Abstract
Staphylococcus aureus produces a wide array of cell surface and extracellular proteins involved in virulence. Expression of these virulence factors is tightly controlled by numerous regulatory loci, including agr, sar, sigB, sae, and arl, as well as by a number of proteins with homology to SarA. Rot (repressor of toxins), a SarA homologue, was previously identified in a library of transposon-induced mutants created in an agr-negative strain by screening for restored protease and alpha-toxin. To date, all of the SarA homologues have been shown to act as global regulators of virulence genes. Therefore, we investigated the extent of transcriptional regulation of staphylococcal genes by Rot. We compared the transcriptional profile of a rot agr double mutant to that of its agr parental strain by using custom-made Affymetrix GeneChips. Our findings indicate that Rot is not only a repressor but a global regulator with both positive and negative effects on the expression of S. aureus genes. Our data also indicate that Rot and agr have opposing effects on select target genes. These results provide further insight into the role of Rot in the regulatory cascade of S. aureus virulence gene expression.
Staphylococcus aureus is an important human pathogen that causes a wide spectrum of diseases ranging from benign skin infections to life-threatening endocarditis and toxic shock syndrome (20, 32). The large array of virulence determinants produced by S. aureus is central to the pathogenesis of this organism. The role of these factors in pathogenesis has been demonstrated mainly by comparing the virulence of a mutant defective in one gene with that of its wild-type parent strain in specific animal models of infection. Recently, the use of signature-tagged mutagenesis (STM) identified several factors that play a role in pathogenesis, as their disruption made the mutants unable to survive within the host (10, 25). These virulence factors can be broadly subdivided into cell surface and secreted proteins. Cell surface proteins include microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), which enable the bacteria to colonize host tissues, and factors that facilitate the establishment of the infection by allowing the bacteria to evade the host immune system. Secreted proteins include various enzymes that degrade host components with antimicrobial activity, as well as different hemolysins that damage the host tissue (32).
Production of S. aureus virulence determinants is controlled by several global regulatory loci, which include agr (1), sar (5), sigB (3, 41), sae (14), arl (13), and six SarA homologues (2, 24). These regulators are parts of an important network modulating the expression of S. aureus virulence genes. One target virulence gene can be under the influence of several regulators that “cross talk” to ensure that the specific gene is expressed only when conditions are favorable. For instance, in vitro studies have demonstrated that agr negatively regulates the expression of spa, encoding protein A (32), whereas SarS binds to the spa promoter and activates its expression (38). Interestingly, agr downregulates sarS expression (6, 38). It has thus been proposed that agr downregulates spa expression by downregulating the expression of its activator, sarS (38). Therefore, virulence gene regulators could affect the expression of target genes directly, by binding to their promoters, or indirectly, via other regulators.
The specific role and/or importance of a virulence factor or a virulence gene regulator in S. aureus pathogenesis may vary from one infection type to another. For instance, fibronectin-binding proteins have been shown to mediate invasion of mammalian cells by S. aureus (36). In contrast, McElroy et al. have demonstrated that fibronectin-binding proteins decrease virulence in S. aureus-induced pneumonia (22). Similarly, the agr locus has been shown to be important in S. aureus pathogenesis in various animal infection models such as the mouse arthritis and abscess models (28, 29). However, a recent study by Yarwood et al. has shown that agr expression is not necessary for the development of toxic shock syndrome (40). Taken together, these observations illustrate the multifactorial aspect of S. aureus pathogenesis. In view of the fact that an increasing number of S. aureus strains are becoming more difficult to treat due to their resistance to multiple antibiotics (39), elucidation of S. aureus pathogenesis at the molecular level is crucial in the battle against this important human pathogen, as it may help in the design of new therapeutic strategies.
Among the regulators described above, SarA and agr are the best characterized. SarA is a transcriptional regulator that binds to a consensus motif in the promoter of its target genes (8). Recent studies have shown that SarA is a member of a large family of transcriptional regulators that share a common amino acid motif, KXRXXXDER (2). The agr system is self-activating in a cell density-dependent manner (29). Expression of the agr effector molecule, RNAIII, correlates with downregulation of select genes encoding cell surface proteins, such as protein A and fibronectin-binding proteins, and upregulation of genes encoding secreted proteins, such as alpha- and beta-toxins, lipases, and proteases (32). In vitro, RNAIII-regulated factors follow a temporal trend, with surface proteins being produced during the early-exponential phase of growth and secreted proteins synthesized predominantly during the post-exponential phase. The sequential expression of these determinants correlates with the production of RNAIII, whose levels peak during the post-exponential phase (20, 32). The mechanism by which RNAIII regulates the expression of these different genes is not completely understood. Several studies have suggested the existence of cofactors that interact directly or indirectly with RNAIII to control the expression of the different target genes (18, 24, 30). These factors include members of the Sar family of transcriptional regulators (2).
Recently, a novel SarA homologue, Rot, was identified as a repressor of toxins based on the following observation. An agr-negative strain produces low levels of alpha-toxin and protease activity. However, in a rot agr double mutant, expression of these proteins is increased due to the lack of repression by Rot. Understanding how Rot regulates these and other virulence factors is important in order to further discern S. aureus virulence gene regulation. Most S. aureus virulence gene modulators have both negative and positive effects on their target genes (8, 32, 38). For these reasons, we hypothesized that Rot may have both up- and downregulatory functions. To identify additional target genes regulated by Rot, we compared the transcriptional profile of a rot agr double mutant to that of its parental agr strain during the late-exponential phase of growth by use of the Affymetrix GeneChip. The GeneChip used in this study covers 86% of the S. aureus COL genome and contains approximately 4,600 DNA fragments, representing 2,339 individual S. aureus genes (12).
In this study, we demonstrate that Rot is a global regulator that negatively regulates the transcription of several known virulence genes. Some of these genes encode secreted proteins such as lipase, hemolysins, and proteases, all of which are postulated to play a role in tissue invasion. Rot also positively regulates the expression of a number of genes including those encoding cell surface adhesins. In addition, Rot modulates the expression of genes not previously implicated in pathogenesis. A relationship between Rot and agr in the regulation of S. aureus virulence genes is proposed.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All S. aureus strains used in this study are listed in Table 1. Bacteria were grown in 0.3 GL agar and CY broth, supplemented with β-glycerophosphate (GP), at 37°C (27). Bacterial cell cultures were grown with rotary agitation at 200 rpm. All mutants were maintained on erythromycin or tetracycline at a concentration of 5 μg ml−1.
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| RN6390 | agr+ laboratory strain | 29 |
| RN6911 | agrΔ::tetM (Tcr) | 29 |
| KT201 | sarH1::pKT200 (Emr) | 38 |
| PM614 | agr-null chr::Tn917::rot (Tcr Emr) | 24 |
| BK9986 | tetM::agr Tn917::rot (Tcr Emr) | This study |
| BK9987 | tetM::agr sarH1::pKT200 (Tcr Emr) | This study |
Construction of rot agr and sarH1 (sarS) agr mutants.
The rot agr double mutant (BK9986) was constructed by bacteriophage φ11-mediated transduction of the rot::Tn917 insertion from strain PM614 into the agr mutant strain RN6911. The sarH1 agr double mutant (BK9987) was made by transduction of the agr::tmn mutation from RN6911 into KT201 (sarH1 mutant). Transfer of the rot and agr mutations into strains BK9986 and B9987 was verified by Southern blot analysis (data not shown).
RNA isolation and Northern blot analysis.
Overnight cultures of S. aureus were diluted 1:50 in CY-GP medium and grown to the late-exponential (5 h) phase of growth. Since our aim was to determine genes whose expression is affected by Rot at any time point, we chose the late-exponential-growth phase because at this time point, we can still observe transcripts of genes expressed early, such as cell surface protein genes, as well as those expressed later, such as genes encoding secreted enzymes and toxins. Cells were harvested by centrifugation, and RNA was isolated by the FastPrep system (developed by Savant [Farmingdale, N.Y.] and Bio 101 [Vista, Calif.]) (4). RNA samples (10 μg) were electrophoresed through a 1.5% agarose-0.66 M formaldehyde gel in morpholinepropanesulfonic acid (MOPS) running buffer. RNA was blotted onto a Hybond N+ membrane (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) by using the VacuGene XL blotting system (Amersham Pharmacia). The transfer was performed with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate),, pH 7.0, for 2 h. After the RNA transfer, the RNA ladder (Invitrogen, Carlsbad, Calif.) was manually marked before the hybridization step. Membranes were hybridized overnight with a PCR-amplified probe derived from the target gene (Table 2). Specific transcripts were detected by using the enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia). In all the Northern blot experiments, the same RNA samples were hybridized with 16S rRNA as an internal control (38). New membranes were used for each Northern blot experiment.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) | GenBank accession no. | Nucleotide positions |
|---|---|---|---|
| hlbF | GGTGCACTTACTGAC | X13404 | 824-838 |
| hlbR | CGCATATACATCCCATGGC | 1678-1660 | |
| 16S rRNAF | GCGTGGGGATCAAACAGG | X68417 | 777-794 |
| 16S rRNAR | CCCCAATCATTTGTCCCACC | 1500-1481 | |
| splAF | CGAAGGAGGTAAAATATGAATAAAAATG | AF271715 | 987-1013 |
| splAR | GTGGTGTGAAATAAACACCGAAATTC | 1670-1645 | |
| sspAF | CAACGAATGGTCATTATGCACCC | AF309515 | 598-620 |
| sspAR | GGTACACCGCCCCAATGAATTCC | 1123-1101 | |
| gehF | GCACAAGCCTCGG | M12715 | 808-820 |
| gehR | GACGGGGGTGTAG | 1280-1268 | |
| ureCF | CTCGTGTAACACGTGATGACGTGAACG | AP003136 | 251461-251487 |
| ureCR | GGCGCATGACCGCCACCAGCACCTTC | 252139-252114 | |
| spaF | AGACGATCCTTCGGTGAGCAAAGA | U54636 | 829-852 |
| spaR | GCTTTTGCAATGTCATTTACTGTAT | 1203-1179 | |
| sarSF | GTCAAGCCTGAAGTCGATATG | AB035454 | 911-931 |
| sarSR | GCATGGTCTTGCTGCGCGTC | 1518-1499 | |
| rotF | GTTTTGGGATTGTTGGGATG | AF189239 | 478-497 |
| rotR | GCATTGCTGTTGCTCTACTTGC | 924-903 |
RNA labeling.
The integrity of RNA preparations was analyzed by electrophoresis in 1.2% agarose-0.66 M formaldehyde gels. Samples with defined 16S and 23S rRNA bands were subjected to a series of reactions that resulted in 5′ biotin-labeled fragmented RNA for GeneChip analysis, as previously described (12). Briefly, total bacterial RNA was first enriched for mRNA. This was accomplished by reverse transcription with rRNA-specific primers, followed by RNase H digestion to remove the rRNA of RNA-cDNA hybrid molecules and DNase I treatment. Enriched mRNA was then chemically fragmented and 5′-thiolated by T4 polynucleotide kinase treatment in the presence of γ-S-ATP. Samples were then biotinylated by addition of polyethylene oxide (PEO)-iodoacetyl-biotin.
GeneChip analysis.
The custom-made S. aureus GeneChips used in this study have been described previously (12). Each chip contains 4,528 open reading frames (ORFs), 12 tRNAs, and 3 rRNAs, with an average of 25 probe sets per ORF. Due to the preliminary state of the genome sequence at the time of design, many genes are represented on the chip as partial, duplicate, or overlapping fragments. Subsequent analyses based on the updated S. aureus genomic sequences suggest that these 4,528 tilings represent approximately 2,700 to 2,900 individual genes and that >86% of the S. aureus COL genome is represented on the chip. The missing genes (14%) include those encoding staphylococcal accessory regulator A (sarA), the sensor histidine kinase SaeS (saeS), d-alanine-d-alanyl carrier protein ligase (dltA), serine protease (sspA), leukotoxin D (lukD), sigma factor B regulators (rsbU, rsbV, and rsbW), and staphylococcal accessory regulator R (sarR).
GeneChip analysis was performed by the method described by Dunman and colleagues (12). Accordingly, to address the issue of reproducibility, two separate RNA samples from each strain were prepared from two different experiments, and for each sample, 1.5 μg of biotinylated RNA was hybridized to at least two separate GeneChips for 14 h at 45°C. GeneChips were then washed and subjected to a series of staining procedures that included the successive binding of streptavidin, biotinylated anti-streptavidin, and phycoerythrin-conjugated streptavidin. Each GeneChip was washed and scanned at a 570-nm wavelength and a 3-μm resolution in an Affymetrix GeneChip scanner. Affymetrix Microarray Suite 4.0 algorithms calculated signal intensities (average difference) and present or absent determinations for each ORF (19). GeneChips were then normalized, and background was defined by using GeneSpring 4.0 (Silicon Genetics) as previously described (12). GeneSpring software was used to further analyze the transcription patterns of genes. Genes negatively regulated by Rot were identified as ORFs with transcript titers at least twofold higher in BK9986 (rot negative) than in RN6911 (rot positive). Genes whose transcript levels were at least twofold higher in RN6911 (rot positive) than in BK9986 (rot negative) were categorized as being positively regulated by Rot. These lists were further filtered to include only genes with signal intensities above background level in the rot agr (rot-negative) strain for Rot-downregulated genes and in the agr (rot-positive) strain for Rot-upregulated genes. Genes represented more than once on the chip are represented only once on the list.
Urease activity.
Urease production was assayed on urea agar slants (Remel, Lenexa, Kans.) as described by the manufacturer. In this test, urease-positive strains show a color change from orange to pink while urease-negative strains have an intact orange color.
RESULTS
Rot is a transcriptional modulator.
GeneChip results indicate that Rot behaves as both a positive and a negative modulator of numerous virulence genes. We have identified 60 genes whose expression is negatively regulated by Rot (Table 3) and 86 genes whose expression is positively regulated by Rot (Table 4). From both categories of regulated genes, we chose seven genes with a well-known or postulated virulence function for further validation (Table 5).
TABLE 3.
Genes downregulated by Rot
| Chip ORF no.a | N315 ORF no.b | N315 geneb | N315 descriptionb | Fold changec | agr or sar effectd | Role categoryd |
|---|---|---|---|---|---|---|
| 4877 | SA1811 | hlb | Truncated beta-hemolysin | 6.9 | ||
| 4218 | SA0143 | adhE | Alcohol-acetaldehyde dehydrogenase | 3.8 | ||
| 1726 | SA0471 | cysK | Cysteine synthase (o-acetylserine sulfhydrylase) homologue | 4.6 | ||
| 1664 | SA2312 | ddh | D-specific d-2-hydroxyacid dehydrogenase | 11.5 | agr, down | Carbohydrate metabolism |
| 904 | SA1267 | ebhA | HP, similar to streptococcal adhesin Emb | 567.4 | ||
| 1885 | SA1268 | ebhB | HP, similar to streptococcal adhesin Emb | Upregulated | agr, up | Virulence factors |
| 1434 | SA1091 | fmhC (eprh) | FmhC protein | Upregulated | ||
| 1527 | SA0309 | geh | Glycerol ester hydrolase | 8.4 | agr, up | Virulence factors |
| 4046 | SA2294 | gntK | Gluconokinase | 3.1 | ||
| 5281 | SA2293 | gntP | Gluconate permease | 4.6 | sar, down | Transport |
| 1928 | SA2209 | hlgB | Gamma-hemolysin component B | 31.9 | agr, up | Virulence factors |
| 1927 | SA2208 | hlgC | Gamma-hemolysin component C | 19.6 | agr, up | Virulence factors |
| 1700 | SA1881 | kdpA | Probable potassium-transporting ATPase A chain | 2.8 | ||
| 4715 | SA1879 | kdpC | Probable potassium-transporting ATPase C chain | 3.7 | ||
| 4583 | SA1090 | lytN | LytN protein | Upregulated | ||
| 5046 | SA0549 | mvaK2 | Phosphomevalonate kinase | Upregulated | ||
| 2268 | SA2334 | mvaS | 3-Hydroxy-3-methylglutaryl CoA synthase | 8.1 | ||
| 3524 | SA2185 | narG | Respiratory nitrate reductase alpha chain | 6.2 | agr, down | Electron transport |
| 431 | SA2435 | pmi | Mannose-6-phosphate isomerase | 3.0 | ||
| 4537 | SA1659 | prsA | Peptidyl-prolyl cis/trans isomerase homolog | 6.5 | ||
| 3551 | SA2326 | ptsG | PTS system, glucose-specific IIABC component | 2.7 | ||
| 1071 | SA1589 | ribD | Riboflavin-specific deaminase | Upregulated | ||
| 5329 | SA1631 | splA | Serine protease SplA (pathogenicity island SaPIn3) | Upregulated | agr, up | Virulence factors |
| 2929 | SA1630 | splB | Serine protease SplB (pathogenicity island SaPIn3) | Upregulated | agr, up | Virulence factors |
| 3742 | SA1629 | splC | Serine protease SplC (pathogenicity island SaPIn3) | 62.5 | ||
| 324 | SA1628 | splD | Serine protease SplD (pathogenicity island SaPIn3) | Upregulated | agr, up | Virulence factors |
| 325 | splE | Serine protease SplE | 133.4 | |||
| 2377 | SA1627 | splF | Serine protease SplF (pathogenicity island SaPIn3) | Upregulated | agr, up | Virulence factors |
| 2174 | SA0900 | sspB | Cysteine protease precursor | 10.4 | agr, up | Virulence factors |
| 2175 | SA0899 | sspC | Cysteine protease | 5.2 | agr, up | Virulence factors |
| 4823 | SA2082 | ureA | Urease gamma subunit | Upregulated | ||
| 4822 | SA2083 | ureB | Urease beta subunit | Upregulated | ||
| 1900 | SA2084 | ureC | Urease alpha subunit | Upregulated | ||
| 4817 | SA2088 | ureD | Urease accessory protein UreD | 2.9 | ||
| 1899 | SA2085 | ureE | Urease accessory protein UreE | 6.7 | ||
| 1898 | SA2086 | ureF | Urease accessory protein UreF | 6.0 | ||
| 1897 | SA2087 | ureG | Urease accessory protein UreG | 23.7 | ||
| 401 | SA0124 | HP, similar to glycosyltransferase TuaA | 5.6 | |||
| 657 | SA2455 | Capsular polysaccharide biosynthesis, CapC | 11.1 | |||
| 849 | SA2414 | HP, similar to glutathione peroxidase | 10.8 | |||
| 1386 | SA0368 | HP, similar to proton/sodium-glutamate symport protein | 6.4 | agr, up | Transport | |
| 1574 | SA2062 | HP | 3.5 | |||
| 1815 | SA2200 | HP, similar to ABC transporter, ATP binding subunit | 3.0 | |||
| 1816 | SA2201 | HP, similar to ABC transporter, permease protein | 3.2 | |||
| 1994 | SA1812 | HP, similar to synergohymenotropic toxin precursor of Staphylococcus intermedius | 3.1 | |||
| 2064 | SA2218 | Reverse complement of HP | 4.4 | |||
| 2067 | SA1437 | Conserved HP | 3.6 | |||
| 2238 | SA2434 | Fructose phosphotransferase system enzyme FruA homolog | 5.4 | |||
| 2543 | SA1725 | Staphopain, cysteine proteinase | 11.9 | |||
| 2612 | SA1093 | DNA topoisomerase I TopA homolog | Upregulated | |||
| 2860 | SA0989 | Conserved HP | 7.2 | |||
| 2926 | SA0663 | HP | 3.0 | |||
| 3017 | SA0127 | HP, similar to capsular polysaccharide synthesis protein 14L | 50.7 | |||
| 3078 | SA0165 | HP, similar to alpha-helical coiled-coil protein SrpF | 2.7 | |||
| 3373 | SA0904 | HP, probable autolysin transcription regulator | Upregulated | |||
| 3768 | SA0850 | HP, similar to oligopeptide ABC transporter oligopeptide- binding protein | Upregulated | |||
| 4061 | SA1007 | Alpha-hemolysin precursor | 24.5 | agr, up | Virulence factors | |
| 4069 | SA0123 | HP, similar to UDP-glucose 4-epimerase (Gale-1) | Upregulated | |||
| 4530 | SA0710 | Conserved HP | Upregulated | |||
| 5131 | SA2297 | HP, similar to GTP-pyrophosphokinase | 2.8 |
S. aureus GeneChip ORF number.
Based on the published sequence of strain N315 (accession no. NC_002745). For genes not present in N315, the gene name and description are from the COL genome, available from The Institute for Genomic Research Comprehensive Microbial Resource website (http://www.tigr.org). HP, hypothetical protein; CoA, coenzyme A; PTS, phosphotransferase system.
Normalized values in the agr rot mutant over values in the agr strain. “Upregulated” denotes genes highly downregulated in the agr strain, such that the transcripts were below detectable levels and the fold change could not be accurately calculated.
As described by Dunman et al. (12). up, upregulated; down, downregulated.
TABLE 4.
Genes upregulated by Rot
| Chip ORF no.a | N315 ORF no.b | N315 geneb | N315 descriptionb | Fold changec | agr or sar effectd | Role categoryd |
|---|---|---|---|---|---|---|
| 3994 | SA1736 | aldH | Aldehyde dehydrogenase | 2.1 | ||
| 5372 | clfB | Clumping factor B | 2.9 | |||
| 2462 | SA2336 | clpL | ATP-dependent Clp proteinase chain ClpL | 2.7 | sar, down | Adaptation |
| 3833 | coa | Coagulase | 4.3 | |||
| 2163 | SA1253 | ctpA | Probable carboxy-terminal processing proteinase CtpA | 2.9 | ||
| 5110 | SA1164 | dhoM | Homoserine dehydrogenase | 2.3 | ||
| 4873 | SA0796 | dltD | Poly(glycerophosphate chain) d-alanine transfer protein | 2.0 | agr, down | Transport |
| 3383 | epiA | Lantibiotic gallidermin precursor EpiA | 2.6 | |||
| 84 | SA0602 | fhuA | Ferrichrome transport ATP-binding protein | 2.1 | ||
| 4551 | SA2172 | gltT | Proton/sodium-glutamate symport protein | 2.2 | ||
| 4042 | SA2288 | gtaB | UTP-glucose-1-phosphate uridyltransferase | 2.4 | ||
| 3984 | SA0189 | hsdR | Probable type I restriction enzyme restriction chain | 2.4 | ||
| 2883 | SA1505 | lysP | Lysine-specific permease | 2.4 | agr, up | Transport |
| 4486 | SA0250 | lytS | Two-component sensor histidine kinase | 2.3 | ||
| 1047 | SA1194 | msrA | Peptide methionine sulfoxide reductase homolog | 2.4 | ||
| 854 | SA2410 | nrdD | Anaerobic ribonucleoside-triphosphate reductase | 2.1 | ||
| 4205 | SA0685 | nrdI | NrdI protein involved in ribonucleotide reductase function | 2.9 | ||
| 4390 | SA0374 | pbuX | Xanthine permease | 2.3 | ||
| 4905 | SA0923 | purM | Phosphoribosylformylglycinamidine cyclo-ligase PurM | 2.2 | sar, down | Nucleotide and nucleic acid metabolism |
| 2800 | SA0676 | recQ | Probable DNA helicase | 2.8 | agr, up | DNA replication |
| 1079 | SA1583 | rot | Repressor of toxins Rot | 12.0 | ||
| 2343 | sdrC | SdrC protein | 2.7 | sar, up | Virulence factors | |
| 2119 | SA0107 | spa | Immunoglobulin G binding protein A precursor | 15.6 | agr, down | Virulence factors |
| 5112 | SA1166 | thrB | Homoserine kinase homolog | 3.9 | ||
| 3239 | SA1165 | thrC | Threonine synthase | 3.8 | ||
| 5075 | SA0373 | xprT | Xanthine phosphoribosyltransferase | 3.0 | ||
| 17 | HP | 2.0 | ||||
| 18 | SA0220 | HP, similar to glycerophosphodiester phosphodiesterase | 2.9 | |||
| 237 | SAS013 | Reverse complement of HP (pathogenicity island SaPIn2) | 2.3 | agr, up | Unknown | |
| 392 | SA0739 | Conserved HP | 2.4 | |||
| 442 | SA0651 | HP | 2.2 | |||
| 457 | SA1613 | Conserved HP | 2.5 | |||
| 477 | SA0428 | Conserved HP | 2.5 | |||
| 493 | SA2378 | Conserved HP | 3.2 | agr, down | Unknown | |
| 534 | SA1717 | Glutamyl-tRNAGln amidotransferase subunit C | 2.6 | |||
| 600 | SA0523 | HP, similar to poly (GP) alpha-glucosyltransferase (TA biosynthesis) | 2.2 | sar, up | Cell wall | |
| 616 | SA2133 | Conserved HP | 2.8 | |||
| 678 | SA0682 | HP, similar to di-tripeptide ABC transporter | 2.1 | |||
| 748 | SA2261 | HP, similar to efflux pump | 3.0 | |||
| 873 | SAS088 | HP | 2.2 | |||
| 1027 | SA2007 | HP, similar to alpha-acetolactate decarboxylase | 2.1 | |||
| 1082 | SA0620 | Secretory antigen SsaA homologue | 2.0 | |||
| 1175 | SA2284 | HP, similar to accumulation-associated protein | 2.5 | agr, up | Virulence factors | |
| 1353 | HP | 2.7 | ||||
| 1356 | SA2170 | HP, similar to general stress protein 26 | 2.1 | |||
| 1357 | SA2171 | HP | 2.0 | |||
| 1398 | SA0914 | Reverse complement of HP, similar to chitinase B | 3.0 | agr, up | Miscellaneous | |
| 1563 | SA0407 | Conserved HP | 2.0 | |||
| 1564 | SA0408 | HP | 2.3 | |||
| 1592 | HP | 3.0 | ||||
| 1597 | HP | 2.0 | ||||
| 1666 | SA2303 | HP, similar to membrane-spanning protein | 3.2 | |||
| 1765 | Epidermin immunity protein F | 4.7 | agr, up | Antibiotic production | ||
| 1991 | SA0439 | HP, similar to lysine decarboxylase | 2.5 | |||
| 2032 | SA1059 | Methionyl-tRNA formyltransferase | 2.4 | agr, up | Aminoacyl tRNA synthetases | |
| 2076 | 2.4 | |||||
| 2077 | SA0291 | HP | 3.1 | |||
| 2078 | SA0292 | HP | 2.5 | |||
| 2103 | SA0271 | Conserved HP | 6.3 | agr, up | Unknown | |
| 2132 | SA2131 | Conserved HP | 4.9 | |||
| 2133 | SA2132 | HP, similar to ABC transporter (ATP-binding protein) | 2.4 | |||
| 2232 | SA2440 | HP | 2.0 | |||
| 2255 | HP | 2.1 | ||||
| 2337 | SA2001 | HP, similar to oxidoreductase, aldo/ketoreductase family | 2.0 | |||
| 2454 | SA1320 | HP | 4.4 | |||
| 2507 | SA0380 | Conserved HP (pathogenicity island SaPIn2) | 3.5 | |||
| 2697 | SA2442 | Preprotein translocase SecA homolog | 2.2 | |||
| 3259 | SA0675 | HP, similar to ABC transporter ATP-binding protein | 3.0 | |||
| 3313 | SA2436 | HP, similar to phage infection protein | 2.1 | sar, up | Bacteriophage related | |
| 3391 | Conserved HP | 2.0 | ||||
| 3426 | SA1924 | HP, similar to aldehyde dehydrogenase | 2.5 | |||
| 3599 | ABC transporter, ATP-binding protein | 2.6 | ||||
| 3698 | SA0173 | HP, similar to surfactin synthetase | 2.9 | agr, up | Antibiotic production | |
| 3888 | SA2339 | HP, similar to antibiotic transport-associated protein | 2.1 | |||
| 4153 | SA0310 | HP | 2.1 | |||
| 4253 | SA0003 | Conserved HP | 2.2 | |||
| 4282 | SA1942 | Conserved HP | 2.5 | |||
| 4525 | SA1016 | Reverse complement of conserved HP | 2.4 | |||
| 4550 | HP | 2.3 | ||||
| 4667 | SA0973 | Phosphopantetheine adenyltransferase homolog | 3.2 | |||
| 4752 | SA1056 | HP | 3.5 | |||
| 5065 | SA2256 | Conserved HP | 15.8 | |||
| 5245 | SA0827 | HP, similar to ATP-dependent nuclease subunit B | 2.3 | |||
| 5437 | SA0246 | Hypotheticl protein, similar to d-xylulose reductase | 2.1 | |||
| 5471 | SA1131 | HP, similar to 2-oxoacid ferredoxin oxidoreductase, alpha subunit | 2.3 | |||
| 5554 | SA1679 | HP, similar to d-3-phosphoglycerate dehydrogenase | 2.0 |
S. aureus GeneChip ORF number.
Based on the published sequence of strain N315 (accession no. NC_002745). For genes not present in N315, the gene name and description are from the COL genome, available from The Institute for Genomic Research Comprehensive Microbial Resource website (NC_002745). For genes not present in N315, the gene name and description are from the COL genome, available from The Institute for Genomic Research Comprehensive Microbial Resource website (http://www.tigr.org). HP, hypothetical protein; TA, teichoic acid.
Normalized values in the agr strain over values in the agr rot double mutant.
As described by Dunman et al. (12). up, upregulated; down, downregulated.
TABLE 5.
Classification of Rot-regulated genes
| Gene/gene producta | Function | Effectb of:
|
|
|---|---|---|---|
| Rot | agr | ||
| Secreted enzymes | |||
| geh/lipasec | Degrades lipids | − | + |
| splA through splF/serine protease operonc | Degrades proteins | − | + |
| sspB, sspC/within serine protease operonc | Degrades proteins | − | + |
| Secreted toxins | |||
| hla/alpha-toxin | Disrupts red blood cells | − | + |
| hlb/β-hemolysinc | Sphingomyelinase activity | − | + |
| Cell surface proteins | |||
| clfB/clumping factor | Binds fibrinogen | + | NA |
| sdrC | Putative cell surface adhesin | + | ND |
| spa/protein A | Binds Fc portion of immuno- globulin | + | − |
| Potential virulence factors | |||
| thrB | Threonine synthase | + | ND |
| ureA through ureGc,d | Urease complex | − | + |
| Regulator, sarS | Virulence gene regulator | + | − |
Genes are classified based on their known or postulated virulence function.
+, activation; −, repression; NA, not affected; ND, not determined.
Validated by Northern blot analysis.
Validated by protein activity.
Virulence genes negatively regulated by Rot.
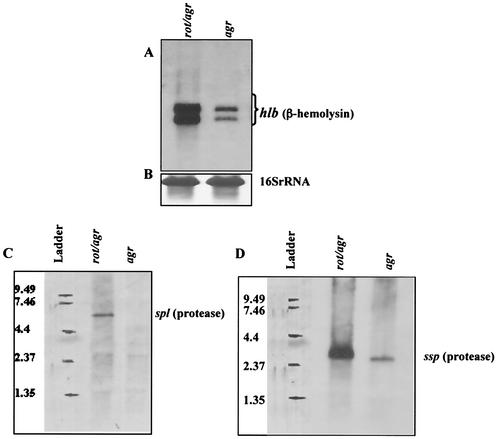
Initial characterization of Rot has indicated that it is a repressor of hla, encoding alpha-toxin (24). Our GeneChip data showed that Rot negatively regulates another toxin gene, hlb, which encodes beta-toxin (Tables 3 and 5). The GeneChip result for hlb was confirmed by Northern blot analysis using the rot agr double mutant (rot negative) and its parental agr strain (rot positive). As shown in Fig. 1A, the hlb transcript is present at higher levels in the rot agr strain than in the agr mutant. The detection of two bands that hybridize with hlb is consistent with previous findings (7, 41). The 16S rRNA transcript (internal control) is present at similar levels in the two strains (Fig. 1B).
FIG. 1.
Northern blot analysis of hlb, spl, and ssp. RNA was isolated from BK9986 (rot agr) and RN6911 (agr) at the late-exponential-growth phase and hybridized with hlb (A), splA (C), or sspA (D). Transcription of 16S rRNA was used as an internal control (B).
Rot was previously shown to have a phenotypic effect on protease production. (24). Unlike alpha-toxin and beta-toxin, which are solely encoded by hla and hlb, respectively, staphylococcal protease activity is associated with the gene products of at least two operons, splABCDEF (33) and sspABC (34). Our GeneChip results demonstrated that Rot negatively regulates both operons (Tables 3 and 5). The sspA gene, encoding V8 protease (34), is not listed in Table 3 because it is not represented on the GeneChip used in this study.
The GeneChip data for spl were confirmed by Northern blot analysis, using splA as a probe. The transcript of the spl operon (5.5 kb) is detectable only in the rot agr strain, BK9986, not in the agr strain, RN6911 (Fig. 1C). We tested ssp operon expression in the two strains by Northern blot analysis, using sspA as a probe, and found that the transcript levels of the ssp operon (2.8 kb) were higher in the rot agr double mutant than in the agr strain (Fig. 1D).
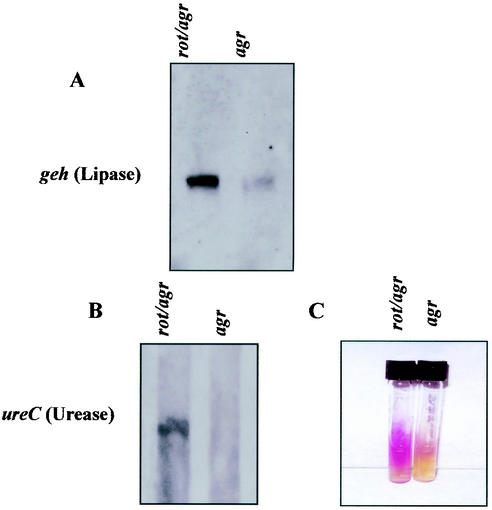
Rot also negatively regulates the expression of geh, encoding lipase (Tables 3 and 5). The negative regulation of geh expression by Rot was confirmed by Northern blot analysis in which RNA samples from the rot agr double mutant and the agr strain taken at late-exponential growth were hybridized with a geh-specific probe. The results demonstrated that geh mRNA is present at higher levels in the rot agr strain than in the agr strain (Fig. 2A).
FIG. 2.
Expression analysis of geh (A) and ureC (B and C). (A) RNA samples from BK9986 (rot agr) and RN6911 (agr) were taken during the late-exponential phase of growth and hybridized with a geh-specific probe. (B) ureC transcripts in BK9986 (rot agr) and its parental agr strain (RN6911) were analyzed. (C) Urease activity in both strains was tested on urea agar slants.
As shown in Table 3, GeneChip data indicated that Rot negatively regulates various urease genes such as ureA, ureB, and ureC, encoding different subunits of the urease enzyme, as well as accessory genes such as ureD and ureE, required for the enzyme activity (17) (GenBank accession number AP003136). We performed Northern blot analysis using the ureC gene as a probe and found that the rot agr strain had higher levels of ureC transcripts than the agr strain (Fig. 2B). One of the urease genes downregulated by Rot, ureD, was previously identified as a potential virulence gene, because a ureD transposon mutant was strongly attenuated in an animal model (10). We also tested for urease activity on urea agar slants for the two strains. As shown in Fig. 2C, the rot agr strain was urease positive but the agr strain was urease negative.
Virulence genes upregulated by Rot.
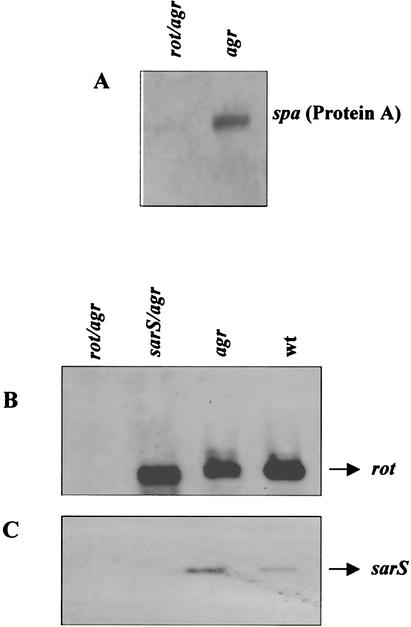
Although Rot was originally identified as being a repressor of certain virulence factors, our results indicate that Rot also acts as a positive modulator of staphylococcal virulence genes. Among the genes upregulated in the presence of Rot are those encoding cell surface proteins. One of these genes is the well-studied spa gene, which encodes protein A (Tables 3 and 5). To validate this result, Northern blot analysis was performed on rot agr and agr strains, and the results demonstrated that spa mRNA is detected only in the agr (rot-positive) strain, not in the rot agr double mutant (rot negative) (Fig. 3A).
FIG. 3.
Northern blot analysis of spa (A), rot (B), and sarS (C). (A) Analysis for the spa transcript in BK9986 (rot agr) and RN6911 (agr) at the late-exponential phase of growth. (B and C) RNA samples from BK9986 (rot agr), BK9987 (sarS agr), RN6911 (agr), and RN6390 (wild-type [wt] strain) at the late-exponential phase of growth were hybridized with rot- and sarS-specific probes, respectively.
Previous studies have demonstrated that SarH1, also called SarS, positively regulates the expression of the protein A gene, spa (6, 38). Since our results indicate that spa is also positively regulated by Rot, we sought to determine if Rot positively regulates spa expression by positively regulating the expression of the spa activator SarS. Using Genespring software analysis, we found that the sarS transcript was considered present in the rot-positive strain (RN6911) but absent in the rot mutant (BK9986), suggesting that Rot may positively regulate sarS expression.
To validate the sarS result, and to analyze the effects of Rot and SarS on each other, we created a sarH1 agr double mutant (BK9987) and compared the transcription of both rot and sarS in the two strains, i.e., BK9986 (rot agr) and BK9987 (sarS agr), as well as in RN6911 (agr) and RN6390 (wild-type strain), in a Northern blot assay. We found that rot is expressed in all the strains tested except the rot mutant, as expected (Fig. 3B). On the other hand, sarS is expressed only in the presence of Rot and in the absence of RNAIII (Fig. 3C). The fact that the sarS transcript is detectable in RN6911 (agr) but not in BK9986 (rot agr) indicates that Rot is indeed required for sarS expression. The higher levels of sarS transcript in RN6911 compared to RN6390 are in good agreement with previous data that showed that agr represses sarS expression (6, 38).
Rot and agr have opposing effects on the expression of target genes.
Our results indicate that Rot-regulated factors follow a general expression pattern opposite that of agr, that is, secreted proteins generally upregulated by agr are negatively regulated by Rot (Table 5). For example, Rot negatively regulates hla and hlb while agr positively regulates them (32). Since Rot negatively regulates the expression and activity of urease, we tested to see if agr will upregulate them. For this, we tested the urease activity of an agr mutant (RN6911) and its parental wild-type strain (RN6390) on urea agar slants and found that indeed the agr-positive strain was urease positive whereas the agr-negative strain was urease negative (data not shown). Conversely, cell surface proteins generally downregulated by agr appear to be positively modulated by Rot (Table 5). For instance, Rot positively regulates spa, encoding protein A, whereas agr negatively regulates it. In addition to structural genes, Rot and agr also have opposing effects on the expression of the virulence gene regulator sarS. Our results indicate that Rot positively regulated the expression of sarS, which has been shown to be repressed by agr (6, 38).
Additional Rot-regulated genes.
Table 3 lists additional virulence genes negatively regulated by Rot as determined by GeneChip analysis, and Table 4 lists all genes shown to be positively modulated by Rot. These tables include genes which encode factors previously shown to promote the growth and survival of S. aureus in an infection model. For instance, Rot downregulates hlgB and hlgC, which encode gamma-hemolysins B and C, respectively. Gamma-hemolysin is an S. aureus virulence factor that has been shown to play a role in S. aureus endophthalmitis and corneal pathogenesis (11, 37). Rot positively regulates thrB, a gene involved in threonine biosynthesis. Disruption of this gene reduces virulence in animal infection models (25). From both lists, it is clear that in addition to virulence genes, Rot also regulates the expression of genes that play different biological roles, including amino acid and carbohydrate metabolism, cell wall biosynthesis, and transport.
DISCUSSION
S. aureus is a versatile human pathogen capable of causing diverse infections in different sites of the body. This versatility can be attributed to its impressive arsenal of virulence factors (20). It is now apparent that S. aureus has numerous regulators that, alone and in combination, control the expression of these factors. Although the list of these virulence factors is quite substantial, it is believed to be incomplete and limited to determinants that are easily detectable by available assays. The recent advent of genomewide expression profiling techniques such as DNA arrays and proteomics provides an unprecedented ability to identify genes regulated in a coordinated manner, thus increasing our understanding of biological processes. In a recent study, Dunman and colleagues used DNA GeneChip technology to demonstrate that the two well-studied virulence gene regulators agr and SarA regulate the expression of known virulence genes as well as novel genes not previously known to play a role in S. aureus pathogenesis (12). Likewise, using a proteomic approach, Ziebandt et al. identified new members of the SarA regulon (41).
Virulence factors whose expression is controlled by the different regulators generally fall into two categories: cell surface proteins and secreted proteins. The agr system generally downregulates cell surface proteins such as protein A and upregulates secreted proteins such as alpha-toxin. By use of DNA array technology, we have clearly demonstrated in this report that Rot does not act exclusively as a repressor. The work presented here indicates that Rot also positively regulates the expression of genes, many of which have previously been determined to be virulence factors. We also demonstrated that Rot and agr have opposing effects on the expression of certain virulence determinants. Collectively, these findings suggest that Rot is a more global virulence regulator than previously recognized. Whether the effects of Rot on its target genes are direct or indirect is currently unknown.
Rot positively regulates the expression of sarS, a transcriptional modulator of virulence gene expression (6, 38). Arvidson and colleagues have shown that SarS binds to the promoter of spa (38), a gene activated by both Rot and SarS. Thus, Rot may affect spa transcription via SarS. Other SarA homologues have also been shown to regulate the expression of virulence gene regulators. For instance, SarR represses sarA expression (21) and SarA represses sarT expression (35). We could postulate that these modulators affect certain target genes by influencing the expression of regulators of these genes.
Besides genes with well-characterized or postulated virulence functions, Rot also regulates genes not previously linked to virulence. These genes could either be potential virulence genes or additional accessory genes with other biological functions. In the latter case, as Dunman and colleagues point out, the regulators involved in the agr-SarA regulatory pathway may be more global than previously recognized and may affect processes that have yet to be established in S. aureus pathogenesis (12). Given the ability of S. aureus to cause a broad range of clinical manifestations, it is likely that many of these gene products are involved in various, as yet unrecognized stages of infection.
Although this study identified numerous members of the Rot regulon, it is very likely that the list is not complete due to the fact that 14% of the staphylococcal genes are missing from the chip. For example, the sspA gene from the serine protease operon is missing from the GeneChip and hence from Table 3. However, Northern blot analysis showed that the whole operon is downregulated by Rot. With the complete genome available, new GeneChips are being designed to cover 100% of the genome, and these will be used in future studies.
Based on the virulence function of Rot-regulated genes, we hypothesize that Rot plays an important role in the early steps of infection. Factors positively modulated by Rot include cell surface adhesins such as ClfB (clumping factor B), which binds fibrinogen (26), and SdrC, a cell surface adhesin (15). Rot also positively regulates the expression of a gene represented by ORF 1175 on the chip, which corresponds to N315 ORF 2284 (Table 4). This ORF contains a cell wall sorting signal motif, LPXTG, which is characteristic of most adhesins (17). Rot positively regulates the production of protein A, a factor involved in evasion of the host immune system (32). Another gene upregulated by Rot is dltD (Table 4). This gene is a member of the dlt operon, which consists of dltA, dltB, dltC, and dltD (31). This operon encodes proteins involved in d-alanine incorporation into teichoic acid in the S. aureus cell wall. Interestingly, dltA is not present on the list because it is among the genes missing from the GeneChip used in this study (see Materials and Methods). In addition, dltB and dltC are not listed because their fold difference (1.3 for dltB and 1.4 for dltC) did not make the cutoff. The dlt operon has been shown to confer resistance to human antimicrobial agents such as defensins (31). A recent study by Collins et al. has demonstrated that dlt mutants are more efficiently killed by human neutrophils than wild-type strains (9). The same study has also shown that mortality and arthritis frequencies were significantly lower in mice infected with dlt mutants than in mice infected with the wild type strains, indicating that the strains lacking the dlt operon were less virulent than dlt-positive strains. All these factors positively modulated by Rot are hypothesized to be required at an early stage of infection. Some factors, such as the adhesins, allow the bacteria to attach to host cells, while others, such as protein A and DltA, -B, -C, and -D facilitate establishment of the infection through inhibition of the host immune response.
Simultaneously, Rot negatively regulates factors that may interfere with the establishment of the infection. For instance, Rot negatively regulates the production of proteases. One role for proteases is to degrade surface proteins such as protein A, fibronectin-binding proteins, and clumping factor B, which facilitate adherence of the bacteria to host tissues. Degradation of these factors will allow the bacteria to detach from the initial site of infection and start the infection at another location (16, 23). Rot also represses the synthesis of hla and hlb, which encode alpha- and beta-hemolysin, respectively. These hemolysins are known to disrupt a variety of mammalian cells, and because of their function, they are classified as virulence factors involved in invasion and tissue penetration (32). Thus, Rot promotes early stages of infection by stimulating factors needed at this stage and by inhibiting those that interfere with the early processes of infection or that are needed for later processes.
During late stages of infection, which correspond to the post-exponential growth phase, there is a peak of the levels of RNAIII, the agr effector molecule (32). Since our results indicate that Rot and agr have opposing effects on the expression of S. aureus virulence genes, it is likely that RNAIII either directly or indirectly exerts an inhibitory effect on Rot. Since rot is transcribed throughout the growth curve (data not shown) and the rot transcript is present in both RNAIII-positive and RNAIII-negative strains (Fig. 3B), Rot is likely to be inhibited by RNAIII posttranscriptionally. The inhibitory effects of RNAIII on Rot result in the repression of the cell surface proteins and the activation of the secreted proteins that promote the spread of the bacteria and allow them to initiate infection at other sites. Given that Rot was identified in an agr-negative strain, we proceeded to determine the extent of S. aureus gene regulation by Rot in the same background. However, preliminary results indicate that Rot effects on select target genes such as ureC, geh, spl, and ssp operons could be validated in an agr-positive background as well.
In conclusion, the present body of work has employed transcription profiling technology to investigate the spectrum of genes that are regulated by the S. aureus repressor of toxin, Rot. The results demonstrate that Rot is a global transcriptional modulator of virulence genes and that its influence on the expression of certain target genes is opposite that of agr. Genes whose expression is influenced by Rot can be grouped in three major categories: (i) genes that have been extensively studied and have well-defined virulence functions, (ii) genes postulated to play a role in virulence, as their disruption results in attenuation of the mutant in a specific infection model, and (iii) genes not previously known to play a role in S. aureus pathogenesis. By unveiling additional regulatory levels, this study gives us better insight into the agr-SarA regulatory network of S. aureus virulence genes. Additional studies are needed to investigate the molecular interaction of Rot with its target genes and with other regulators, as well as the role of the novel genes regulated by Rot in S. aureus virulence.
Acknowledgments
We are indebted to Barun Mathema, Suzanne Lutwick, and Lefa Alksne for critical reading of the manuscript and for helpful suggestions. We are also grateful to David Shlaes and the Wyeth antimicrobial research department for providing us with the necessary materials for GeneChip experiments.
T.J.F. and F.M.M. acknowledge the Health Research Board of Ireland for support.
REFERENCES
- 1.Abdelnour, A., S. Arvidson, T. Bremell, C. Ryden, and A. Tarkowski. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., and P. Ying. 1994. Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J. Bacteriol. 176:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 9.Collins, L. V., S. A. Kristian, C. Weidenmaier, M. Faigle, K. P. Van Kessel, J. A. Van Strijp, F. Gotz, B. Neumeister, and A. Peschel. 2002. Staphylococcus aureus strains lacking d-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 186:214-219. [DOI] [PubMed] [Google Scholar]
- 10.Coulter, S. N., W. R. Schwan, E. Y. Ng, M. H. Langhorne, H. D. Ritchie, S. Westbrock-Wadman, W. O. Hufnagle, K. R. Folger, A. S. Bayer, and C. K. Stover. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30:393-404. [DOI] [PubMed] [Google Scholar]
- 11.Dajcs, J. J., M. S. Austin, G. D. Sloop, J. M. Moreau, E. B. Hume, H. W. Thompson, F. M. McAleese, T. J. Foster, and R. J. O'Callaghan. 2002. Corneal pathogenesis of Staphylococcus aureus strain Newman. Investig. Ophthalmol. Vis. Sci. 43:1109-1115. [PubMed] [Google Scholar]
- 12.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 15.Josefsson, E., K. W. McCrea, D. Ni Eidhin, D. O'Connell, J. Cox, M. Hook, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144:3387-3395. [DOI] [PubMed] [Google Scholar]
- 16.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 18.Li, S., S. Arvidson, and R. Mollby. 1997. Variation in the agr-dependent expression of alpha-toxin and protein A among clinical isolates of Staphylococcus aureus from patients with septicaemia. FEMS Microbiol. Lett. 152:155-161. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 20.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 21.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McElroy, M. C., D. J. Cain, C. Tyrrell, T. J. Foster, and C. Haslett. 2002. Increased virulence of a fibronectin-binding protein mutant of Staphylococcus aureus in a rat model of pneumonia. Infect. Immun. 70:3865-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGavin, M. J., C. Zahradka, K. Rice, and J. E. Scott. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect. Immun. 65:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 26.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 27.Novick, R. P., and R. Brodsky. 1972. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J. Mol. Biol. 68:285-302. [DOI] [PubMed] [Google Scholar]
- 28.Novick, R. P., and T. W. Muir. 1999. Virulence gene regulation by peptides in staphylococci and other Gram-positive bacteria. Curr. Opin. Microbiol. 2:40-45. [DOI] [PubMed] [Google Scholar]
- 29.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papakyriacou, H., D. Vaz, A. Simor, M. Louie, and M. J. McGavin. 2000. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181:990-1000. [DOI] [PubMed] [Google Scholar]
- 31.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 32.Projan, S., and R. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In G. Archer and K. Crossley (ed.), Staphylococci in human diseases. Churchill Livingstone, New York, N.Y.
- 33.Reed, S. B., C. A. Wesson, L. E. Liou, W. R. Trumble, P. M. Schlievert, G. A. Bohach, and K. W. Bayles. 2001. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect. Immun. 69:1521-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha, B., P. P. Francois, O. Nusse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell Microbiol. 1:101-117. [DOI] [PubMed] [Google Scholar]
- 37.Supersac, G., Y. Piemont, M. Kubina, G. Prevost, and T. J. Foster. 1998. Assessment of the role of gamma-toxin in experimental endophthalmitis using a hlg-deficient mutant of Staphylococcus aureus. Microb. Pathog. 24:241-251. [DOI] [PubMed] [Google Scholar]
- 38.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 39.Tenover, F. C., J. W. Biddle, and M. V. Lancaster. 2001. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 7:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Hoper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]