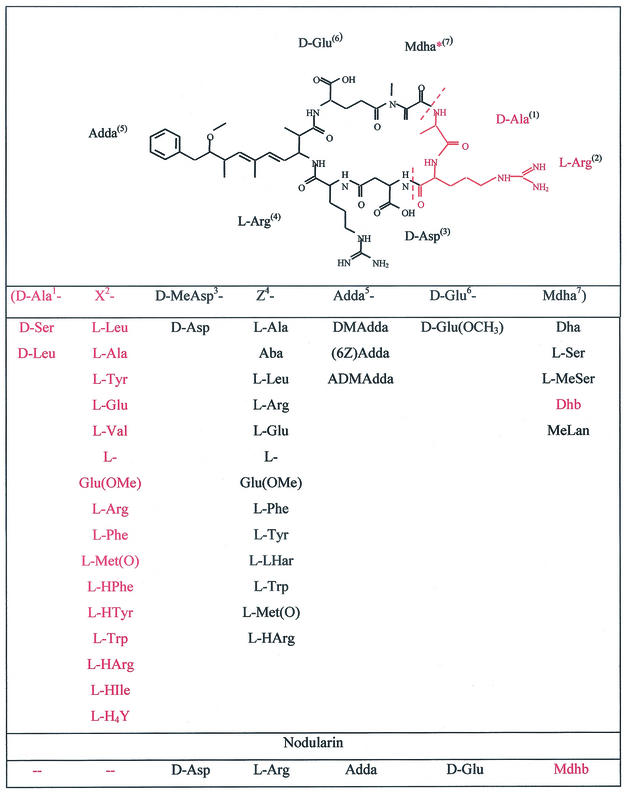
Microcystins are cyclic heptapeptides and share the common structure cyclo(-Adda-d-Glu-Mdha-d-Ala-l-X-d-MeAsp-l-Z), where X and Z represent variable l-amino acids, Adda is 3-amino-9-methoxy-2,6,8-trimethyl-10-phenyl-4,6-decadienoic acid, d-MeAsp is 3-methylaspartic acid, and Mdha is N-methyl-dehydroalanine. More than 60 isoforms of microcystin have been described (Fig. 1) (4). These variations occur between strains of the same genus, as well as between different genera. Microcystins are produced by a wide variety of different planktonic cyanobacteria, including unicellular colony-forming Microcystis, the filamentous motile Planktothrix, and the filamentous heterocystous Anabaena. Each of these genera commonly form blooms in natural water bodies during the summer months. Due to the hepatotoxic character of the microcystins, such mass occurrences lead to serious human health concerns (12).
FIG. 1.
Microcystin and nodularin isoforms. (For a review, see reference 4.) The structure of d-Asp-MCYST-RR is given as an example. Amino acids missing in the nodularin structure are indicated in red. The asterisk indicates the Mdha moiety that is replaced by Mdhb in the nodularin structure. Abbreviations: Aba, amino-isobutyric acid; Dha, dehydroalanine; Mdha, N-methyl-dehydroalanine; Met(O), methionine-S-oxide; Glu(OMe), glutamate methyl ester; (H4)Y, 1,2,3,4-tetrahydrotyrosine; DMAdda, desmethyl-Adda; Dhb, dehydrobutyric acid; MeLan, N-methyl-lanthionin; ADMAdda, O-acetyl-Adda.
The biosynthesis of microcystin has been elucidated for two strains of Microcystis aeruginosa (26, 28, 33). Microcystin is synthesized nonribosomally via a giant enzyme complex comprising peptide synthetases, polyketide synthases (PKSs), and additional modifying enzymes. Nonribosomal peptide synthetases (NRPSs), which catalyze the formation of peptides by a thiotemplate mechanism, are found in both prokaryotes and lower eukaryotes. They are involved in the synthesis of linear, cyclic, and branched-cyclic peptides, including potent drugs, such as penicillin, vancomycin, and cyclosporine (18, 23). NRPSs possess a modular structure, with each module being responsible for the activation, thiolation, modification, and condensation of one specific substrate amino acid. A minimal module consists of condensation (C), adenylation (A), and peptidyl carrier protein (PCP) domains. NRPS products are structurally highly diverse due to the incorporation of many unusual residues, such as d- and N-methylated amino acids or hydroxy acids, in addition to the proteinogenic amino acids (18). Similarly, modular PKSs (PKS type I) are multifunctional megasynthases organized into repeated functional units. PKSs assemble acyl coenzyme A monomers by using the core domains ketosynthase, acyltransferase, and acyl carrier protein. Structural variety is generated by integrated reactions, including ketoreduction, dehydration, or enoyl reduction (2). Among the vast number of polyketides are numerous important pharmaceuticals, including the macrolide antibiotic erythromycin (16).
The current investigation has dealt with the biosynthesis of microcystin in Planktothrix agardhii. Individual P. agardhii strains may produce up to six different nonribosomal peptides simultaneously (9; unpublished data). Furthermore, microcystin production in Planktothrix differs from that in Microcystis in the assortment of microcystin isoforms produced and in the cellular production rates of microcystin, which have been found to be higher in the filamentous strains in field studies (7, 11). Whereas Microcystis is usually characterized by a coexistence of different microcystin (MCYST) isoforms, with considerable variation at the X and Z positions and MCYST-LR, MCYST-YR, and MCYST-RR as predominant toxins (4), Planktothrix is very often dominated by one demethyl-variant of MCYST-RR (7; unpublished data).
Comparative analyses of related NRPSs as performed in the present study has been carried out for lipopeptide antibiotics such as the iturin group (34), the surfactin group (17, 29), and the fengycin-plipastatin group (20). Structural variations within these groups have been proposed to be generated by relaxed specificity of A domains for hydrophobic amino acids (17) but have also been explained by the occurrence of recombination events (34). However, microcystins represent an extraordinarily large family of related peptides (Fig. 1), and amino acid substitutions include various chemically unrelated amino acids.
Here we report the sequence of the mcy gene cluster coding for microcystin biosynthesis in P. agardhii CYA 126. We describe the first genetic manipulation of a Planktothrix strain and demonstrate that mutagenesis of one of the mcy genes resulted in the production of a new microcystin variant. Based on comparison of sequence and organization of the mcy genes of Planktothrix and Microcystis, we discuss the evolution of mcy gene clusters.
MATERIALS AND METHODS
Cyanobacterial strains and culture.
The axenic strain P. agardhii CYA126/8 was provided by K. Sivonen (University of Helsinki, Helsinki, Finland). It was found to contain mainly [d-Asp]-MCYST-RR, aeruginosin, anabaenopeptin, and microviridin (unpublished data).
The strain was cultured in Z8 medium under continuous white light at 15 μEm−2 s−1 with aeration or shaking at 40 rpm. Culture was also performed under the same light conditions on 0.5% agarose plates (Difco, Detroit, Mich.).
Cloning and sequencing of the microcystin synthetase operon.
Chromosomal DNA was isolated from P. agardhii CYA 126/8 as described by Franche and Damerval (8). A lambda ZAP library (Stratagene, La Jolla, Calif.) was constructed, and screenings were performed according to the protocol supplied by the manufacturer. DNA sequencing was performed on both strands with the dye terminator cycle sequencing kit (ABI, Weiterstadt, Germany) with an automatic sequencer (ABI).
Construction of a deletion vector for mcyJ.
The phagemid pBK-CMVJ, carrying the 3′ end of mcyC, mcyJ, and a neighboring IS genetic element was incubated with XmnI. A 486-bp fragment, including the 5′ start of mcyJ, was deleted, and the remainder was religated. The 1.4-kb BsaAI fragment from pACYC184 containing the chloramphenicol resistance determinant was inserted into the AgeI site located in the IS element flanking the mcy gene cluster. Clones bearing the antibiotic resistance determinant in both orientations (pBK-CMVΔJF and pBK-CMVΔJR) were identified by restriction analysis and used for homologous recombinational deletion of mcyJ.
Transformation of P. agardhii CYA 126/8.
pBK-CMVΔJF and pBK-CMVΔJR constructs (20 μg of plasmid DNA) were linearized by restriction with BamHI at its unique site in the multiple cloning region of the vector pBK-CMV. Restricted DNA was column purified (QiaQuick; Qiagen, Hilden, Germany), further incubated for 10 min at 95°C, and then immediately transferred onto ice. A 50-ml sample of a log-phase culture of P. agardhii grown under the conditions described above was centrifuged. The cell pellets obtained were washed three times with 1 mM HEPES (30) and then subjected to electroporation with 10 μg of the recombinant plasmid DNA (1.0 kV, 25 μF, 200 Ω).
Cells were spread on a sterile HATF membrane (Millipore Corp., Bedford, Mass.) covering a nonselective Z8 plate solidified with 0.5% agarose. After 72 h the membrane was transferred onto a plate of solidified Z8 containing 0.5 μg of chloramphenicol/ml. After 4 weeks the membranes were again transferred onto a fresh Z8 antibiotic plate. Transformants moved toward the light source and were collected from the plate after 8 weeks of cultivation under the described culture conditions.
Microcystin analysis.
Microcystins were extracted with 75% aqueous methanol from lyophilized cell material and analyzed by reversed-phase high-performance liquid chromatography (HPLC) incorporating photodiode array detection according to a procedure described previously (7). UV spectra were obtained from 200 to 300 nm, and microcystins were identified by their characteristic absorption spectra (19). Peaks showing the respective microcystin spectra were isolated and further analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS). Positive-ion mass spectra were recorded by using a MALDI-TOF mass spectrometer (Voyager DE-PRO; PerSeptive Biosystems, Framingham, Mass.) as described previously (7). After determination of monoisotopic mass values, post-source decay (PSD) measurements were performed directly from the template plate (33).
Microcystins were identified by means of characteristic fragment ions obtained from PSD analysis. [d-Asp]MCYST-RR and [DMAdda, d-Asp]MCYST-RR were isolated and quantified by HPLC by using standard Microcystin-LR (Calbiochem) as the external standard. The purity of these microcystins was >92% as determined by HPLC-photodiode array detection and MALDI-TOF MS.
PP2A inhibition assay.
For the PP2A inhibition assay, 3-μg MCYST-LR equivalents of [d-Asp]MCYST-RR and [DMAdda, d-Asp]MCYST-RR were purified by HPLC as described above. The two samples were dissolved in 1 ml of 50% aqueous methanol and analyzed three times with PP2A after a 20-fold dilution in 50% aqueous methanol (22).
RESULTS
Identification of the microcystin synthetase gene cluster.
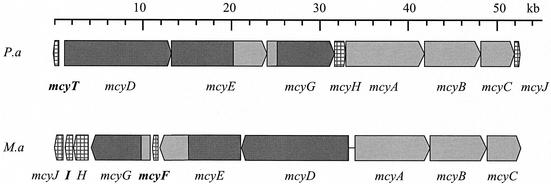
In order to isolate DNA fragments encoding peptide synthetase adenylation domains, we performed a PCR with the primer pair MTF2 and MTR2 (25) by using total genomic DNA of the microcystin-producing strain P. agardhii CYA 126/8 as a template. The resulting amplicon was cloned in the pGEM-T vector, and 20 clones were randomly sequenced. Four different peptide synthetase gene fragments were obtained, one of which showed 75% identity on amino acid level to McyA of M. aeruginosa. This fragment was used to screen a genomic library of P. agardhii CYA 126/8. The initial phagemid clone encoded a protein with homology to a larger part of McyA from M. aeruginosa, including the very characteristic condensation domain. Subsequently, 60 kb were sequenced from overlapping phagemid clones spanning the putative microcystin biosynthesis gene cluster and flanking regions (Fig. 2). The sequence has been deposited in the EMBL database (accession number AJ441056).
FIG. 2.
Organization of the gene clusters for microcystin biosynthesis in P. agardhii and M. aeruginosa. Genes coding for nonribosomal peptide synthetases or PKSs are indicated in light and dark gray, respectively. Hatched regions indicate genes with putative microcystin tailoring functions. Genes which are unique in each cluster are in boldface (33).
Sequence analysis of the mcy region revealed a 55-kb cluster of 9 genes presumably involved in microcystin biosynthesis in P. agardhii. Eight of these genes (mcyA, -B, -C, -D, -E, -G, -H, and -J) show significant similarity to the mcy genes from M. aeruginosa encoding peptide synthetases, PKSs, and modifying enzymes. At the 5′ end of the gene cluster and transcribed in the opposite direction, an additional open reading frame (ORF) was found showing homology to genes and gene domains, respectively, encoding thioesterases. The ORF was designated mcyT (Fig. 2). It is absent from the mcy gene cluster of Microcystis. Two ORFs present in Microcystis are missing: the racemase gene mcyF and mcyI, an ORF whose predicted product is similar to d-3-phosphoglycerate dehydrogenases genes. The arrangement of the mcy genes differs from that in Microcystis. In Microcystis, mcyA-C and McyD-J form two operons that are transcribed bidirectionally from an internal promoter region (13). In contrast, the mcy genes of Planktothrix (except mcyT [see above]) are all on the same strand and may be transcribed as a single operon (Fig. 2).
The Planktothrix mcy gene cluster is flanked by a gene with homology to cyanobacterial sensor kinases at the 5′ end and by an IS element with homology to an IS element of Nostoc PCC 7120 at the 3′ end. It remains to be determined whether the sensor kinase could be involved in the regulation of the mcy gene cluster in Planktothrix.
Function of the mcy genes.
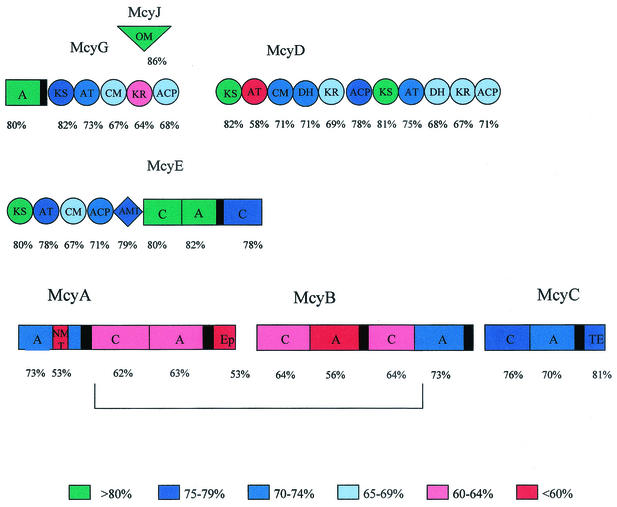
Sequence similarities to genes in the data banks and, in particular, comparison with the homologous genes of Microcystis permits prediction of the functions of the proteins presumptively encoded by eight of the Planktothrix mcy ORFs. Thus, McyD of Planktothrix is a bimodular PKS of 436 kDa with high identity (72%) to McyD of M. aeruginosa. Both the number and the arrangement of catalytic domains are identical to those for Microcystis. McyE (392 kDa), a PKS-peptide synthetase hybrid enzyme is encoded by a gene only 71 bp downstream of the mcyD stop codon and shares 78% identity with McyE from M. aeruginosa. McyE of Planktothrix has the characteristic domain with homology to glutamate semialdehyde aminotransferases, which is supposed to be involved in the transfer of an amino group to the side chain of the Adda moiety. No mcyF was found. A 294-kDa, peptide synthetase-PKS hybrid encoded by a gene downstream from mcyE is 72% identical on an amino acid level to, and shows the same order and characteristics of domains as, McyG from Microcystis and is therefore denoted McyG. The start codon of mcyH follows 99 bp downstream of mcyG, an ORF presumptively encoding an ABC transporter composed of a membrane domain and a cytosolic ATP-binding domain that is most likely involved in an active export of microcystin. McyH of Planktothrix is 73% identical to McyH from Microcystis. Another close homologue (62% identical amino acids) is NosG from Nostoc GSV 224 (part of the nostopeptolide gene cluster, accession number AAF17285). McyA, a bimodular peptide synthetase with 61% identity to its Microcystis counterpart, is encoded by a gene 53 bp downstream of mcyH. This enzyme, together with McyB, another bimodular peptide synthetase, whose gene is 77 bp downstream of mcyA and shows 63% identity to the Microcystis homologue, have the lowest mark of identity between the two cyanobacterial genera. McyC, a peptide synthetase comprising an integrated thioesterase domain is encoded by a gene 74 bp downstream of mcyB. Finally, the gene for McyJ, located at the 3′ end of the gene cluster, shows 86% identity on amino acid level to its M. aeruginosa homologue. McyJ is a putative O-methyltransferase that is suggested to transfer a methyl group to the side chain of the characteristic Adda moiety of microcystins (33).
We have analyzed the percentage of identity between the Microcystis and Planktothrix Mcy proteins on a domain basis. The modules and domains of the mcy gene cluster show very different levels of conservation, ranging from 47 to 88% identity on the amino acid level (Fig. 3). Interestingly, the second module of McyA and the first module of McyB, but not the complete proteins, show an overall low conservation (53 to 64% identity). This region has a distinctly higher GC content (45%) compared to any other part of the microcystin synthetase gene cluster (35 to 38%). The only exception is mcyT, with a GC content of 46%.
FIG. 3.
Domainwise comparison of microcystin synthetases from M. aeruginosa PCC 7806 and P. agardhii CYA 126. Colors indicate the degree of similarity.
Deletion of mcyJ.
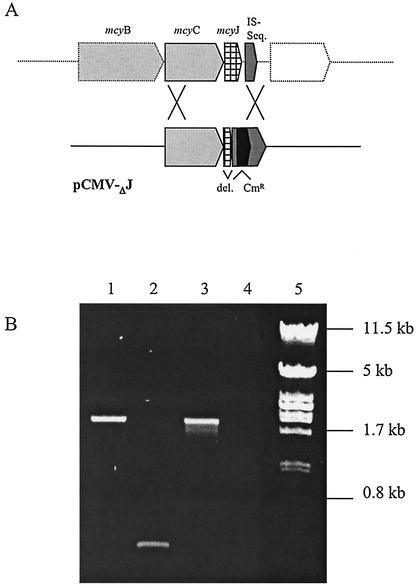
To confirm the proposed role of McyJ for the O-methylation of the Adda moiety during microcystin synthesis (1) and thereby to provide evidence for the involvement of the cloned gene cluster in the biosynthesis of microcystin, we deleted this gene by homologous recombination. Since P. agardhii is motile, no distinct colonies can be obtained. Therefore, we had to develop a new strategy for finding transformed clones. The deletion construct pBK-CMVΔJF was introduced by electroporation. Afterward, cells were transferred to a filter and placed on agarose (containing chloramphenicol) in petri dishes illuminated from one side. We expected only chloramphenicol-resistant transformants to be motile under this condition. During the first week the filaments on the membrane bleached out completely. After 6 weeks of growth on selective medium, a cluster of green putative transformants was observed, heading toward the light source. Single filaments were isolated from this area and grown in liquid medium containing chloramphenicol. PCR amplification of mcyJ from the chloramphenicol-resistant cells demonstrated the stable integration of the pBK-CMVΔJF construct at the expected position within the mcy gene cluster due to a double homologous crossover recombination event (Fig. 4). There was no indication of integration at another position.
FIG. 4.
Inactivation of the O-methyltransferase gene (mcyJ). (A) Schematic diagram of the deletional mutation of the mcyJ gene by homologous recombination. (B) PCR amplification with the DNA from the mutant (lane 1), wild type (lane 2), construct (lane 3), and negative control (lane 4) and the primer pair mcyJF and mcyISR. Lambda/PstI marker (lane 5) shows sizes in kilobases. The amplicon with mutant DNA is of the same size as that of the construct, which includes the chloramphenicol resistance marker (Cmr).
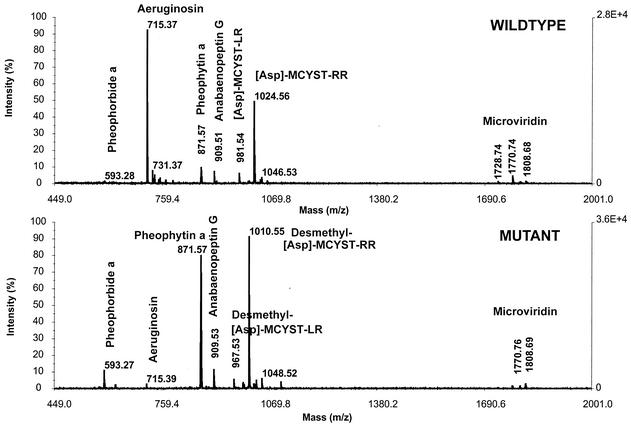
The peptide composition of wild-type and mutant cells was compared by MALDI-TOF MS. P. agardhii CYA 126/8 wild-type produced several nonribosomal peptides in addition to microcystin, including aeruginosin, anabaenopeptin, and microviridin (9; unpublished data). The major microcystin variant produced was [d-Asp]-MCYST-RR (m/z 1,024) (Fig. 5). The mcyJ mutant showed a nearly identical mass pattern compared to the wild type, i.e., it produced the same peptides. The only exception was the mass peak at m/z 1,024, which was replaced in the mutant by a mass peak of m/z 1,010, indicating the lack of a methyl group in microcystin (Fig. 5). The same result was observed for putative transformants by using the pBK-CMVΔJR construct (data not shown). To determine precisely the position site of this methyl group within the microcystin structure, we used the PSD mode of the mass spectrometer. The mass peak of m/z 1,010 showed the typical [d-Asp] MCYST-RR fragmentation and new fragments that could be identified based on its masses as DMAdda fragments (data not shown). Subsequently the novel microcystin was purified by HPLC and tested against protein phosphatase 2A. Microcystins were quantified as microcystin-LR equivalents by HPLC. An equivalent of 3 μg of MCYST-LR of the novel microcystin (DMAdda-d-Asp-microcystin-RR) showed 68% of the inhibition compared to 3 μg of MCYST-LR equivalents of the corresponding wild-type microcystin (d-Asp-MCYST-RR).
FIG. 5.
MALDI-TOF MS analysis of whole cells from the wild type (upper spectrum) and the mcyJ mutant (lower spectrum). Major peaks in the wild type include [d-Asp]-MCYST-RR (m/z 1024), whereas this peak in the mutant is replaced by [DMAdda-d-Asp]-MCYST-RR (m/z 1010).
DISCUSSION
We have identified a cluster of genes in the microcystin-producing P. agardhii strain CYA 126 that shows remarkable similarity to, but also striking differences from, the two completely sequenced mcy gene clusters of M. aeruginosa (26, 28, 33). It is an intriguing question how microcystin biosynthesis has evolved. Microcystins are produced by entirely different cyanobacteria, including unicellular, multicellular filamentous, heterocystous, and nonheterocystous genera. Many strains produce several microcystins simultaneously, although only one or two of these are dominant in any single strain. On the other hand, most cyanobacteria do not synthesize microcystins, and even within the microcystin-producing species many closely related strains do not produce microcystins (4). Therefore, microcystin synthetases are suitable for studying evolutionary aspects of NRPSs and PKSs in cyanobacteria, as well as for investigating the structure-function relationships of variable domains. The identification of microcystin synthetase genes in the different cyanobacterial genera further provides a powerful tool for the detection of certain toxigenic cyanobacterial strains in the environment (25).
General organization of the mcy gene cluster.
Most of the mcy genes detected in Planktothrix have counterparts in the mcy clusters of Microcystis. They include genes coding for peptide synthetases, PKSs, and hybrid enzymes. Presumptively, microcystin biosynthesis in both genera follows the same pathway starting with the synthesis of Adda by the activity of PKSs, followed by the stepwise incorporation of the remaining six amino acids by peptide synthetases. Specific domains of these large multifunctional proteins, as well as enzymes encoded by separate genes, have additional tailoring functions (33).
There are several interesting differences between the mcy gene clusters of Microcystis and Planktothrix, including the general arrangement and the transcriptional orientation of the mcy genes. These differences could be explained by deletion or rearrangement of several genes (mcyF and mcyI are lacking in the Planktothrix cluster, mcyT is missing in the Microcystis cluster) and by inversion of others. Whereas the mcy genes in Microcystis are organized in two operons transcribed in opposite directions, all Planktothrix mcy genes, except mcyT, are transcribed from the same DNA strand and may form a single operon. Recent studies showing similar transcription rates for the two separate mcyA-C and mcyD-J operons in M. aeruginosa PCC 7806 (14) indicate that transcriptional coordination of the two biosynthetic operons in Microcystis (13) is not a limiting factor for the formation of a functional microcystin synthetase complex. In contrast to the coding regions, the putative mcy promoter region of Planktothrix does not show any obvious similarity to the bidirectional promoter region in Microcystis, which otherwise is 99% conserved between the two sequenced mcy gene clusters from M. aeruginosa PCC 7806 and K139 (26, 33).
Genetic manipulation of O methylation.
mcyJ is one of the genes suggested to code for modifying or tailoring functions in microcystin biosynthesis. Based on sequence similarity, it has been proposed to be responsible for O methylation of the Adda moiety. We obtained evidence for the suggested function of McyJ by generating and analyzing a Planktothrix mutant without McyJ activity. Mutant cells produced a variant of microcystin that lacks the specific methyl group proposed to be transferred to Adda by McyJ. This is the first example of a cyanobacterial nonribosomal peptide variant generated by genetic manipulation. Furthermore, the mcyJ deletion mutant is the first mcy mutant which still produces detectable levels of microcystin. Previous mutations in mcyA, -B, -D, -E, and -F of Microcystis resulted in a complete loss of microcystin biosynthesis by the cells (6, 26, 27, 28, 33). Although transcripts were detected, no translation products of the mcy genes were found in the mcyB and mcyA mutant cells (33; unpublished data). Deletion of two key components, McyA and -B, of the Mcy complex, but not of the tailoring enzyme McyJ, leads to the absence of the complete enzyme complex.
Thioesterases.
The thioesterase domain of McyC in Microcystis and Planktothrix is supposed to be needed for cleavage of the peptide from the enzyme complex as well as for the cyclization. The Planktothrix mcy cluster contains a gene (mcyT) encoding a distinct thioesterase in addition to the integrated thioesterase domain of McyC. While this has been shown for many prokaryotic NRPS systems (31), the few cyanobacterial NRPSs which have been characterized so far, including the microcystin synthetase gene clusters in Microcystis, lack this type of gene. Recently, Schneider and Marahiel (31) demonstrated the importance of both thioesterases for tyrocidine biosynthesis by deletion experiments. Only the cooperation of both thioesterases appears to ensure efficient production of tyrocidine. It remains to be shown whether this is also true for Planktothrix. If so, Planktothrix CYA 126 might have a higher capacity for microcystin production than the two investigated Microcystis strains that seem to lack a second thioesterase. Alternatively, a second thioesterase might also exist in Microcystis, and its gene may be localized in another region of the genome distant from the mcy cluster.
Specific features of NRPS domains.
In both Microcystis and Planktothrix three of the seven A domains show considerable variations in the core motifs A1 to A10 defined by Marahiel et al. (23) (Fig. 6). One example is McyG-A, which clusters with acyl coenzyme A synthetases. This domain presumably activates phenylacetate, which has been shown to be one of the precursors of the unusual Adda moiety (24). McyG-A is highly conserved between Microcystis and Planktothrix. Accordingly, Adda is 100% conserved within the microcystin structure of both genera (4).
FIG. 6.
Alignment of the adenylation (A) domain core motifs (23) of microcystin synthetases from P. agardhii and M. aeruginosa (rows P.a. and M.a., respectively). The consensus sequences for motifs A1 to A10 of the peptide synthetase adenylation domains are indicated above. Amino acids in P. agardhii and M. aeruginosa that are identical to the consensus sequences are indicated in red. Boldface type indicates amino acids that are the same in P. agardhii and M. aeruginosa but differ from the consensus.
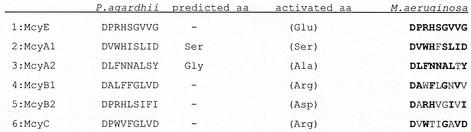
The other two remarkable domains are McyE-A and the second A domain of McyB (McyB-A2). These domains may activate d-Glu and d-Asp, respectively. Alternatively, these amino acids may be first activated as the L-form and then racemized at the peptidyl- or amino-acyl stage by an external racemase, as has been suggested for the biosynthesis of syringomycin (10). A candidate protein present in Microcystis is McyF, which shows homology to aspartate/glutamate racemases of archaea (33). Glutamate racemization by McyF has been shown by complementation experiments in E. coli (27). This protein is not part of the mcy gene cluster in Planktothrix but may be located at another locus. Glutamate racemases are common in other cyanobacteria, including Synechocystis sp. strain PCC 6803 (15), a strain that does not synthesize nonribosomal peptides (5). Usually, the α-carboxyl group is involved in peptide bonds. In microcystins, however, glutamate and aspartate provide their β- and γ-carboxyl groups, respectively, for the peptide bond. It is tempting to speculate, therefore, that the altered core motifs of these two domains, at least in part, are due to the requirements for activation of another type of substrate. A basic difference between Microcystis and Planktothrix relating to one of these domains is the fact that Planktothrix strictly incorporates d-Asp, whereas Microcystis usually introduces d-MeAsp and, in rarer cases, d-Asp into the microcystin structure (7). Nonetheless, the corresponding A domains of Microcystis sp. strain PCC 7806 and Planktothrix sp. strain CYA 126 are very similar, including the specificity pockets (Fig. 7 and data not shown). The activation of either d-Asp or d-MeAsp may therefore depend on other factors not encoded in the mcy gene clusters of Microcystis and Planktothrix.
FIG. 7.
Comparison of the putative specificity-conferring codes of adenylation domains (3, 32) in microcystin biosynthesis from P. agardhii and M. aeruginosa. Residues conserved in both genera are indicated in boldface type (http://raynam.chm.jhu.edu/∼nrps/).
PCP domains of NRPSs are classified into two types. One type belongs to the ordinary C-A-PCP modules comprising the [GGHSL] motif for phosphopantetheine (Ppant) binding, whereas the second type belongs to A-PCP-E modules that contain an altered [GGDSI] motif. Recently, Linne et al. (21) have shown by site-directed mutagenesis that the latter motif is essential for the epimerization process. Both M. aeruginosa and P. agardhii contain this modified PCP motif as part of the first and the second modules of McyA. These modules are expected to introduce Mdha and d-Ala, respectively, into the microcystin structure. The reason for the presence of the modified PCP motif in the second case is obvious, since the second module of McyA comprises a classic epimerization domain. In contrast, the first module of McyA appears to be unique, since a C domain, not an epimerization domain, follows the PCP domain. Since serine is activated by this A domain (6), a dehydration reaction must occur prior to or during the condensation reaction. The C domain of McyA shows deviations from the core motifs in both Microcystis and Planktothrix (33). This points to an active role of the C domain in the dehydration process. Hence, the PCP domain with the GGDSI motif might be important not only for epimerization (21) but also for the interaction with the specific C domain in dehydration.
In Microcystis, an enzyme with similarity to d-3-phosphoglycerate dehydrogenases is encoded as part of the mcy gene cluster (McyI). Although this enzyme is essential for serine biosynthesis, one might speculate that it also could play a role in the predicted serine dehydration reaction. The mcyI gene is not part of the Planktothrix mcy gene cluster. Nevertheless, the Mdha moiety is contained in the microcystin produced by Planktothrix CYA126, and it is thus expected that this ORF is encoded at another locus in the genome.
Domain duplication.
A closer inspection of the adenylation (A) domains of mcy genes in Microcystis and Planktothrix points to duplication and recombination events leading to the addition of new NRPS modules and thus contributing to the evolution of mcy gene clusters. A substantial part of the structural variations within naturally occurring microcystins is due to a variation of individual amino acid positions within the microcystins. The microcystin synthetase complex comprises seven NRPS A domains as part of McyA, -B, -C, -E, and -G. The substrate specificity-conferring residues of A domains are known and have been used to establish a specificity-conferring code of adenylation domains (3, 32). As in M. aeruginosa PCC 7806 (33), only two of the six amino acid adenylation domains in P. agardhii CYA 126 show the same or a similar code that is like those of the corresponding adenylation domains from other organisms (McyA-A1 and McyA-A2; see Fig. 7). The unusual codes of McyE-A and McyB-A2 may be explained by the fact that their substrates are activated in the β- and γ-configurations, respectively (see above and Fig. 6). The codes of the remaining two adenylation domains (McyB-A1 and McyC-A) show considerable variation between the two genera. Nevertheless, McyC-A activates l-Arg in both strains. Although the McyB-A1 pocket of M. aeruginosa PCC 7806 shows a clear leucine code when we applied the algorithm of Challis et al. (3), the P. agardhii CYA 126 McyB-A1 pocket does not fit into the specificity rules (Fig. 7).
The existence of the different microcystin isoforms in single microcystin-producing strains may indicate the existence of multispecific domains that allow for incorporation of many different amino acids, in particular at the X position of MCYST-XZ (see the introduction and Fig. 1). This phenomenon may reflect the process of adaptation after recent recombination events. The evolution from monospecific McyB-A1 binding pockets to multispecific pockets or vice versa has to be studied in more detail by comparing strains that produce high amounts of the different microcystin isoforms simultaneously.
Genes for nodularin biosynthesis as potential ancestors of mcy genes.
Upon comparing the identical amino acids of mcy domains in Planktothrix and Microcystis, we found a low similarity of the continuous sequence, including the second NRPS module of McyA and the first module of McyB is striking (Fig. 3). There is a sharp boundary to the flanking regions, with distinctly higher identity between the two genera. This finding suggests an evolutionary origin of the two modules that is different from the remaining NRPS modules and the PKS part of the mcy cluster. Exactly these two less-conserved modules are expected to be missing from the genome of Nodularia spumigena strains producing the hepatotoxin nodularin. Nodularins are microcystin-related pentapeptides with the structure Adda-d-Glu-Mdhb-d-Asp-Arg (1) (Fig. 1). Thus, the biosynthesis of this peptide should require biosynthetic genes similar to those needed for microcystin biosynthesis, except for the second module of McyA and the first module of McyB, the two modules showing the lowest similarity between Microcystis and Planktothrix. Hybridization of microcystin synthetase genes with DNA from nodularin-producing Nodularia strains demonstrated the expected similarity (25). Hence, we hypothesize that the mcy gene clusters originated from the nodularin biosynthesis genes by the addition of the C-terminal module of McyA (for integration of d-Ala in most microcystins) and the N-terminal module of McyB (for integration of l-Arg, l-Leu, and other l-amino acids). The low level of identity of these modules suggests that their addition occurred more than once by independent events in Microcystis and Planktothrix. To prove or disprove this hypothesis, we need to know the sequence of the respective Nodularia genes and of more mcy gene clusters.
Acknowledgments
We thank Kaarina Sivonen for providing the axenic strain P. agardhii CYA 126 and Noureddine Bouaicha for performing protein phosphatase inhibition assays. We thank Hans von Döhren, Melanie Kaebernick, and two anonymous reviewers for critical reading of the manuscript.
This work was financially supported by grants from the EU (CYANOTOX/ENV4-CT98-0802) to T.B. and J.F. and from the Fond der Chemischen Industrie to T.B.
REFERENCES
- 1.Annila, A., J. Lehtimaki, K. Mattila, J. E. Eriksson, K. Sivonen, T. T. Rantala, and T. Drakenberg. 1996. Solution structure of nodularin: an inhibitor of serine/threonine-specific protein phosphatases. J. Biol. Chem. 271:16695-16702. [DOI] [PubMed] [Google Scholar]
- 2.Cane, D. E., and C. T. Walsh. 1999. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 6:R319-R325. [DOI] [PubMed] [Google Scholar]
- 3.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 4.Chorus, I., and J. Bartram. 1999. Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management. E&FN Spon, Lndon, United Kingdom.
- 5.Christiansen, G., E. Dittmann, L. Via Ordorika, R. Rippka, M. Herdman, and T. Börner. 2001. Nonribosomal peptide synthetase genes occur in most cyanobacterial genera as evidenced by their distribution in axenic strains of the PCC. Arch. Microbiol. 176:452-458. [DOI] [PubMed] [Google Scholar]
- 6.Dittmann, E., B. A. Neilan, M. Erhard, H. von Döhren, and T. Börner. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26:779-784. [DOI] [PubMed] [Google Scholar]
- 7.Fastner, J., M. Erhard, W. W. Carmichael, F. Sun, K. L. Rinehart, H. Ronicke, and I. Chorus. 1999. Characterization and diversity of microcystins in natural blooms and strains of the genera Microcystis and Planktothrix from German freshwaters. Arch. Hydrobiol. 45:147-163. [Google Scholar]
- 8.Franche, C., and T. Damerval. 1988. Tests on nif probes and DNA hybridizations. Methods Enzymol. 167:803-808. [Google Scholar]
- 9.Fujii, K., K. Sivonen, E. Naganawa, and K. Harada. 2000. Non-toxic peptides from toxic cyanobacteria, Oscillatoria agardhii. Tetrahedron 56:725-733. [Google Scholar]
- 10.Guenzi, E., G. Galli, I. Grgurina, D. C. Gross, and G. Grandi. 1998. Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. J. Biol. Chem. 273:32857-32863. [DOI] [PubMed] [Google Scholar]
- 11.Henriksen, P. 1996. Microcystin profiles and content in Danish populations of cyanobacteria/blue-green algae as determined by HPLC. Phycologia 35(Suppl.):102-110. [Google Scholar]
- 12.Jochimsen, E. M., W. W. Carmichael, J. S. An, D. M. Cardo, S. T. Cookson, C. E. Holmes, M. B. Antunes, D. A. de Melo Filho, T. M. Lyra, V. S. Barreto, S. M. Azevedo, and W. R. Jarvis. 1998. Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N. Engl. J. Med. 38:873-878. [DOI] [PubMed] [Google Scholar]
- 13.Kaebernick, M., E. Dittmann, T. Börner, and B. A. Neilan. 2002. Multiple alternate transcripts direct the biosynthesis of microcystin, a cyanobacterial nonribosomal peptide. Appl. Environ. Microbiol. 68:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaebernick, M., B. A. Neilan, T. Börner, and E. Dittmann. 2000. Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl. Environ. Microbiol. 66:3387-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, T. C. Takeuchi, Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3(Suppl.):185-209. [DOI] [PubMed] [Google Scholar]
- 16.Katz, L. 1997. Manipulation of modular polyketide syntheses. Chem. Rev. 97:2557-2575. [DOI] [PubMed] [Google Scholar]
- 17.Konz, D., S. Doekel, and M. A. Marahiel. 1999. Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J. Bacteriol. 181:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konz, D., and M. A. Marahiel. 1999. How do peptide synthetases generate structural diversity? Chem. Biol. 6:R39-R48. [DOI] [PubMed] [Google Scholar]
- 19.Lawton, L. A., C. Edwards, and G. A. Codd. 1994. Extraction and high-performance liquid chromatographic method for the determination of microcystins in raw and treated waters. Analyst 119:1525-1530. [DOI] [PubMed] [Google Scholar]
- 20.Lin, T. P., C. L. Chen, L. K. Chang, J. S. Tschen, and S. T. Liu. 1999. Functional and transcriptional analyses of a fengycin synthetase gene. fenC, from Bacillus subtilis. J. Bacteriol. 181:5060-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linne, U., S. Doekel, and M. A. Marahiel. 2001. Portability of epimerization domain and role of peptidyl carrier protein on epimerization activity in nonribosomal peptide synthetases. Biochemistry 40:15824-15834. [DOI] [PubMed] [Google Scholar]
- 22.Maatouk, I., N. Bouaïcha, D. Fontan, and Y. Levi. Seasonal variation of microcystin concentrations in the Saint-Caprais reservoir (France) and their removal in a small full-scale treatment plant. Water Res., in press. [DOI] [PubMed]
- 23.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in peptide synthesis. Chem. Rev. 97:26511-26573. [DOI] [PubMed] [Google Scholar]
- 24.Moore, R. E., J. L. Chen, B. S. Moore, G. M. L. Patterson, and W. W. Carmichael. 1991. Biosynthesis of microcystin-LR. Origin of the carbons in the Adda and Masp units. J. Am. Chem. Soc. 113:5083-5084. [Google Scholar]
- 25.Neilan, B. A., E. Dittmann, L. Rouhiainen, R. L. Bass, V. Schaub, K. Sivonen, and T. Börner. 1999. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 181:4089-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishizawa, T., M. Asayama, K. Fujii, K. Harada, and M. Shirai. 1999. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 126:520-529. [DOI] [PubMed] [Google Scholar]
- 27.Nishizawa, T., M. Asayama, and M. Shirai. 2001. Cyclic heptapeptide microcystin biosynthesis requires the glutamate racemase gene. Microbiology 147:1235-1241. [DOI] [PubMed] [Google Scholar]
- 28.Nishizawa, T., A. Ueda, M. Asayama, K. Fujii, K. Harada, K. Ochi, and M. Shirai. 2000. Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J. Biochem. 127:779-789. [DOI] [PubMed] [Google Scholar]
- 29.Peypoux, F., J. M. Bonmatin, and J. Wallach. 1999. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51:553-563. [DOI] [PubMed] [Google Scholar]
- 30.Rouhiainen, L., L. Paulin, S. Suomalainen, H. Hyytiainen, W. Buikema, R. Haselkorn, and K. Sivonen. 2000. Genes encoding synthetases of cyclic depsipeptides, anabaenopeptilides, in Anabaena strain 90. Mol. Microbiol. 37:156-167. [DOI] [PubMed] [Google Scholar]
- 31.Schneider, A., and M. A. Marahiel. 1998. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis. Arch. Microbiol. 169:404-410. [DOI] [PubMed] [Google Scholar]
- 32.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 33.Tillett, D., E. Dittmann, M. Erhard, H. von Döhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 34.Tsuge, K., T. Akiyama, and M. Shoda. 2001. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]