Abstract
Nucleotide sequence analysis of the Streptococcus salivarius clpP locus revealed potential binding sites for both the CtsR and HrcA repressors. Dual regulation by HrcA and CtsR was demonstrated by using Bacillus subtilis as a heterologous host, and CtsR was shown to bind directly to the clpP promoter sequence. This is the first example of a clpP gene under the control of HrcA.
Streptococcus salivarius is a widespread human commensal colonizing the oral cavity and upper respiratory tract. A member of the Streptococcus viridans group, this gram-positive bacterium is also an opportunistic pathogen responsible for endocarditis and was recently diagnosed as the pathogen in several cases of meningitis (12, 29).
Stress resistance genes play an important role in the virulence of several pathogens. clpP, clpC, and clpE of Listeria monocytogenes were shown to participate in intracellular parasitism (6), cell adhesion (20), and virulence (19); clpX of Staphylococcus aureus (16) and clpE of Streptococcus pneumoniae (24) were identified by signature-tagged mutagenesis; and the synthesis of DnaK and GroESL in S. aureus was shown to be induced during infection of human epithelial cells (25).
Stress-induced proteins are mainly molecular chaperones or proteases acting to refold or degrade misfolded or denatured proteins (9). Among these, the Clp ATP-dependent protease is composed of Clp ATPase subunits, which confer substrate specificity to the proteolytic subunit ClpP. ClpP has been shown to play a central role in stationary-phase adaptive responses of Bacillus subtilis (18), in the degradation of SsrA-tagged proteins in Escherichia coli (8) and B. subtilis (28), in the modulation of virulence gene expression in Yersinia enterocolitica (23), and in the biofilm formation of Pseudomonas fluorescens (22).
In gram-positive bacteria, stress genes have been grouped into four regulatory classes (4, 26). Class I genes, encoding classical chaperones (DnaK, GroES, and GroEL), are controlled by the HrcA repressor, which recognizes the controlling inverted-repeat chaperone expression (CIRCE) operator sequence. This highly conserved element is composed of a well-conserved 9-bp inverted repeat sequence separated by 9 bp (TTAGCACTCX9GAGTGCTAA) and is always found in association with dnaK or groEL genes (21). Class II genes encode general stress proteins, and their expression requires the σB stress sigma factor. Class III heat shock genes are controlled by the CtsR repressor, a DNA-binding protein which recognizes a tandemly repeated heptad operator sequence (GGTCAAAXAXGGTCAAA) (4). Class IV genes are defined as those devoid of the CIRCE or CtsR operator sequences and whose induction by heat shock or general stress conditions is σB independent.
Sequencing of the upstream and downstream regions of the ftf gene of S. salivarius ATCC 25975 (GenBank accession number LO7793) revealed a gene whose product is highly similar to ClpP of E. coli (7). In this work, we report an analysis of the clpP promoter of S. salivarius, showing evidence for a direct control at the transcriptional level of this gene by a putative ortholog of CtsR. We also demonstrate that HrcA negatively controls clpP expression as well, which is the first example of a clp gene under dual heat shock repression.
Expression of the S. salivarius clpP gene is controlled by CtsR.
Analysis of the S. salivarius clpP promoter region revealed a potential CtsR binding site (4). In order to demonstrate regulation by CtsR, we used B. subtilis as a heterologous host, since the S. salivarius genome sequence is not available. A transcriptional fusion between the S. salivarius ATCC 9758 clpP promoter region and the Bacillus stearothermophilus bgaB gene, which encodes a thermostable β-galactosidase (11), was first constructed by cloning a PCR-generated fragment (positions −295 to −12 with respect to the position of the translation initiation codon) into plasmid pDK (2) and then introduced into B. subtilis.
Derivatives of strain QB8068, in which the endogenous ctsR gene was deleted (2), were then constructed in several steps. The final strain, QB8081 [trpC2 ΔctsR amyE::(clpP′-bgaB aphA3) ΔhrcA::cat thrC::(PxylA-B. subtilis ctsR spec)], carries the S. salivarius clpP′-bgaB transcriptional fusion integrated as a single copy at the amyE locus, a deletion-replacement of the endogenous hrcA gene (17), and a copy of the B. subtilis ctsR gene integrated at the thrC locus under the control of the PxylA xylose-inducible promoter.
Strain QB8081 was grown in Luria-Bertani medium until an optical density at 600 nm (OD600) of 0.3 was achieved, and β-galactosidase activities were assayed during growth at 37°C in the presence or absence of 20 mM xylose and expressed as nanomoles of o-nitrophenyl per minute per milligram of protein.
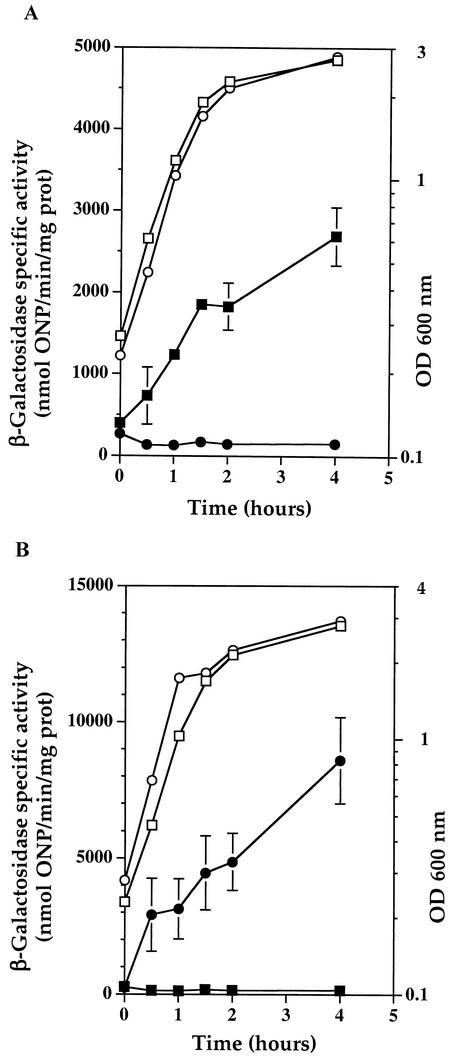
As shown in Fig. 1A, induction by xylose of the B. subtilis ctsR gene led to an up to 16-fold repression of clpP′-bgaB expression. In order to test whether repression by CtsR was relieved during heat shock, strains were grown at 48°C in the presence of xylose. Expression levels of clpP′-bgaB were found to be fully derepressed in the presence of CtsR during growth at 48°C (Fig. 1B). These results strongly suggest that the S. salivarius clpP gene is subjected to negative regulation by an ortholog of CtsR and that this repression no longer occurs during growth at high temperature, leading to heat shock induction of clpP.
FIG. 1.
(A) Expression of S. salivarius clpP is repressed by CtsR. Levels of expression of clpP′-bgaB (strain QB8081) in the presence (○, •) or absence (□, ▪) of xylose are shown. Cultures were grown in Luria-Bertani medium at 37°C until the OD600 was 0.3, and xylose was added to one-half of the culture at a final concentration of 20 mM. (B) Repression of S. salivarius clpP expression by CtsR is abolished by heat shock. The expression of clpP′-bgaB (QB8081) was measured in cells expressing ctsR during growth at 37°C (□, ▪) or 48°C (○, •). Cells were grown in Luria-Bertani medium at 37°C until the OD600 was 0.3, 20 mM xylose was added, and one-half of the culture was shifted to 48°C. Open symbols indicate OD600s. Solid symbols indicate β-galactosidase specific activities expressed as nanomoles of o-nitrophenyl per minute per milligram of protein, and values are the means ± standard deviations of results from two independent assays. ONP, o-nitrophenyl.
Purified CtsR of B. subtilis binds specifically to the heptad direct repeat.
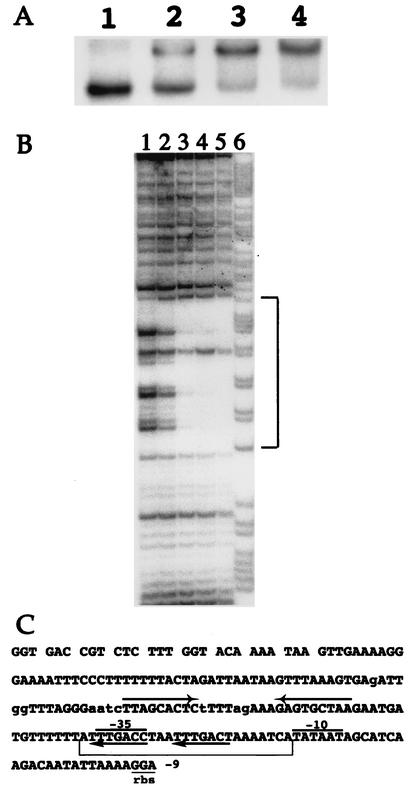
An in vitro approach was used to confirm the direct interaction of CtsR with the target site of S. salivarius. For this purpose, the CtsR protein of B. subtilis was overproduced in E. coli with a carboxy-terminal histidine tag and purified on a Ni-nitrilotriacetic acid agarose (Qiagen) column as previously described (4). The purified protein was then used in gel mobility shift DNA-binding assays. A radiolabeled DNA fragment, corresponding to the S. salivarius clpP promoter region (positions −142 to +44 with respect to the position of the translation initiation codon), was incubated with increasing amounts of CtsR of B. subtilis in the presence of an excess of nonspecific competitor DNA [1 μg of poly(dI-dC)]. Radiolabeling, DNA binding, and gel electrophoresis mobility shift assays were performed as previously described (4). As shown in Fig. 2A, CtsR bound specifically, forming a single protein-DNA complex, with a complete displacement of the DNA fragment occurring at the highest CtsR concentration.
FIG. 2.
(A) CtsR binds specifically to the S. salivarius clpP promoter region. In gel mobility shift experiments, a radiolabeled DNA fragment (10,000 cpm) corresponding to the S. salivarius clpP promoter region (positions −142 to +44 with respect to the translation initiation codon) was incubated with increasing amounts of purified B. subtilis CtsR protein. Lanes 1 to 4, 0, 20, 40, and 80 ng of CtsR, respectively. (B) CtsR protects the directly repeated heptad sequence. In DNase I footprinting analyses of CtsR binding, a radiolabeled DNA fragment (50,000 cpm) corresponding to the S. salivarius clpP promoter region (positions −142 to +44 with respect to the translation initiation codon) was incubated with increasing amounts of purified CtsR. Lanes 1 to 5, 0, 100, 200, 400, and 800 ng of CtsR, respectively; lane 6, G+A Maxam and Gilbert reaction of the corresponding DNA fragment. The region protected by CtsR is shown by the bracket. (C) Nucleotide sequence of the S. salivarius ATCC 9758 clpP promoter region. Potential −35 and −10 promoter sequences are overlined; the CtsR heptad direct-repeat operator sequences are indicated by arrows; the CIRCE operator sequence is indicated by facing arrows; the region protected by CtsR in DNase I footprint experiments is indicated by a bracket; lowercase letters indicate differences in the nucleotidic sequence from that of the ATCC 25975 strain. Positions are numbered relative to the translation initiation codon.
A DNase I footprinting assay was performed on the S. salivarius DNA fragment carrying the clpP promoter region to determine the extent of the protected region and the precise location of the potential CtsR binding site (Fig. 2B). Labeling and DNase I treatment were performed as previously described (4). When the nontemplate strand of the clpP DNA fragment was end labeled, CtsR protected a region extending from positions −63 to −39 relative to the translational start site (Fig. 2B and C). The region protected by CtsR contains the tandem heptanucleotide repeat recognition sequence and the −35 sequence of the promoter in agreement with sequence analysis predictions (Fig. 2C). These results indicate that clpP of S. salivarius is controlled directly by CtsR.
Analysis of the S. salivarius clpP promoter region reveals a potential CIRCE sequence.
During the analysis of the published S. salivarius clpP nucleotide sequence (7), we noted the existence of a 9-bp palindromic sequence upstream from the CtsR binding site, sharing 100% identity with the CIRCE consensus sequence. Surprisingly, the two repeats were separated by 8 bp, whereas in a compilation of 70 CIRCE sequences, the spacer region was invariably 9 nucleotides (10). Since all CIRCE sequences described to date are exclusively associated with the dnaK or groEL operon, it was therefore tempting to consider this noncanonical CIRCE sequence as a cryptic sequence.
In order to test whether HrcA does indeed play a role in controlling expression of the clpP gene, we first sequenced the clpP promoter region of S. salivarius ATCC 9758. As described above, a DNA fragment generated by PCR with chromosomal DNA from the reference strain of S. salivarius ATCC 9758 was cloned into plasmid pDK. The nucleotide sequence of two DNA fragments resulting from independent PCRs was determined (GenBank accession number AY137346), revealing numerous differences with the nucleotide sequence of strain ATCC 25975 (indicated in Fig. 2C). Interestingly, in strain ATCC 9758, the inverted repeat sequences of the CIRCE motif are separated by the consensus 9-bp spacer instead of 8 bp. This reinforced the hypothesis of a potential role for HrcA in the regulation of clpP and suggests that the reported 8-nucleotide spacer sequence from strain ATCC 25975 (7) may be due to a sequencing error.
clpP of S. salivarius is controlled by HrcA.
To evaluate the role of HrcA and this potential CIRCE sequence in S. salivarius clpP regulation, we used B. subtilis as a heterologous host as detailed above.
A DNA fragment corresponding to the coding sequence of B. subtilis hrcA was generated by PCR and cloned between the HindIII and EcoRI sites of plasmid pXT (3), placing hrcA under the control of the xylose-inducible promoter (PxylA). The construct was then integrated as a single copy at the thrC locus of B. subtilis. The resulting strain, QB8083 [trpC2 ΔctsR amyE::(clpP′-bgaB aphA3) ΔhrcA::cat thrC::(PxylA-B. subtilis hrcA spec)], carries the S. salivarius clpP′-bgaB transcriptional fusion integrated as a single copy at the amyE locus, as well as a deletion-replacement of the endogenous hrcA gene (17).
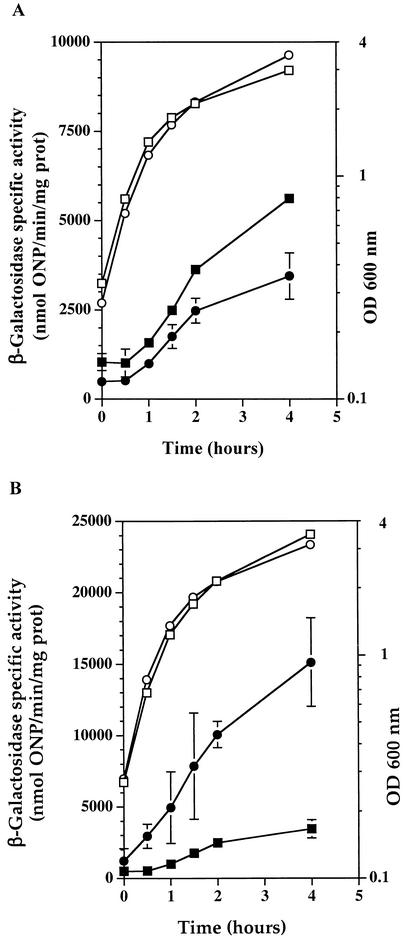
β-Galactosidase assays were performed during growth at 37°C in the presence or absence of xylose. Expression of clpP′-bgaB was repressed twofold in the presence of HrcA (Fig. 3A), whereas expression in the same background was repressed up to 16-fold by CtsR (Fig. 1A). When strain QB8083 was grown at 48°C in the presence of xylose, expression levels of the bgaB fusion were fully derepressed (Fig. 3B). These results indicate that the S. salivarius clpP gene is also repressed by HrcA, albeit weakly but reproducibly, and that this repression is abolished during heat shock, leading to induction of this gene.
FIG. 3.
(A) The expression of clpP of S. salivarius is repressed by HrcA. Levels of expression of clpP′-bgaB (QB8083) in the presence (○, •) or absence (□, ▪) of xylose were monitored as indicated in the legend to Fig. 2A. (B) The repression of S. salivarius clpP expression by HrcA is abolished by heat shock. Levels of expression of clpP′-bgaB (QB8081) were measured in cells expressing hrcA during growth at 37°C (□, ▪) and 48°C (○, •) as indicated in the legend to Fig. 2B. ONP, o-nitrophenyl.
The small effect of HrcA may be due to a weak interaction between the B. subtilis protein and the S. salivarius promoter since HrcA proteins of different bacteria are poorly conserved (10). However, it is worth noting in this case that the CIRCE operator is placed 17 bp upstream from the −35 sequence of the promoter (Fig. 2C), which may explain the weak repression by HrcA. Indeed, it was shown with B. subtilis that increasing the distance between the transcriptional start site and a downstream CIRCE motif progressively decreased the negative regulatory effect (30), and expression of groE′-bgaB was totally abolished when the distance reached 21 bp. Although we cannot exclude the possibility of the existence of a second promoter upstream from the CIRCE sequence, we suggest that HrcA contributes to the repression of clpP expression by interfering with RNA polymerase binding to the downstream promoter. Although the large majority of HrcA-controlled promoters have a CIRCE motif located downstream from the transcriptional start site (21, 27), we note that in the streptococcal group, the CIRCE operator sequence is present twice within the promoter region of the dnaK operon. One CIRCE sequence is found a couple of bases downstream from the transciptional start site, which has been characterized in some cases (13-15). The second CIRCE sequence is located 17 to 18 bp upstream from the −35 promoter sequence in Streptococcus mutans, Streptococcus agalactiae, S. pneumoniae, Streptococcus pyogenes, and Enterococcus faecalis at the same position as the single CIRCE sequence upstream from the clpP gene in S. salivarius. In Lactococcus lactis, there is only a single CIRCE sequence upstream from the dnaK operon and it is located upstream from the potential promoter (5). Although these upstream HrcA recognition sites have not been characterized, their perfect conservation strongly suggests that these sequences are not cryptic and that they must play a role in the regulation of stress gene expression.
clpP of S. salivarius is the first example of a clp gene controlled by both CtsR and HrcA.
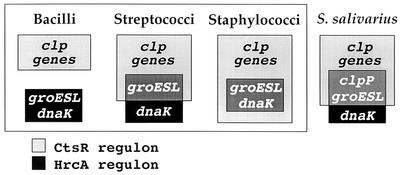
Using both in vivo and in vitro approaches, we have shown that the S. salivarius clpP gene is efficiently repressed by CtsR from B. subtilis, strongly suggesting the existence of a CtsR regulon in this bacterium. Furthermore, we have presented evidence indicating a likely repression of clpP expression by the transcriptional regulator HrcA as well. Although the genome of S. salivarius has not been sequenced, the incomplete genome sequence of the closely related bacterium Streptococcus thermophilus is available (http://www.biol.ucl.ac.be/gene/genome), and we were able to identify potential genes encoding orthologs of both HrcA and CtsR. Inspection of the nucleotide sequence preceding the S. thermophilus clpP gene revealed the same tandem arrangement of conserved CIRCE and CtsR binding sites, suggesting that dual regulation of clpP by both HrcA and CtsR also occurs in S. thermophilus. We recently reported the existence in S. aureus of a significant regulatory overlap between class I and class III stress response genes, since the entire HrcA regulon (consisting of the dnaK and groESL operons) is embedded within the CtsR regulon (Fig. 4), with both operons being preceded by tandemly arranged operator sites for the two repressors (1). Furthermore, comparative genome analysis allowed us to predict the existence in many gram-positive bacteria of a partial overlap between class I and class III genes, particularly in the streptococcal group (S. pneumoniae, S. pyogenes, S. mutans, S. agalactiae, and L. lactis) (Fig. 4), in which only the groESL operon presents both the highly conserved CIRCE-HrcA recognition sequence and the CtsR target site organized in tandem (2). This dual regulation is probably not redundant, since we have shown that in S. aureus, CtsR and HrcA act together synergistically to maintain low levels of expression of the dnaK and groESL operons in the absence of stress (1).
FIG. 4.
Dual regulation by CtsR and HrcA in different gram-positive bacteria. In bacilli the two regulons are distinct, whereas in streptococci they partially overlap, and the HrcA regulon is entirely embedded within the CtsR regulon in staphylococci. clpP of S. salivarius is the first example of a clp gene that is dually regulated by both HrcA and CtsR.
The S. salivarius clpP promoter has an original structure with a classical CtsR binding site present in most clpP promoters of low-G+C-content gram-positive bacteria and an upstream CIRCE motif more characteristic of the dnaK promoters of streptococci (Fig. 2C and 4). Nevertheless, clpP of S. salivarius is the first example of a clp gene associated with a CIRCE sequence. Dual regulation by both repressors may play a role in the fine-tuning of clpP expression in S. salivarius as well as closely coordinating synthesis of the classical chaperones (GroESL and DnaK) with that of the Clp ATP-dependent protease during the stress response.
Acknowledgments
We are grateful to Isabelle Derré for the gift of B. subtilis CtsR protein and many helpful discussions and to Georges Rapoport for critical reading of the manuscript. We thank Marc Galimand for the gift of S. salivarius ATCC 9758 chromosomal DNA. We thank the Institute for Genomic Research for generously providing access to unfinished microbial genome sequences and Pascal Hols and Benoît Grossiord (Université Catholique de Louvain, Louvain, Belgium) for the nucleotide sequences of stress genes from Streptococcus thermophilus LMG1831.
This work was supported by research funds from the European Commission (grant QLRT-1999-01455); the Centre National de la Recherche Scientifique, Institut Pasteur, Université Paris 7; the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires of the Ministère de la Recherche; and the Délégation Générale pour l'Armement. Arnaud Chastanet was the recipient of a Ph.D. thesis fellowship from the Ministère de la Recherche.
REFERENCES
- 1.Chastanet, A., J. Fert, and T. Msadek. Comparative genomics reveal novel heat-shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol, in press. [DOI] [PubMed]
- 2.Chastanet, A., M. Prudhomme, J.-P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derré, I., G. Rapoport, and T. Msadek. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol. Microbiol. 38:335-347. [DOI] [PubMed] [Google Scholar]
- 4.Derré, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol. Microbiol. 31:117-132. [DOI] [PubMed] [Google Scholar]
- 5.Eaton, T., C. Shearman, and M. Gasson. 1993. Cloning and sequence analysis of the dnaK gene region of Lactococcus lactis subsp. lactis. J. Gen. Microbiol. 139:3253-3264. [DOI] [PubMed] [Google Scholar]
- 6.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 7.Giffard, P. M., C. Rathsam, E. Kwan, D. W. Kwan, K. L. Bunny, S. P. Koo, and N. A. Jacques. 1993. The ftf gene encoding the cell-bound fructosyltransferase of Streptococcus salivarius ATCC 25975 is preceded by an insertion sequence and followed by FUR1 and clpP homologues. J. Gen. Microbiol. 139:913-920. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman, S., E. Roche, Y. N. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815-823. [DOI] [PubMed] [Google Scholar]
- 10.Hecker, M., W. Schumann, and U. Völker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 11.Hirata, H., T. Fukazawa, S. Negoro, and H. Okada. 1986. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Idigoras, P., A. Valiente, L. Iglesias, P. Trieu-Cout, and C. Poyart. 2001. Meningitis due to Streptococcus salivarius. J. Clin. Microbiol. 39:3017.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 14.Laport, M. S., A. C. de Castro, A. Villardo, J. A. Lemos, M. C. Bastos, and M. Giambiagi-deMarval. 2001. Expression of the major heat shock proteins DnaK and GroEL in Streptococcus pyogenes: a comparison to Enterococcus faecalis and Staphylococcus aureus. Curr. Microbiol. 42:264-268. [DOI] [PubMed] [Google Scholar]
- 15.Lemos, J. A. C., Y.-Y. M. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 17.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Msadek, T., V. Dartois, F. Kunst, M.-L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 19.Nair, S., C. Frehel, L. Nguyen, V. Escuyer, and P. Berche. 1999. ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol. Microbiol. 31:185-196. [DOI] [PubMed] [Google Scholar]
- 20.Nair, S., E. Milohanic, and P. Berche. 2000. ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect. Immun. 68:7061-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 23.Pederson, K. J., S. Carlson, and D. E. Pierson. 1997. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol. Microbiol. 26:99-107. [DOI] [PubMed] [Google Scholar]
- 24.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qoronfleh, M. W., C. A. Bortner, P. Schwartzberg, and B. J. Wilkinson. 1998. Enhanced levels of Staphylococcus aureus stress protein GroEL and DnaK homologs early in infection of human epithelial cells. Infect. Immun. 66:3024-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumann, W., M. Hecker, and T. Msadek. 2002. Regulation and function of heat-inducible genes in Bacillus subtilis, p. 359-368. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 27.Segal, R., and E. Z. Ron. 1996. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiol. Lett. 138:1-10. [DOI] [PubMed] [Google Scholar]
- 28.Wiegert, T., and W. Schumann. 2001. SsrA-mediated tagging in Bacillus subtilis. J. Bacteriol. 183:3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaniv, L. G., and I. Potasman. 2000. Iatrogenic meningitis: an increasing role for resistant viridans streptococci? Case report and review of the last 20 years. Scand. J. Infect. Dis. 32:693-696. [DOI] [PubMed] [Google Scholar]
- 30.Yuan, G., and S.-L. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the σA-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]