Abstract
DivIVA is involved in Bacillus subtilis cell division and is located at the cell poles. Previous experiments suggested that the cell division proteins FtsZ and PBP 2B are required for polar targeting of DivIVA. By using outgrowing spores, we show that DivIVA accumulates at the cell poles independent of the presence of FtsZ or PBP 2B.
To proliferate, bacterial cells divide into two equal-sized daughter cells. Cell division initiates with polymerization of the tubulin-like protein FtsZ into a ring-like structure, the contractile Z ring, on which the cytokinesis apparatus assembles (7, 10). For efficient propagation it is essential that the Z ring be formed in the middle of the parent cell, as a polar division would result in the formation of anucleate mini-cells. Rod-like bacteria prevent polymerization of FtsZ too close to the cell poles by means of the conserved protein couple MinC/MinD (14). In Bacillus subtilis, MinC and MinD are located at the cell poles and inhibit FtsZ polymerization in that area of the cell. The polar accumulation of MinC and MinD is established by the pole-located protein DivIVA (11, 12). In Escherichia coli, MinC and MinD oscillate between the extremities of the cell and in this way prevent polar division (15). E. coli does not contain a DivIVA homologue, but this protein seems to be conserved in gram-positive bacteria (6, 17).
During cell division, DivIVA is recruited to the newly formed Z ring, and after septation is completed and the Z ring dissolves, DivIVA remains attached to the newly formed cell poles (5). FtsZ is an essential protein, but in a B. subtilis mutant in which the concentration of FtsZ can be regulated, depletion of FtsZ prevents accumulation of DivIVA at the potential division site, as no Z rings are formed (12). Under these conditions, DivIVA remains present at the cell poles. On the basis of this experiment, it was postulated that FtsZ is involved in recruiting DivIVA to the newly formed septum and therefore is required for polar positioning of DivIVA. In such a scenario, DivIVA is retained at the poles by a mechanism independent of the presence or absence of FtsZ. However, since DivIVA was already present at the poles, this experiment did not prove that FtsZ itself is essential for the polar positioning of DivIVA. It remains possible that a certain factor associated with the polar regions of the cell is the determinant for the localization of DivIVA and that this factor depends on the formation of a polar region, a process that normally depends on cell division and therefore on the presence of FtsZ. To address this issue, we needed a system in which the presence of a cell pole is not directly linked to the preceding formation of a Z ring. In B. subtilis, such a situation is found in the germinating spore. Spores are very compact structures developed to survive extremely inhospitable environmental conditions. The chromosome is compressed into a ring-like structure, and no replication or Z ring or other cytokinesis apparatus is likely to be present in the dormant spore (16). When spores germinate, they start to swell and burst out of their rigid spore coats. They then elongate to normal-sized cells before cell division starts. Since outgrowing spores divide like normal vegetatively growing cells, DivIVA (and the MinC/MinD protein couple) should be present at the cellular extremities from an early stage. However, there is no Z ring present at the poles of germinating spores to which DivIVA can attach. Thus, the normal division of outgrowing spores suggests that FtsZ is not essential for the polar positioning of DivIVA. We set out to investigate this supposition by monitoring the localization of a DivIVA-green fluorescent protein (GFP) fusion during germination of spores from a wild-type B. subtilis strain and a strain in which FtsZ can be depleted.
Localization of DivIVA in outgrowing spores.
Before we could start with fluorescence microscopy studies of germinating B. subtilis spores, we had to overcome an important obstacle, namely, the strong autofluorescence of the spore coat. The spore coat tends to snap in half and stay attached to the poles of the outgrowing spore, as a consequence of which the fluorescence of pole-localized GFP fusion proteins is masked. We tried several extraction procedures to reduce the autofluorescence of the spore coat but with no success. However, treatment of spores with sodium dodecyl sulfate (SDS) under strong reducing conditions, a procedure normally used to extract certain spore coat proteins, appeared to be useful (19). After such extraction, outgrowing spores detached much more easily from the enveloping spore coat. Unfortunately, such extraction decreases the efficiency of germination. We discovered that the addition of the detergent Tween 20 to the growth medium substantially stimulated germination and countered the adverse effects of SDS extraction. With these adjustments, the complete procedure for spore isolation, extraction, and germination was as follows. Schaeffer's sporulation medium (8) was inoculated with B. subtilis strain 1803 (containing a divIVA-gfp reporter fusion under the transcriptional control of the divIVA promoter) (Table 1). This strain was used in previous studies to examine the localization of DivIVA (17). Spores were collected by centrifugation and washed in TES buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 1 M NaCl). The pellet was resuspended in water and incubated with lysozyme (0.1 mg/ml) for 1 h at 37°C. After this, DNase (20 μg/ml), RNase (20 μg/ml), and SDS (0.5%) were added and incubation was continued for another hour. Next, spores were washed once with TES buffer and three times with 0.01% Tween 20 and stored at −80°C. Washed spores were extracted in a solution of 10 mM Tris-HCl, 0.5% SDS, 10 mM dithiothreitol, and 100 mM NaCl for 1 h at 37°C. After this extraction, the spores were washed six times with 0.01% Tween 20. Spores were diluted in germination medium to an optical density at 600 nm (OD600) of 1. The germination medium consisted of S medium (chosen for its low-fluorescence background signal) supplemented with the germinant alanine (10 mM) (8) and Tween 20 (0.002%) (12). To increase the germination efficiency, spores were heat shocked for 30 min at 70°C (19). After heat shock, the spore mixture was incubated at 30°C under continuous shaking. At different time intervals, samples were taken for fluorescence microscopy examination. Outgrowing spores were viewed on agarose slides, and images were obtained as described by Marston and Errington (11). Figure 1 shows a compilation of the different stages of outgrowing spores as observed by fluorescence microscopy. DivIVA-GFP appears as fluorescent dots concentrated at the cell poles. During growth of the cell, more DivIVA-GFP spots emerge at the periphery of the cell, and when cytokinesis starts, DivIVA-GFP concentrates at the site of division. The localized concentrations of DivIVA-GFP are presumably a consequence of oligomerization of DivIVA (13). Thomaides et al. (17) have reported that DivIVA is not targeted to the asymmetric sporulation septum. Since DivIVA-GFP is observed at both cell poles of germinating spores, the possibility that small amounts of old DivIVA molecules, remaining from the prespore, serve as polar attachment sites for newly synthesized DivIVA is unlikely.
TABLE 1.
Bacterial strains
| B. subtilis strain | Relevant genotypea | Construction or reference (antibiotic[s])b |
|---|---|---|
| 1803 | divIVA::(PdivIVA-gfp divIVA+cat) | 17 |
| 1801 | chr::(Pspac-ftsZ ble) | 12 |
| 2060 | amyE::(Pxyl-c-myc-mreBCD spc) mreB::neo | 9 |
| 2505 | mbl::spc | 9 |
| 3295 | chr::(Pspac-pbpB neo) | R. Daniel, unpublished data |
| 3291 | divIVA::(PdivIVA-gfp divIVA+cat) chr::(Pspac-ftsZ ble) | 1801 transformed with 1803 DNA (Cm) |
| 3292 | divIVA::(PdivIVA-gfp divIVA+cat) amyE::(Pxyl-c-myc-mreBCD spc) mreB::neo | 1803 transformed with 2060 DNA (Sp, Km) |
| 3293 | divIVA::(PdivIVA-gfp divIVA+cat) mbl::spc | 1803 transformed with 2505 DNA (Sp) |
| 3294 | divIVA::(PdivIVA-gfp divIVA+cat) chr::(Pspac-pbpB neo) | 1803 transformed with 3295 DNA (Km) |
All strains carry the trpC2 marker.
Antibiotics used for selection are indicated (Cm, chloramphenicol; Sp, spectinomycin; Km, kanamycin).
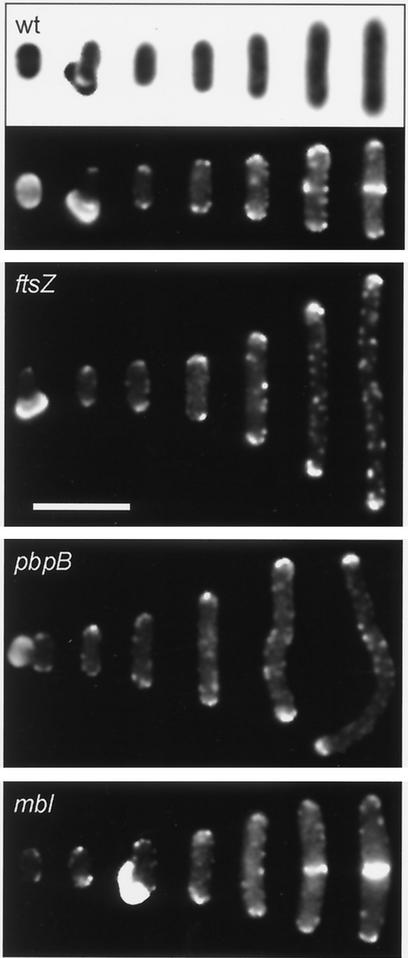
FIG. 1.
Localization of DivIVA-GFP in germinating B. subtilis spores. The lower half of the top panel (wt) shows a composite fluorescence image of frames depicting successive stages of germination of wild-type B. subtilis spores (corresponding phase-contrast images are shown in the upper half of the panel). In the first two frames from the left, the spore coat is still present. Escape from the spore coat occurs about 2 h after inoculation, and the first cell division takes place about 1 h later. The upper-middle panel (ftsZ) shows a composite fluorescence image of frames depicting germinating B. subtilis 3291 spores in which FtsZ was absent. In the leftmost frame, a spore coat is visible. Scale bar, 5 μm. The lower-middle panel (pbpB) shows a composite fluorescence image of frames depicting germinating B. subtilis 3294 spores in which PBP 2B was absent. In the leftmost frame, a spore coat is visible. The bottom panel (mbl) shows a composite fluorescence image of frames depicting germinating B. subtilis 3293 spores in which Mbl was absent. In the third frame from the left, a spore coat is visible.
Localization of DivIVA in the absence of FtsZ or PBP 2B.
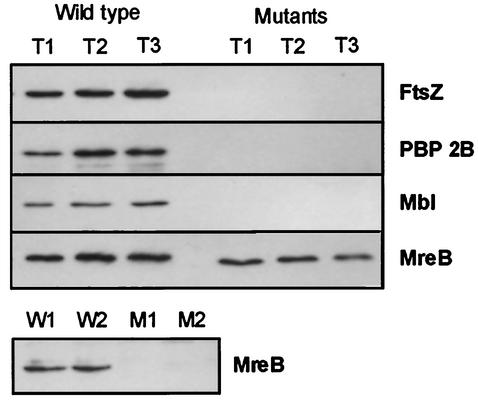
As indicated in the lower half of the top panel of Fig. 1, DivIVA-GFP appears to be present at the cell poles during the early stages of germination prior to Z-ring formation, suggesting that polar positioning of DivIVA does not depend on the presence of FtsZ. To corroborate this supposition, we isolated spores from B. subtilis strain 3291, which contains an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible allele of ftsZ. This strain was constructed by chromosomal transformation, and the genetic characteristics of this strain are described in Table 1. Spore isolation, extraction, and germination were performed as described above, except that 1 mM IPTG was present in the sporulation medium but absent from the germination medium. No FtsZ could be detected in germinating spores when analyzed by Western blotting using polyclonal anti-FtsZ serum (3) (Fig. 2). The subsequent germination stages displayed in Fig. 1 indicate that repression of FtsZ does not hamper the outgrowth of the spores, but it did completely block cell division. Again, DivIVA-GFP accumulated strongly at the cell poles, indicating that FtsZ is indeed not essential for polar disposition of DivIVA.
FIG. 2.
Western blot analyses of different germinating B. subtilis strains. Samples of wild-type (strain 1803) and mutant spores were taken 2 h (T1), 2.5 h (T2), and 3 h (T3) after inoculation. The antisera used are indicated in bold at the right of the panels. In the top panel of the four adjoining panels, germinating spores of strain 3291 (FtsZ−) were analyzed; in the upper-middle panel, germinating spores of strain 3294 (PBP 2B−) were analyzed; in the lower-middle panel, germinating spores of strain 3293 (Mbl−) were analyzed; and in the bottom panel of the four adjoining panels, germinating spores of strain 3292 (MreB−) were analyzed. The separate panel at the bottom of the figure shows the results of Western blot analyses of cell extracts (taken 2 h [W1 and M1] and 2.5 h [W2 and M2] after inoculation) from the wild-type strain (W) and strain 3292 (M) grown under MreB-depleted conditions (experiments performed in duplicate).
In addition to FtsZ, recruitment of DivIVA to the site of division requires other septal division proteins such as DivIB and DivIC (12). Depletion of PBP 2B, which catalyses the final stages of peptidoglycan synthesis in the septum, also prevents targeting of DivIVA to the site of division (H. B. Thomaides and J. Errington, unpublished data). The finding that targeting of PBP 2B to the cell division site depends on the formation of the Z ring, and the observation that some PBP 2B remains present at the newly formed cell poles, led Daniel et al. (4) to suggest that PBP 2B might be the immediate target for DivIVA. To test this hypothesis, we introduced an IPTG-inducible allele of pbpB into strain 1803 (creating strain 3294) (Table 1). Spores were isolated and germinated as described for strain 3291. Immunodetection using polyclonal anti-PBP 2B serum was used to check the absence of detectable amounts of PBP 2B in germinating spores from the mutant strain (Fig. 2). The findings presented in the lower-middle panel in Fig. 1 indicate that it is unlikely that PBP 2B is the immediate target for DivIVA, as DivIVA still accumulated at the extremities of outgrowing spores in aseptate filaments containing undetectable levels of PBP 2B. Therefore, polar targeting of DivIVA appears to be not directly dependent on the presence of either FtsZ or PBP 2B; the normal requirement for these proteins probably arises only through their role in formation of the new cell pole.
Localization of DivIVA in the absence of the cytoskeletal element Mbl or MreB.
Several mechanisms can be envisioned as explanations of how DivIVA accumulates at the cell poles. Passive diffusion combined with a polar capture mechanism is one possibility. Alternatively, DivIVA might migrate to the cell poles by an active mechanism, much like that operating in eukaryotes in which the cytoskeleton guides proteins to their proper locations in the cell. With the recent discovery of two new cytoskeletal elements in B. subtilis, Mbl and MreB, the possibility that an active mechanism is involved became more likely (9). Mbl forms a double-helix structure along the periphery of the cell, and the helical filaments run through almost the entire length of the cell. MreB forms helical filaments that encircle the cell at midcell. Mbl and MreB are similar in primary structure, and both show some homology with the actin superfamily (2, 9, 18). It is assumed that these two proteins form an actin-like cytoskeletal structure which determines the shape of the bacterium, since absence of one of the two results in a strong deformation in cell shape. Both proteins are conserved in rod-shaped bacteria, but they are missing in the cocci. How these cytoskeletal components control cell shape is unknown. By analogy with the function of actin in yeast, Jones et al. (9) postulated that the Mbl and MreB helical filaments control the spatial distribution of the cell-wall-synthesizing machinery. Potentially, these cytoskeletal elements might also control polar positioning of DivIVA.
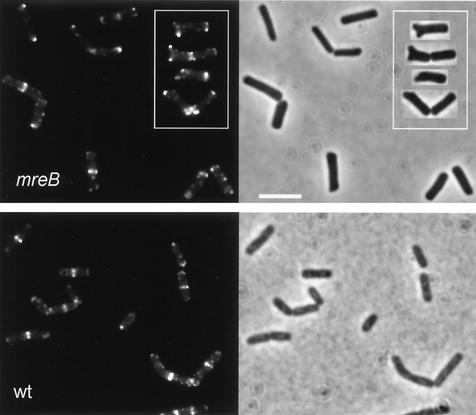
By means of chromosomal transformation, we constructed an mbl-knockout mutant containing a divIVA-gfp reporter fusion (strain 3293) (Table 1). The absence of Mbl from this strain was checked by Western blotting using polyclonal anti-Mbl serum (Fig. 2) (9). Strain 3293 is still able to sporulate (1), and spores were isolated and treated as described above. The results depicted in Fig. 1 indicate that spores from an mbl-knockout mutant germinate normally, and accumulation of DivIVA-GFP at the cell poles was not affected by the mutation. The characteristic cell deformation linked to an mbl mutation was not very evident in the germination medium we used. A proportion of cells started to show a bulging appearance (e.g., rightmost image in the bottom panel of Fig. 1) only after cell division had taken place. This distortion of cell shape did not appear to influence DivIVA-GFP localization. A comparable result was found when B. subtilis cells were depleted of MreB. Since MreB is an essential protein, we had to resort to the use of a B. subtilis strain containing mreB under transcriptional control of the repressible Pxyl promoter (strain 3292) (Table 1). However, Western blotting analysis using polyclonal anti-MreB serum indicated that repression of the Pxyl promoter was insufficient during the early stages of germination (Fig. 2). Therefore, the dependence of polar DivIVA positioning on the presence of MreB could not be studied in outgrowing spores, and we employed an MreB-depletion protocol essentially as described by Jones et al. (9). B. subtilis strain 3292 was grown overnight on a nutrient agar plate containing 1% xylose. A colony from this plate was used to inoculate Spizizen's minimal medium without xylose (8). To ensure that cells went through several rounds of cell division in the absence of MreB, the culture was diluted another 100-fold after reaching an OD600 of about 0.5. When the diluted culture reached an OD600 of about 1, cells were collected for Western blotting and fluorescence microscopy analyses. No MreB was detectable by immunodetection (Fig. 2), and many cells showed a deformed morphology, as shown in Fig. 3. Despite the absence of normal concentrations of MreB, DivIVA-GFP accumulated at the division septa and cell poles even when the poles were deformed. Thus, polar as well as septal positioning of DivIVA does not seem to depend on the presence of MreB or Mbl.
FIG. 3.
Localization of DivIVA-GFP under MreB-depletion conditions. The left upper panel (mreB) shows fluorescence images of B. subtilis strain 3292 cells grown in Spizizen's minimal medium without xylose. The inset shows some examples of cells with deformed poles. Scale bar in right half of panel, 5 μm. The left lower panel (wt) shows fluorescence images of wild-type B. subtilis (strain 1803) grown under the same conditions. Corresponding phase-contrast images are shown in the right halves of the panels.
In conclusion, we have shown that the formation of septa is not an essential step in the polar positioning of DivIVA. It is therefore unlikely that the positioning process can be simply subdivided into two steps, namely, binding of DivIVA to a component of the cell division septum followed by a mechanism that retains DivIVA at the newly formed poles. Our new results favor a mechanism in which DivIVA is directly recruited to the (newly formed) cell poles. Edwards et al. (6) showed that polar targeting of DivIVA is in fact a rather promiscuous process and that it can be observed in E. coli and even in the fission yeast Schizosaccharomyces pombe. The mechanism behind the apparent general affinity of DivIVA for cell poles remains unclear; at least, the cytoskeletal proteins MreB and Mbl appear not to be involved.
Acknowledgments
We thank the members of our group, in particular Richard Daniel, for helpful advice and discussions.
This research was supported by an EMBO long-term fellowship awarded to L. Hamoen and a grant from the U.K. Biotechnology and Biological Science Research Council.
REFERENCES
- 1.Abhayawardhane, Y., and G. C. Stewart. 1995. Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene. J. Bacteriol. 177:765-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel, R. A., and J. Errington. 2000. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol. Microbiol. 36:278-289. [DOI] [PubMed] [Google Scholar]
- 4.Daniel, R. A., E. J. Harry, and J. Errington. 2000. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 35:299-311. [DOI] [PubMed] [Google Scholar]
- 5.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905-915. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, D. H., H. B. Thomaides, and J. Errington. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 19:2719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hale, C. A., and P. A. de Boer. 2002. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. John Wiley & Sons Ltd, Chichester, West Sussex, United Kingdom.
- 9.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 10.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 11.Marston, A. L., and J. Errington. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33:84-96. [DOI] [PubMed] [Google Scholar]
- 12.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muchova, K., E. Kutejova, D. J. Scott, J. A. Brannigan, R. J. Lewis, A. J. Wilkinson, and I. Barak. 2002. Oligomerization of the Bacillus subtilis division protein DivIVA. Microbiology 148:807-813. [DOI] [PubMed] [Google Scholar]
- 14.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raskin, D. M., and P. A. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Setlow, B., K. A. McGinnis, K. Ragkousi, and P. Setlow. 2000. Effects of major spore-specific DNA binding proteins on Bacillus subtilis sporulation and spore properties. J. Bacteriol. 182:6906-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomaides, H. B., M. Freeman, M. El Karoui, and J. Errington. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 15:1662-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 19.Vary, J. C. 1973. Germination of Bacillus megaterium spores after various extraction procedures. J. Bacteriol. 116:797-802. [DOI] [PMC free article] [PubMed] [Google Scholar]