Abstract
The membrane energetics of the intestinal pathogen Vibrio cholerae involves both H+ and Na+ as coupling ions. The sequence of the c subunit of V. cholerae F0F1 ATPase suggested that this enzyme is H+ specific, in contrast to the results of previous studies on the Na+-dependent ATP synthesis in closely related Vibrio spp. Measurements of the pH gradient and membrane potential in membrane vesicles isolated from wild-type and ΔatpE mutant V. cholerae show that the F1F0 ATPase of V. cholerae is an H+, not Na+, pump, confirming the bioinformatics assignments that were based on the Na+-binding model of S. Rahlfs and V. Müller (FEBS Lett. 404:269-271, 1999). Application of this model to the AtpE sequences from other bacteria and archaea indicates that Na+-specific F1F0 ATPases are present in a number of important bacterial pathogens.
Transmembrane circulation of Na+ ions plays a significant role in the physiology of many bacteria and archaea (14, 15). In the case of the halotolerant intestinal pathogen Vibrio cholerae, Na+ transport is apparently linked to virulence gene expression (13). In addition to the primary Na+-translocating pump, NADH:ubiquinone oxidoreductase (NQR), the V. cholerae membrane contains an Na+-driven flagellar motor (13, 22), a battery of Na+/H+ antiporters (8, 15, 36), and an Na+-dependent multidrug efflux pump (2). However, the issue of the energy requirements of oxidative phosphorylation in Vibrio species is still unresolved. The marine bacterium Vibrio alginolyticus has been shown to use sodium motive force to energize ATP synthesis (5). Na+-coupled ATP synthesis driven by respiration or an artificial sodium ion gradient has been also reported in the closely related species Vibrio parahaemolyticus (29, 30). In Propionigenium modestum and Acetobacterium woodii, F0F1-type ATPases have been shown to transport Na+ ions (17, 24), which has led to the suggestion that the vibrial enzyme might also be Na+ translocating (6).
Studies of the mechanism of H+ (and Na+) translocation through the F0 portion of the F1F0 ATPase (9, 11) have demonstrated the key role of Asp61 of subunit c (AtpE) of the Escherichia coli enzyme in this process. The acidic (Asp or Glu) residue in this position is conserved among c subunits of both H+-dependent and Na+-dependent F1F0 ATPases from various bacteria, as well as among the equivalent K subunits of the archaeal- and vacuolar-type (A/V-type) ATPases (reviewed in reference 1) (Table 1). In Na+-conducting c and K subunits, however, the Glu residue is followed by a hydroxyl-containing (Ser or Thr) residue, which apparently provides additional liganding groups, which are essential for binding alkali cations (20, 27). The presence of conserved Pro and Gln residues on the adjacent transmembrane segment and the overall membrane topology of the c subunit have also been implicated in the determination of the cation selectivity of the enzyme (19, 20, 27). Combining the available data, Rahlfs and Müller (27) proposed that there are two determinants of Na+ specificity for the F1F0 ATPase of A. woodii: (i) an enlargement of the C terminus of subunit c and (ii) the presence of the Na+-binding motif of P25, Q29, E62, and T63 (Table 1).
TABLE 1.
Partial protein sequence alignment of the membrane fragments of c subunits (AtpE) of F1F0-type ATPases and K subunits (NtpK) of the A/V-type ATPases
| Enzyme | Organism or sourcea | Sequenceb | Total length (amino acids) of protein |
|---|---|---|---|
| F1F0-type H+ ATPase | E. coli | 19LAAIGAAIGIGILGG-19-FFIVMGLVDAIPMIAVGL70 | 79 |
| V. cholerae | 18LCAVGTAIGFAVLGG-19-MFIIAGLLDAVPMIGIVI69 | 85 | |
| V. alginolyticus | 18LASLGTAIGFALLGG-19-MFIIAGLLDAVPMIGIVI69 | 84 | |
| B. subtilis | 12LGALGAGIGNGLIVS-19-MFMGIALVEALPIIAVVI63 | 70 | |
| E. hirae | 12GAAIGAGYGNGQVIS-19-MFIGVALVEAVPILGVVI63 | 71 | |
| Yeast mitochondria | 17IGLLGAGIGIAIVFA-19-AILGFALSEATGLFCLMV68 | 74 | |
| Human mitochondria | 82VGVAGSGAGIGTVFG-19-AILGFALSEAMGLFCLMV143 | 136 | |
| F1F0-type Na+ ATPase | A. woodii c3 | 20IAGVGPGIGQGFAAG-19-MLLGAAVAETTGIYGLIV71 | 82 |
| A. woodii c1 | 44VAGVGPGIGQGFAAG-19-MLLGAAVAETSGIFSLVI88 | 182 | |
| 120IAGIGPGTGQGYAAG-19-MLLGQAVAQTTGIYALIV171 | 182 | ||
| P. modestum | 23IAGIGPGVGQGYAAG-19-MVLGQAIAESTGIYSLVI74 | 89 | |
| T. maritima | 26IGAIGPGIGEGNIGA-19-MLLADAVAETTGIYSLLI77 | 85 | |
| F1F0-type ATPase unknown cation | Mycoplasma genitalium | 41IAGSTVGIGQGYIFG-19-IFIGSAVSESTAIYGLLI92 | 102 |
| Mycoplasma pneumoniae | 44VGGATVGLGQGYIFG-19-IFIGSAISESSSIYSLLI95 | 105 | |
| U. urealyticum | 50LAAGAVGLMQGFSTA-19-MIVGLALAEAVAIYALIV81 | 89 | |
| S. pyogenes | 9LACFGVSLAEGFLMA-19-MILGVAFIEGTFFVTLVM60 | 65 | |
| A/V-type H+ ATPase | Halobacterium salinarum | 16LAALAAGYAERGIGS-15-GLILTVLPETLVILALVV63 | 71 |
| Sulfolobus acidocaldarius | 45LAAIGAGVAVGMAAA-15-ILIFVAIGEGIAVYGILF92 | 101 | |
| Yeast VMA11 | 30LSCLGAAIGTAKSGI-15-SLIPVVMSGILAIYGLVV76 | 164 | |
| 107FACLSSGYAIGMVGD-15-IVLILIFSEVLGLYGMIV154 | 164 | ||
| A/V-type Na+ ATPase | E. hirae | 24FSGIGSAKGVGMTGE-15-ALILQLLPGTQGLYGFVI72 | 156 |
| 101FTGLFSGIAQGKVAA-15-GIIFAAMVETYAILGFVI148 | 156 | ||
| C. trachomatis | 14LAMIGSAVGCGMAGV-15-IIGLSAMPSSQSIYGLIF62 | 141 | |
| 89SALLLSAFMQGKCCV-15-SFASIGIVESFALFAFVF136 | 141 | ||
| S. pyogenes | 26LSGMGSAYGVGKGGQ-15-ALILQLLPGSQGIYGFAI74 | 159 | |
| 103IVGYFSAKHQGNVSV-15-GVILAAMVETYAILAFVV150 | 159 | ||
| T. pallidum | 14ISAVGSALGLALAGQ-19-LLAFAGAPLTQTIYGFLL65 | 140 | |
| 88LGIAASALSQGRAAA-15-YLTIVGLCETVALLVMVF135 | 140 |
The organisms, sequence accession numbers in the NCBI protein database, and the references for experimentally studied proteins are as follows: E. coli P00844 (26), V. cholerae AAF95908, V. alginolyticus P12991 (23), B. subtilis P37815 (31), E. hirae P26682 (33) and BAA04271 (21), yeast mitochondria P00841 (25), human mitochondria P05496 (7), A. woodii AAF01475 (27) and AAF01474 (28), P. modestum CAA46895 (20), T. maritima AAD36682, M. genitalium P47644, M. pneumoniae AAC43654, U. urealyticum AAF30542, S. pyogenes AAK33697 (AtpE) and AAK33254 (NtpK), H. salinarium BAA13179 (18), S. acidocaldarius AAA72703 (4), yeast vacuole P32842 (35), C. trachomatis AAC67897, and T. pallidum AAC65416.
Residues involved in cation binding are underlined. The Gly23 and Gly27 residues, creating the cavity for Asp61 in the E. coli enzyme (11), are shown in boldface type.
An inspection of the AtpE sequences from V. alginolyticus (23) and V. cholerae (16) showed that they share 92% identity and are very similar to the H+-conducting c subunits of the F1F0 enzymes from E. coli, Bacillus subtilis, Enterococcus hirae, and mitochondria of Saccharomyces cerevisiae and humans (Table 1). Although vibrial AtpE subunits had longer C-terminal fragments than did the H+ ATPases from E. coli and B. subtilis and the Na+ ATPases from A. woodii and P. modestum, they clearly lacked the predicted Na+-binding motif (Table 1). This apparent contradiction of the two previously established criteria of ATPase cation specificity (27) prompted us to investigate the nature of the coupling ion in V. cholerae F1F0 ATPase in more detail and find out whether V. cholerae uses the sodium motive or proton motive force in oxidative phosphorylation.
Growth of wild-type and ΔatpE cells.
This study used V. cholerae strain O395N1 (12) and its isogenic ΔatpE derivative, carrying a deletion of the entire c subunit (proteolipid) of the F1F0 ATPase. The atpE deletion was generated by PCR-based amplification of the genomic DNA by using primer 1 (GGACTAGTCTCCGGCTCGAATAATAA) and primer 2 (GGAATTCCACTTTAGGGGGTAG) for the region downstream of the atpE gene and primer 3 (GGAATTCTCCAAAGATTCAATGGGTATTA) and primer 4 (AATGGTCGACATCTCGTTTTAT) for the region upstream of atpE. Novel EcoRI sites were introduced at the 5′ ends of primers 2 and 3 to allow ligation of the two regions, resulting in a complete deletion of the atpE gene. Novel SpeI and SalI sites were introduced into primers 1 and 4, respectively, to allow direct cloning of the PCR products into the suicide vector pWM91. The DNA was introduced into the chromosome of V. cholerae strain O395N1 following sucrose selection as described previously (12). Genetic elimination of the c subunit allowed the inactivation of the F1F0 ATPase without creating undesirable ion leakage through the mutant enzyme. Growth measurements showed that while the wild-type cells were able to grow in M9 minimal medium supplemented with glucose (2%), succinate (1.2%) or glycerol (2%), the ΔatpE mutant grew only on the fermentable substrate (glucose), thus displaying a classical unc phenotype (data not shown). Very low (3 to 5 μM) concentrations of the protonophore uncoupler, carbonyl cyanide m-chlorophenylhydrazone (CCCP), completely arrested the growth on nonfermentable substrates at both pH 7.5 and 8.5 (data not shown), suggesting that proton acts as the coupling ion in oxidative phosphorylation in V. cholerae.
The transmembrane pH gradient (ΔpH) and membrane potential (Δψ) in inside-out membrane vesicles of V. cholerae were measured by fluorescence quenching and dequenching of 0.5 μM acridine orange (32) and 1.0 μM Oxonol V (34), respectively, as described previously (8). For vesicle preparation, both wild-type and ΔatpE strains of V. cholerae were grown aerobically to mid-logarithmic phase at 37°C in standard Luria-Bertani medium. After the cell suspension was passed through a French press, the vesicles were collected by differential centrifugation and then washed once with and resuspended in isolation buffer containing 10 mM MOPS (morpholinepropanesulfonic acid)-Tris (pH 7.5), 10% (wt/vol) glycerol, 0.2 M K2SO4, 25 mM MgSO4, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 0.2 μg of pepstatin A/ml.
Hydrolysis of ATP results in the formation of ΔpH in inside-out vesicles of wild-type V. cholerae.
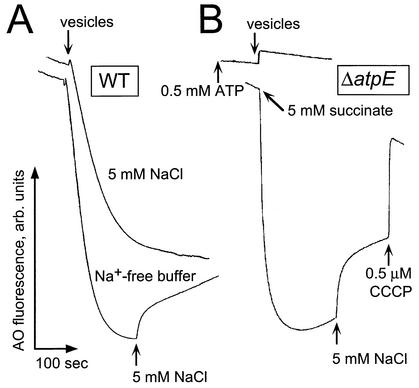
Addition of inside-out vesicles to an experimental buffer containing 0.5 mM Tris-ATP and 0.05 μM valinomycin (added to maximize the formed ΔpH by dissipating the concomitant Δψ) resulted in an immediate proton uptake reflected by the rapid quenching of acridine orange fluorescence (Fig. 1A). No such effect was observed when ATP was not added (data not shown). Na+ was not required for ATP-dependent ΔpH formation. Moreover, in the presence of 5 mM NaCl, the formation of ΔpH was slower and lower in magnitude (Fig. 1A, upper trek) than that in Na+-free buffer (Fig. 1A, lower trek), apparently because of the secondary Na+/H+ antiport. Indeed, the addition of 5 mM NaCl to the mixture after the ΔpH had been established caused a partial dissipation of ΔpH (Fig. 1A, lower trek), which is a typical response of bacterial membranes capable of Na+/H+ antiport. Vesicles isolated from the ΔatpE mutant of V. cholerae lost the ability to generate ΔpH in response to the addition of ATP (Fig. 1B, upper trek) but not the respiratory substrate, succinate (Fig. 1B, lower trek). Furthermore, secondary Na+/H+ exchange was not affected by the deletion (Fig. 1B). The addition of CCCP after the addition of NaCl collapsed the ΔpH completely (Fig. 1B, lower trek). Therefore, hydrolysis of ATP by the F1F0 ATPase of V. cholerae appeared to be directly coupled to uphill proton movement across the membrane.
FIG. 1.
Formation of the ATP-dependent ΔpH in the inside-out subbacterial vesicles from V. cholerae. Aliquots of vesicles (300 μg of protein) were resuspended in 2.5 ml of the isolation buffer (see the text), with MOPS-Tris replaced by Tris-sulfate (pH 7.5 or 8.5 as indicated). The experimental buffer did not contain protease inhibitors and was supplemented with 0.5 μM acridine orange. The resulting quenching of acridine orange fluorescence was monitored in a Shimadzu RF-1501 spectrofluorometer with excitation at 492 nm and emission at 528 nm. (A) Wild-type (O395N1) V. cholerae. Tris-ATP (0.5 mM) was added to the reaction mixture prior to the addition of the vesicles. (B) ΔatpE mutant. Formation of the respiratory ΔpH was initiated by the addition of 5 mM succinate to the experimental mixture containing subbacterial vesicles. In the case of the ATP-dependent ΔpH, 0.5 mM Tris-ATP was added to the reaction mixture prior to the addition of the vesicles.
Effects of protonophore and Na+ on ATP-dependent Δψ in membrane vesicles from V. cholerae.
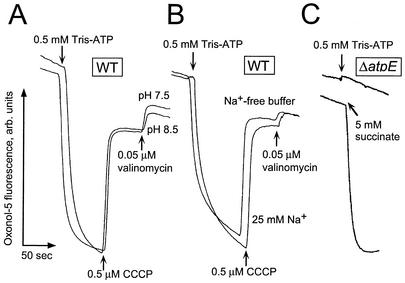
Addition of ATP to the wild-type vesicles resulted in a rapid generation of Δψ (“plus” in vesicular interior) at pH 7.5 and 8.5 (Fig. 2A). Similar to the ATP-dependent ΔpH formation, this process did not require Na+ (Fig. 2B). The protonophore uncoupler collapsed the generated Δψ, so the subsequent addition of valinomycin was without effect (Fig. 2A and B). These observations strongly suggest that the ion translocated by the ATPase was proton, not sodium. The magnitudes of the ATP-dependent Δψ were the same at pH 7.5 and 8.5 (Fig. 2A). Vesicles of the ΔatpE mutant were unable to generate Δψ in response to the addition of ATP, while a respiratory substrate provoked rapid formation of the Δψ (Fig. 2C). Thus, the F1F0 ATPase of V. cholerae displayed behavior typical of proton-translocating ATPases of this type (9). These results indicated that hydrolysis of ATP by this enzyme is coupled to the formation of the proton motive, but not sodium motive, force.
FIG. 2.
Measurements of the ATP-dependent Δψ in subbacterial vesicles of wild-type (O395N1) (A and B) and ΔatpE (C) V. cholerae. Oxonol V (1.0 μM) was used instead of acridine orange. Excitation was at 595 nm, and emission was monitored at 630 nm. All other experimental conditions were as described in the legend to Fig. 1.
Interplay of Na+ and H+ cycles in Vibrio spp.
The data reported in this work show that in V. cholerae, the central membrane-related bioenergetic process, oxidative phosphorylation, is mediated by an H+-dependent F1F0 ATPase. The similarity between the AtpE subunits of V. cholerae and another Vibrio species, V. alginolyticus (Table 1), indicates that the latter enzyme is also H+ dependent. The reason(s) for the previously observed Na+-dependent ATP synthesis in V. alginolyticus (5, 6) and V. parahaemolyticus (29, 30) is not clear at the present time. One possible explanation is that the addition of Na+ ions to whole cells could generate a temporary proton motive force that would not be dissipated immediately by the uncoupler. Such a generation of proton motive force could be due to the activity of any of the several Na+/H+ antiporters present in the cells of Vibrio spp. Another possible explanation is that artificially imposed Na+ gradient could drive reverse electron transport, leading to a substrate-level phosphorylation in the cell cytoplasm, or stimulate some other biochemical process that would result in a temporary boost of ATP levels. It should be noted that one cannot exclude the possible existence of an alternative Na+ ATPase in V. cholerae, which could be repressed under the growth conditions used in this study. An inducible, two-gene ABC-type system extruding Na+ ions, NatAB, has been reported in Bacillus subtilis (3). This transport system supposedly expels toxic Na+ from the cytoplasm and stimulates K+ uptake when the barrier function of the cytoplasmic membrane is affected by uncouplers or alcohols (3). A number of genes encoding putative ABC-type transporters can be found in the V. cholerae genome, but none of them shows significant similarity to the bacillar natAB genes. These putative traffic ATPases of V. cholerae await biochemical characterization.
Na+ and H+ conductance rules.
The data presented here show that of the two determinants of Na+ specificity of the A. woodii F1F0 ATPase identified by Rahlfs and Müller (27), the first, i.e., the length of the C-terminal extension of AtpE, did not seem to correlate with the cation specificity of the enzyme. In contrast, the absence of the likely Na+-binding motif Px3Qx28,32ET (Table 1) led to the correct identification of the V. cholerae enzyme as an H+ ATPase, suggesting that this motif is a reliable predictor of Na+ conductance. Indeed, the presence of a similar sequence motif in the AtpE subunit from Thermotoga maritima suggests that its F1F0 ATPase is Na+ dependent, which is consistent both with the transport data (10) and with the presence in the T. maritima genome of two Na+ pumps, the NQR and the Na+-translocating oxaloacetate decarboxylase (15).
Na+ ATPases in other bacterial pathogens.
Verification of the Na+-binding motif as a reliable predictor of ATPase cation specificity allows one to classify various bacterial F1F0 and A/V-type ATPases into Na+ ATPases and H+ ATPases. Sequence alignment of c and K subunits of F1F0 and A/V ATPases, respectively, shows the presence of the Na+-binding motif in ATPases from such pathogens as Chlamydia trachomatis, Treponema pallidum, and Streptococcus pyogenes (Table 1), which also have primary Na+ pumps and have been predicted to rely on Na+ circulation for their energy metabolism (15). There are some surprises, too. The causative agent of Lyme disease, Borrelia burgdorferi, for example, encodes a vacuolar-type ATPase that is very similar to the one from T. pallidum and also contains a typical Na+-binding motif (data not shown). Remarkably, the genome of B. burgdorferi does not encode any (known) primary H+ or Na+ pump, except for two NQR subunits, NqrA and NqrB, fused into a single polypeptide chain (BB0072). Therefore, it appears that this organism uses its Na+ ATPase for ATP hydrolysis and depends on its two NhaC-type Na+/H+ antiporters (BB0637 and BB0638) for the generation of proton motive force.
The absence of experimental data on the role of the Pro residue in the Na+-binding motif described by Rahlfs and Müller prevents us from predicting the nature of the coupling ion for mycoplasmal F1F0 ATPases (Table 1). The conservation of other residues in the Na+-binding site suggests that these organisms should be able to utilize Na+ as a coupling ion. Remarkably, another species of the Mycoplasmataceae, Ureaplasma urealyticum, appears to have lost the critical Ser residue of the motif and probably has a strictly H+-dependent ATPase.
In conclusion, the results of this work show that in spite of the importance of Na+ circulation for the membrane energetics of Vibrio cholerae and related microorganisms, these organisms still rely on the proton motive force for oxidative phosphorylation. The situation might be different for less versatile bacterial pathogens with smaller genomes that do not possess such a variety of membrane ionic pumps (15).
Acknowledgments
This study was supported in part by a Cancer Center support grant (CA 21765) and an ALSAC (American Lebanese Syrian Associated Charities) grant to C.C.H. J.D. and P.D. were supported by NSERC (Natural Sciences and Engineering Research Council of Canada) operating grant no. 227414-00.
REFERENCES
- 1.Blair, A., L. Ngo, J. Park, I. T. Paulsen, and M. H. Saier, Jr. 1996. Phylogenetic analyses of the homologous transmembrane channel-forming proteins of the F0F1-ATPases of bacteria, chloroplasts and mitochondria. Microbiology 142:17-32. [DOI] [PubMed] [Google Scholar]
- 2.Chen, J., Y. Morita, M. N. Huda, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2002. VmrA, a member of a novel class of Na+-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J. Bacteriol. 184:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, J., A. A. Guffanti, and T. A. Krulwich. 1997. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol. Microbiol. 23:1107-1120. [DOI] [PubMed] [Google Scholar]
- 4.Denda, K., J. Konishi, T. Oshima, T. Date, and M. Yoshida. 1989. A gene encoding the proteolipid subunit of Sulfolobus acidocaldarius ATPase complex. J. Biol. Chem. 264:7119-7121. [PubMed] [Google Scholar]
- 5.Dibrov, P. A., R. L. Lazarova, V. P. Skulachev, and M. L. Verkhovskaya. 1986. The sodium cycle. II. Na+-coupled oxidative phosphorylation in Vibrio alginolyticus cells. Biochim. Biophys. Acta 850:458-465. [DOI] [PubMed] [Google Scholar]
- 6.Dibrov, P. A., R. L. Lazarova, V. P. Skulachev, and M. L. Verkhovskaya. 1989. A study on Na+-coupled oxidative phosphorylation: ATP formation supported by artificially imposed delta pNa and delta pK in Vibrio alginolyticus cells. J. Bioenerg. Biomembr. 21:347-357. [DOI] [PubMed] [Google Scholar]
- 7.Dyer, M. R., and J. E. Walker. 1993. Sequences of members of the human gene family for the c subunit of mitochondrial ATP synthase. Biochem. J. 293:51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dzioba, J., E. Ostroumov, A. Winogrodzki, and P. Dibrov. 2002. Cloning, functional expression in Escherichia coli and primary characterization of a new Na+/H+ antiporter, NhaD, of Vibrio cholerae. Mol. Cell. Biochem. 229:119-124. [DOI] [PubMed] [Google Scholar]
- 9.Fillingame, R. H. 1990. Molecular mechanism of ATP synthesis of the F1F0 type H+-transporting ATP synthases, p. 345-391. In T. A. Krulwich (ed.), The bacteria, vol. 12. Academic Press, New York, N.Y.
- 10.Galperin, M. Y., K. M. Noll, and A. H. Romano. 1996. The glucose transport system of the hyperthermophilic anaerobic bacterium Thermotoga neapolitana. Appl. Environ. Microbiol. 62:2915-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girvin, M. E., V. K. Rastogi, F. Abildgaard, J. L. Markley, and R. H. Fillingame. 1998. Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase. Biochemistry 37:8817-8824. [DOI] [PubMed] [Google Scholar]
- 12.Häse, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Häse, C. C., and J. J. Mekalanos. 1999. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 96:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Häse, C. C., and B. Barquera. 2001. Role of sodium bioenergetics in Vibrio cholerae. Biochim. Biophys. Acta 1505:169-178. [DOI] [PubMed] [Google Scholar]
- 15.Häse, C. C., N. D. Fedorova, M. Y. Galperin, and P. A. Dibrov. 2001. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 65:353-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heise, R., V. Müller, and G. Gottschalk. 1992. Presence of a sodium-translocating ATPase in membrane vesicles of the homoacetogenic bacterium Acetobacterium woodii. Eur. J. Biochem. 206:553-557. [DOI] [PubMed] [Google Scholar]
- 18.Ihara, K., S. Watanabe, K. Sugimura, I. Katagiri, and Y. Mukohata. 1997. Identification of proteolipid from an extremely halophilic archaeon Halobacterium salinarum as an N,N′-dicyclohexyl-carbodiimide binding subunit of ATP synthase. Arch. Biochem. Biophys. 341:267-272. [DOI] [PubMed] [Google Scholar]
- 19.Kaim, G., and P. Dimroth. 1995. A double mutation in subunit c of the Na+-specific F1F0-ATPase of Propionigenium modestum results in a switch from Na+ to H+-coupled ATP synthesis in the Escherichia coli host cells. J. Mol. Biol. 253:726-738. [DOI] [PubMed] [Google Scholar]
- 20.Kaim, G., F. Wehrle, U. Gerike, and P. Dimroth. 1997. Molecular basis for the coupling ion selectivity of F1F0 ATP synthases: probing the liganding groups for Na+ and Li+ in the c subunit of the ATP synthase from Propionigenium modestum. Biochemistry 36:9185-9194. [DOI] [PubMed] [Google Scholar]
- 21.Kakinuma, Y., S. Kakinuma, K. Takase, K. Konishi, K. Igarashi, and I. Yamato. 1993. A gene encoding the 16-kDa proteolipid subunit of Enterococcus hirae Na+-ATPase complex. Biochem. Biophys. Res. Commun. 195:1063-1069. [DOI] [PubMed] [Google Scholar]
- 22.Kojima, S., K. Yamamoto, I. Kawagishi, and M. Homma. 1999. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 181:1927-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krumholz, L. R., U. Esser, and R. D. Simoni. 1992. Characterization of the genes coding for the F1F0 subunits of the sodium dependent ATPase of Propionigenium modestum. FEMS Microbiol. Lett. 70:37-41. [DOI] [PubMed] [Google Scholar]
- 24.Laubinger, W., and P. Dimroth. 1987. Characterization of the Na+-stimulated ATPase of Propionigenium modestum as an enzyme of the F1F0 type. Eur. J. Biochem. 168:475-480. [DOI] [PubMed] [Google Scholar]
- 25.Macino, G., and A. Tzagoloff. 1979. Assembly of the mitochondrial membrane system. The DNA sequence of a mitochondrial ATPase gene in Saccharomyces cerevisiae. J. Biol. Chem. 254:4617-4623. [PubMed] [Google Scholar]
- 26.Nielsen, J., F. G. Hansen, J. Hoppe, P. Friedl, and K. von Meyenburg. 1981. The nucleotide sequence of the atp genes coding for the F0 subunits a, b, c and the F1 subunit delta of the membrane bound ATP synthase of Escherichia coli. Mol. Gen. Genet. 184:33-39. [DOI] [PubMed] [Google Scholar]
- 27.Rahlfs, S., and V. Müller. 1997. Sequence of subunit c of the Na+-translocating F1F0 ATPase of Acetobacterium woodii: proposal for determinants of Na+ specificity as revealed by sequence comparisons. FEBS Lett. 404:269-271. [DOI] [PubMed] [Google Scholar]
- 28.Rahlfs, S., S. Aufurth, and V. Müller. 1999. The Na+-F1F0-ATPase operon from Acetobacterium woodii. Operon structure and presence of multiple copies of atpE which encode proteolipids of 8- and 18-kDa. J. Biol. Chem. 274:33999-34004. [DOI] [PubMed] [Google Scholar]
- 29.Sakai, Y., C. Moritani, M. Tsuda, and T. Tsuchiya. 1989. A respiratory-driven and an artificially driven ATP synthesis in mutants of Vibrio parahaemolyticus lacking H+-translocating ATPase. Biochim. Biophys. Acta 973:450-456. [DOI] [PubMed] [Google Scholar]
- 30.Sakai-Tomita, Y., M. Tsuda, and T. Tsuchiya. 1991. Na+-coupled ATP synthesis in a mutant of Vibrio parahaemolyticus lacking H+-translocating ATPase activity. Biochem. Biophys. Res. Commun. 179:224-228. [DOI] [PubMed] [Google Scholar]
- 31.Santana, M., M. S. Ionescu, A. Vertes, R. Longin, F. Kunst, A. Danchin, and P. Glaser. 1994. Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 176:6802-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuldiner, S., H. Rottenberg, and M. Avron. 1972. Determination of pH in chloroplasts. 2. Fluorescent amines as a probe for the determination of pH in chloroplasts. Eur. J. Biochem. 25:64-70. [DOI] [PubMed] [Google Scholar]
- 33.Shibata, C., T. Ehara, K. Tomura, K. Igarashi, and H. Kobayashi. 1992. Gene structure of Enterococcus hirae (Streptococcus faecalis) F1F0-ATPase, which functions as a regulator of cytoplasmic pH. J. Bacteriol. 174:6117-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, J. C. 1990. Potential-sensitive molecular probes in membranes of bioenergetic relevance. Biochim. Biophys. Acta 1016:1-28. [DOI] [PubMed] [Google Scholar]
- 35.Umemoto, N., Y. Ohya, and Y. Anraku. 1991. VMA11, a novel gene that encodes a putative proteolipid, is indispensable for expression of yeast vacuolar membrane H+-ATPase activity. J. Biol. Chem. 266:24526-24532. [PubMed] [Google Scholar]
- 36.Vimont, S., and P. Berche. 2000. NhaA, an Na+/H+ antiporter involved in environmental survival of Vibrio cholerae. J. Bacteriol. 182:2937-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]