Abstract
The disulfide oxidoreductase, DsbA, mediates disulfide bond formation in proteins as they enter or pass through the periplasm of gram-negative bacteria. Although DsbA function has been well characterized, less is known about the factors that control its expression. Previous studies with Escherichia coli demonstrated that dsbA is part of a two-gene operon that includes an uncharacterized, upstream gene, yihE, that is positively regulated via the Cpx stress response pathway. To clarify the role of the yihE homologue on dsbA expression in Salmonella enterica serovar Typhimurium, the effect of this gene (termed rdoA) on the regulation of dsbA expression was investigated. Transcriptional assays assessing rdoA promoter activity showed growth phase-dependent expression with maximal activity in stationary phase. Significant quantities of rdoA and dsbA transcripts exist in serovar Typhimurium, but only extremely low levels of rdoA-dsbA cotranscript were detected. Activation of the Cpx system in serovar Typhimurium increased synthesis of both rdoA- and dsbA-specific transcripts but did not significantly alter the levels of detectable cotranscript. These results indicate that Cpx-mediated induction of dsbA transcription in serovar Typhimurium does not occur through an rdoA-dsbA cotranscript. A deletion of the rdoA coding region was constructed to definitively test the relevance of the rdoA-dsbA cotranscript to dsbA expression. The absence of RdoA affects DsbA expression levels when the Cpx system is activated, and providing rdoA in trans complements this phenotype, supporting the hypothesis that a bicistronic mechanism is not involved in serovar Typhimurium dsbA regulation. The rdoA null strain was also shown to be altered in flagellar phase variation. First it was found that induction of the Cpx stress response pathway switched flagellar synthesis to primarily phase 2 flagellin, and this effect was then found to be abrogated in the rdoA null strain, suggesting the involvement of RdoA in mediating Cpx-related signaling.
Bacterial proteins located in or in transit through the periplasm undergo some degree of folding into their native conformations. Periplasmic foldases such as disulfide oxidoreductases and peptidyl-prolyl isomerases accelerate this folding process (4). The best-studied protein folding pathway is the Dsb system in Escherichia coli that catalyzes disulfide bond formation, reviewed in references 3, 4, 13, 18, 42, and 53). DsbA, a 21-kDa periplasmic disulfide oxidoreductase, plays a pivotal role in this system by transferring its disulfide bond to substrates as they are assembled (17, 20, 65). Homologues to DsbA have been identified throughout gram-negative and gram-positive bacteria (24, 29, 35, 49, 58-60, 63). The thiol-disulfide chemistry of the active site of DsbA, consisting of the sequence motif Cys30-Pro31-His32-Cys33 with its donor intrapeptide disulfide bond, has been thoroughly investigated and is highly reactive, providing this enzyme with a high oxidizing potential (20, 22, 30, 42, 62, 65). dsbA mutants exhibit pleiotropic phenotypes, since they alter cellular activities depending on the presence of disulfide bond-containing enzymes. Processes such as motility, assembly of pili, resistance to reducing agents, and the production of virulence determinants are all severely affected, making DsbA important for growth and survival (4, 10, 31, 42, 59, 67).
Whereas the functional properties of DsbA have been investigated in detail, less is known about the factors that influence the expression of this catalyst. Most studies concerning dsbA regulation were carried out with E. coli, where dsbA is part of a two-gene operon that includes an uncharacterized (yihE) gene found immediately upstream of the dsbA coding region (7). Two promoters controlling dsbA transcription have been identified for E. coli. The proximal (dsbA) promoter is located in the 3′ portion of the yihE coding region, and the distal (yihE) promoter is upstream of the yihE gene. Transcription from the proximal promoter is proposed to be relatively low and constitutive. Belin and Boquet constructed yihE null strains that showed a reduction in DsbA levels compared to the wild type, although this reduced level of DsbA expression showed no phenotype (7). When they examined a plasmid-borne yihE-dsbA::TnphoA translational fusion containing a frameshift mutation in the yihE gene in a yihE null background, the expression of the TnphoA fusion was reduced by one-third. The presence in trans of the wild-type yihE gene did not restore the wild-type levels of the fusion. The authors concluded that the frameshift mutation conferred a polar transcriptional defect on dsbA expression.
Pogliano et al. (51) showed that the activation of the two-component Cpx pathway, through a CpxR binding site located in the yihE promoter region, increased DsbA protein levels. This correlated with increased levels of transcripts originating from the yihE promoter as determined by nuclease protection assays (12, 51). Members of the Cpx regulon are part of a stress response system involved in sensing and responding to cell envelope protein folding defects (9, 52, 55). Cpx-activating signals include overproduction of the outer membrane lipoprotein NlpE (55), high-pH conditions (43), and an alteration in the phospholipid composition of the bacterial membrane (40).
In contrast to reports from E. coli, the proximal promoter region of Salmonella enterica serovar Typhimurium (herein referred to as serovar Typhimurium) dsbA showed more complex regulation of expression. Two dsbA-specific transcripts were identified for this organism and mapped to two separate promoters proximal to the dsbA coding region (19). Growth phase-dependent promoter activity, with high levels of activity in stationary phase, was demonstrated using a dsbA transcriptional fusion containing both dsbA-proximal promoters, and growth conditions, such as lowered pH and oxygen levels, were shown to affect dsbA transcription (19). In its natural habitats, serovar Typhimurium often encounters a variety of stressful conditions, including fluctuations in pH, hypoxia, oxidative stress, and nutrient limitation (6, 48, 57). The organism can actively adapt to survive under these conditions by inducing systems (for example, the acid tolerance response) and/or specific genes, including virulence determinants, that enhance cellular resistance and provide cross-protection to additional stresses (36, 57).
To study the possible effect of the upstream gene on the regulation of dsbA expression, the DNA sequence further upstream of serovar Typhimurium dsbA was determined and the transcripts encoded by this region were analyzed. While the sequence corresponding to serovar Typhimurium dsbA and the upstream open reading frame (ORF), termed rdoA (regulator of disulfide oxidoreductase A), is homologous to the E. coli yihE-dsbA operon, differences were identified in their respective control of dsbA expression. Our results indicate that the rdoA-dsbA cotranscript does not play a significant role in Cpx-mediated induction of dsbA transcription, unlike the situation proposed to occur with E. coli, although RdoA does influence DsbA protein levels. This study also showed that RdoA also affects flagellin phase variation in a Cpx-dependent manner, suggesting a role for RdoA in relaying Cpx signals.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Table 1 describes all bacterial strains and plasmids used in this study. For β-galactosidase assays, overnight cultures were diluted 1:100 either in fresh Luria-Bertaini (LB) broth or in minimal E-media (pH 7.6 or 4.6) (61) supplemented with 0.4% glucose and amino acids (to a final concentration of 50 μg/ml). Cells were grown at 37°C with aeration. For β-galactosidase assays and RNA preparation of strains containing pND18 or pBAD18 (control) vectors, overnight cultures were diluted 1:100 and grown to an optical density at 600 nm (OD600) of approximately 0.3 in LB with 0.2% glucose (repressing the arabinose promoter in pND18 and pBAD18 vectors). Cultures were centrifuged, and the supernatant was aspirated off. Cell pellets were resuspended in fresh LB media containing either 0.4% arabinose (inducing the arabinose promoter in the pBAD18-based vectors) or 0.2% glucose (repressing the arabinose promoter). Cultures were subsequently assayed for β-galactosidase activity at various time points or, in the case of RNA extraction, grown for 1 h before harvesting of RNA. When antibiotics were required, the medium was supplemented with 30 μg of chloramphenicol/ml or 50 μg of ampicillin/ml. The growth of the bacterial cultures was monitored by measuring the OD600.
TABLE 1.
E. coli or serovar Typhimurium strains and plasmids used in this studya
| Strain or plasmid | Laboratory reference | Description | Source or reference |
|---|---|---|---|
| E. coli | |||
| DH5α | NLM264 | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 23 |
| MG1655 | NLM28 | Wild type, derivative of K-12 | Lab collection |
| MG1655 dsbA | NLM1 | MG1655 lacZ::Tn10(TetS) dsbA::kan | Lab collection |
| MC4100 cpxR | NLM367 | MC4100 cpxR::Ω(spec) | 12 |
| Serovar Typhimurium | |||
| SL1344 | NLM2217 | his Strr | 64 |
| SL1344 | NLM2218 | NLM2217 containing pPrdoA | This study |
| SL1344 dsbA | NLM267 | SL1344 dsbA::kan | 59 |
| SL1344 dsbA | NLM2219 | NLM267 containing pPrdoA | This study |
| SL1344 rdoA | NLM2214 | NLM2217 with a rdoA deletion of 766 bp containing pMP190 | This study |
| SL1344 rdoA | NLM2239 | NLM2214 cured of pMP190 | This study |
| SL1344 | NLM2244 | NLM2217 containing pBAD18 | This study |
| SL1344 | NLM2245 | NLM2217 containing pPrdoA, pND18 | This study |
| SL1344 | NLM2246 | NLM2217 containing pPrdoA, pBAD18 | This study |
| SL1344 | NLM2247 | NLM2217 containing pND18, pMEG2 | This study |
| SL1344 | NLM2248 | NLM2217 containing pBAD18, pMEG2 | This study |
| SL1344 rdoA | NLM2253 | NLM2239 containing pPrdoA, pND18 | This study |
| SL1344 rdoA | NLM2254 | NLM2239 pPrdoA, pBAD18 | This study |
| SL1344 rdoA | NLM2255 | NLM2239 containing pND18, pMEG2 | This study |
| SL1344 rdoA | NLM2256 | NLM2239 containing pBAD18, pMEG2 | This study |
| SL1344 rdoA/rdoA+ | NLM2269 | NLM2238 containing pLMN20 | This study |
| SL1344 cpxR | NLM2274 | NLM2217 transduced with cpxR::Ω from NLM367 containing pND18 | This study |
| Plasmids | |||
| pMEG2 | pMP190 with a 258-bp fragment from immediately upstream of serovar Typhimurium dsbA SD sequence | 19 | |
| pPrdoA | pMP190 with a 236-bp fragment from immediately upstream of serovar Typhimurium rdoA SD sequence | This study | |
| pND18 | pBAD18 which contains the E. coli nlpE gene under the control of the PBAD promoter of the araBAD (arabinose) operon | 55 | |
| pMP190 | 15-kb transcriptional fusion vector containing a promoterless lacZ, Cmr | 56 | |
| pMEG1 | pBAD24 with 6.9-kb NcoI fragment from pLAFR2-31 containing dsbA and rdoA | 19 | |
| pLMN20 | pACYC184 containing a 1.5-kb SacII fragment from pMEG1 coding for rdoA | This study |
SD, Shine-Dalgarno.
Construction of the rdoA::lacZ transcriptional fusion (pPrdoA).
The primers Nm64 (5′TAACTCGAGCAAACTGGAAAGATTTAC3′) containing an XhoI restriction site (underlined) and Nm57 (5′TTAAGATCTATACTACGAATGATTCAG3′) containing a BglII restriction site (underlined) were used to PCR amplify a 236-bp rdoA putative promoter fragment (position, 228 bp upstream to 15 bp upstream of the RdoA start codon) using the template pMEG1. This rdoA promoter region was cloned into pMP190, predigested with SalI (compatible to the XhoI site of the PCR amplicon) and BglII. This construct, called pPrdoA, containing the putative promoter, transcriptional start site, and sequence up to but not including the ribosome binding site is similar to the dsbA::lacZ transcriptional fusion vector, pMEG2.
Assays.
Transcriptional activity from the rdoA::lacZ promoter fusion construct was measured in all serovar Typhimurium strains by assaying β-galactosidase activity as described by Miller (41). Aliquots of cultures were assayed at time intervals throughout the growth of the culture or at specific growth phases. In addition, assays were preformed on one in five dilutions of aliquots, once the OD600 values of cultures reached a value of 1. Each experiment was performed at least twice.
RNA preparation and RT-PCR.
Total RNA was isolated using Trizol (Life Technologies, Gibco BRL) from 1 ml of culture of S. enterica serovar Typhimurium SL1344 and E. coli MG1655 strains containing pND18 (inducible NlpE-overproducing plasmid) or control vector (pBAD18) after 1 h of induction with 0.4% arabinose or repression with 0.2% glucose. Approximately 20 μg of total RNA was subjected to DNase treatment (RQ1 RNase-free DNase; Promega) and subsequent purification using RNeasy columns (RNeasy Mini kit; Qiagen). Multiplex reverse transcription (RT) reactions (Retroscript kit; Ambion) using 100 pmol of internal control primer and the serovar Typhimurium-specific target primers Nm78 (5′ CGCATCAACGAACACTTTACGG 3′) and Nm88 (5′ CCAACGACGAATCAACCAGG 3′) or the E. coli target primers Nm103 (5′ CCTGCGTTGATAAATACATCGC 3′) and Nm105 (5′ CCAACGCCGCATTAGCCAGG 3′) were performed on approximately 1 to 3 μg of purified RNA with minor changes to the manufacturer's instructions. Changes to the protocol include longer heat denaturation of the RNA template (6 min at 80°C) and elevated incubation temperatures (56°C). In addition, RT negative control reactions were performed where water was added instead of the RT enzyme (data not shown). Multiplex PCR amplification was performed using an internal control primer pair and specific target primer pairs: for serovar Typhimurium, Nm88 and Nm89 (5′ GGCAACATTCTATGGCGTG 3′); and for E. coli, Nm104 (5′ GGAATATTCTCTGGCGCGATGGTCC 3′) and Nm105 (5′ CCAACGCCGCATTAGCCAGG 3′). Fifty-microliter PCRs were carried out using 2.5 U of Taq DNA polymerase (Life Technologies, Gibco BRL), 5 μl of RT reaction sample as a template, 50 pmol of each PCR primer pair, 1× PCR buffer, 0.2 mM (each) deoxynucleoside triphosphate, and 1.5 mM MgCl2. Template cDNA was denatured for 2 min at 94°C before Taq DNA polymerase was added. A 25- to 30-cycle PCR (94°C for 45 s, 60°C for 30 s, and 72°C for 1 min with a final extension reaction of 72°C for 7 min) was carried out. To aid in qualitative analysis, specific target RT-PCR products were normalized to an established endogenous internal control transcript (tsf encoding the elongation factor EF-Tsf), the expression of which is relatively constant in serovar Typhimurium at the growth phases studied in this work (26). RT-PCR primers used in the detection of tsf transcript are described by Holmstrom et al. (26). PCR products of both internal control transcript (563 bp) and target (246 bp for serovar Typhimurium rdoA-dsbA cotranscript only or rdoA monocistronic and cotranscript; 241 bp for E. coli yihE-dsbA cotranscript only or yihE specific and cotranscript) were visualized by agarose gel electrophoresis on a 1.2% agarose gel containing 1 μg of ethidium bromide/ml.
Northern blot analysis.
Approximately 30 μg of total RNA from serovar Typhimurium SL1344 or E. coli MG1655 strains were separated on a 1.2% formaldehyde-agarose gel and transferred to a nylon membrane (positively charged; Boehringer Mannheim) as described by Fourney et al. (16). Membranes were initially probed with a serovar Typhimurium rdoA DNA probe or an E. coli yihE DNA probe and subsequently stripped and reprobed with a serovar Typhimurium dsbA probe or an E. coli dsbA DNA probe. Each of the probes was generated by PCR and gel purified using a Qiaquick gel purification kit (Qiagen): the N-terminus-specific serovar Typhimurium dsbA probe (position, 107 to 400 bp downstream of the dsbA ATG start site) using primers Nm86 (5′ CTGGCGAACCCCAGGTACTG 3′) and Nm78; a full-length rdoA probe (position, 25 to 855 bp downstream of the rdoA ATG start site) using primers Nm99 (5′ ACGCTACACCCGGAAACCATC 3′) and Nm89; an N-terminus-specific E. coli dsbA probe (position, 107 to 404 bp downstream of the dsbA ATG start site) using primers Nm102 (5′ CTGGCGCGCCGCAAGTGCTG 3′) and Nm103; and a C terminus E. coli yihE probe (position, 614 to 855 bp downstream of the yihE ATG start site) using primers Nm104 (5′ GGAATATTCTCTGGCGCGATGGTCC 3′) and Nm105 (5′ CCAACGCCGCATTAGCCAGG 3′). Fifty nanograms of all probes were random prime labeled with an oligolabeling kit and [α-32P]dCTP (3,000 Ci/mmol; Amersham Pharmacia Biotech) according to the manufacturer's instructions. The efficiency of labeling was monitored following procedure C of the manufacturer's protocol, and labeled probes were separated from unincorporated 32P-labeled nucleotides using a QIAquick Spin column and the QIAquick nucleotide removal protocol (Qiagen). All prehybridizations, hybridizations, Northern washes, and removal of labeled probes from membranes were performed according to the basic protocol described by Kingston (33). Washed membranes were subsequently exposed to a storage phosphor-imaging screen (Kodak). Phosphorescence signals were captured and quantitated using the Personal Molecular Imager FX (Bio-Rad) and Quantity One (version 4.0; Bio-Rad) quantitation software. For comparisons, the Northern signals were normalized to the 16S rRNA band of the specific RNA preparations loaded.
Gel electrophoresis and Western blotting.
Periplasmic protein preparations were separated using a sodium dodecyl sulfate-12% polyacrylamide gel. Briefly, periplasmic protein isolation involved growing overnight serovar Typhimurium SL1344 (pND18) or control vector cells in 0.2% glucose (repressed) or 0.4% arabinose (induced) to an OD600 of 1. One milliliter of culture was centrifuged and resuspended in 250 μl of Tris-HCl (0.2 M). Subsequently, a solution containing 1 M sucrose in 0.2 M Tris-HCl, 2.5 μl of EDTA (0.1 M), 7.5 μl of lysozyme (4 mg/ml), and 500 μl of distilled water was added and incubated on ice for 2 min. A 30-min incubation was carried out after adding 20 μl of MgCl2 (1 M), and proteins were isolated from the supernatant by trichloroacetic acid extraction.
Two-dimensional (2D) gel electrophoresis was carried out following the manufacturer's suggested conditions using immobilized pH gradient (Bio-Rad) strips of pH ranges 3 to 10, 3 to 6, or 4 to 7 in the first dimension. The samples were run in the second dimension on 12% acrylamide gels using the Protean II system (Bio-Rad). Gels were silver stained (66) and digitized using a transmission flatbed scanner. All data analysis was carried out using the BioImage software package (Genomic Solutions). A minimum of three sample replicates were carried out for each strain or growth condition examined.
Western immunoblots were carried out as previously described by Klapper et al. (34) using polyclonal anti-DsbA sera, peroxidase-conjugated secondary antibody, and the SuperSignal West chemiluminescence detection system (Pierce).
Peptide mass fingerprinting.
Protein spots of interest were excised from the acrylamide gels using a Bio-Rad spot cutter, digested with trypsin, and extracted from the gel pieces using a MicroMass MassPrep digestion robot. Mass spectrometric analysis was carried out using a MicroMass M@LDI mass spectrometer. The spectrum data were analyzed using the MASCOT software (50).
Creation of an rdoA deletion strain.
Primer Nm114 (5′ GGAATTCGTCAACTGGGGCTCAACG 3′) containing an EcoRI restriction site (underlined) and primer Nm115 (5′ CGGGATCCTTGTCGTTCATCCCATCATCCGG 3′) containing a BamHI restriction site (underlined) were used to PCR amplify an 881-bp DNA fragment (position, 855 bp upstream of the rdoA start codon to 11 bp downstream of the rdoA start codon) using the template pMEG1. This fragment (left flank) includes the first 11 nucleotides of the rdoA coding region and enough upstream sequence to allow for homologous recombination. The primers Nm116 (5′ CGGGATCCACGGCTGAAATTGGTCTCA 3′) containing a BamHI restriction site (underlined) and Nm117 (5′ GGAATTCTGCTGCTCGTATCCATGC 3′) containing an EcoRI restriction site (underlined) were used to PCR amplify an 806-bp fragment (position, 778 to 1,568 bp downstream of the rdoA start codon) using the template pMEG1. This fragment (right flank) includes the last 70 codons of the rdoA coding region and enough downstream sequence to allow for homologous recombination. The left and right flank amplicons were gel purified using the QIAquick gel purification kit (Qiagen Inc.), digested with BamHI, and ligated together. PCR amplification using 5 μl of the ligation reaction as a template and the outer EcoRI restriction site containing primers Nm114 and Nm117 was performed to amplify a 1.69-kb DNA fragment containing 11 bp of the N terminus and 210 bp of the C terminus of the rdoA ORF but lacking the intervening 766 bp of the rdoA coding region. This 1.69-kb rdoA deletion fragment was gel purified using the QIAquick spin column (Qiagen Inc.), digested with EcoRI, and subcloned into the ampicillin-resistant and streptomycin-sensitive counter-selectable suicide vector pKAS32 (21). Ligants were transformed into E. coli MC4100λpir strains and selected on ampicillin plates. Plasmid purified from these colonies was then electroporated into the conjugative E. coli Sm10λpir strain. This donor strain was conjugated with the streptomycin- and chloramphenicol-resistant serovar Typhimurium SL1344 recipient strain containing the plasmid pMP190. Transconjugants were selected on LB plates containing chloramphenicol (to select against the donor E. coli strain) and ampicillin (to select for the plasmids). Twenty colonies from each of the conjugations were then restreaked on streptomycin plates, permitting selection of single-crossover clones of the rdoA deletion fragment. Approximately 250 individual colonies were picked and patched onto streptomycin and ampicillin plates. Colonies in which a double crossover event occurred by homologous recombination of the rdoA deletion fragment grew on streptomycin plates but did not grow on ampicillin plates. One such colony (designated NLM 2214) was discovered, and colony PCR amplification confirmed the presence of the rdoA deletion fragment in the chromosome of serovar Typhimurium SL1344. Colonies were then screened for loss of the plasmid pMP190 by repeated subculturing in the absence of the selective antibiotic chloramphenicol followed by screening for lack of growth on chloramphenicol-containing plates generating NLM2239.
Cloning the rdoA gene.
The coding region of the rdoA gene was cloned into the SacII site of pACYC184 by isolation of a SacII rdoA-containing fragment from pMEG1. This construct (pLMN20) includes the native rdoA promoter region and the first 60 bases of the dsbA coding region downstream of rdoA but does not encode functional DsbA.
RESULTS
The rdoA promoter activity is growth phase dependent, and its activity changes in response to the lack of DsbA, minimal medium, and acid shock conditions.
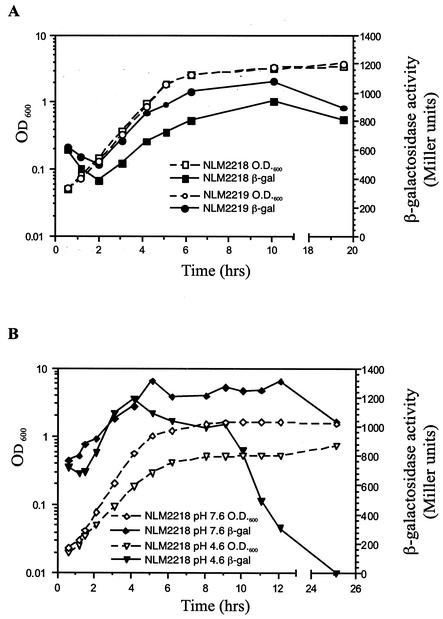
A 236-bp rdoA putative promoter region, immediately upstream of the ribosome binding site of rdoA, was fused in front of the reporter lacZ gene in the low-copy-number vector pMP190 to form the transcriptional fusion plasmid pPrdoA. The behavior of this putative promoter region in rich medium was examined by measuring β-galactosidase activity for wild-type serovar Typhimurium SL1344 and a dsbA null background strain (Fig. 1). In rich medium, the putative rdoA promoter region was functional and exhibited growth-phase-dependent expression, with maximum activity occurring in stationary phase. Initially, after 1:100 subculturing from overnight culture, rdoA promoter activity was quite high (approximately 600 Miller units) and declined rapidly (Fig. 1A). Promoter activity then increased to a maximum of 900 Miller units. After 20 h of growth, activity remained elevated and comparable to earlier stationary phase expression levels. An increase in rdoA transcript levels in stationary phase was also detected using RT-PCR (data not shown). An elevated expression of the rdoA promoter (approximately 200 Miller units) was observed in a dsbA null background at every stage of growth (Fig. 1A), indicating that a lack of disulfide bond-forming ability (provided by DsbA) in the periplasm has a positive feedback effect on rdoA transcription.
FIG. 1.
Expression of the rdoA::lacZ promoter (pPrdoA) in wild-type (NLM2218) and dsbA null (NLM2219) serovar Typhimurium SL1344 background strains throughout the growth curve in rich medium (A) or wild-type (NLM2218) serovar Typhimurium growth in minimal E-glucose medium at pH 7.6 or 4.6 (B). Each point in a curve is the average activity of two samples, and the standard deviation for all data points is less than 5% of the mean. The β-galactosidase activity curves of pPrdoA are representative of two to three separate trials.
The effect of minimal medium and acid shock conditions on rdoA expression (situations that serovar Typhimurium is likely to encounter in the environment of the host [57]) was measured in wild-type SL1344 cells containing pPrdoA that were grown in minimal E glucose media (pH 7.6 and pH 4.6) (Fig. 1B). Growth-phase-dependent expression of the rdoA promoter was again displayed in minimal medium; however, in comparison to rich medium conditions, activity was elevated (by 300 to 400 Miller units) at all stages of growth. The rdoA promoter activity fell sharply in stationary phase SL1344 cells grown in acidic-pH media. Plating of cells to assess viability showed no significant difference in viable cell counts from 9 to 14 h (data not shown). This result indicated that the rdoA promoter is turned off in stationary phase under acidic growth conditions.
The rdoA and dsbA promoters are induced at different points in the growth phase.
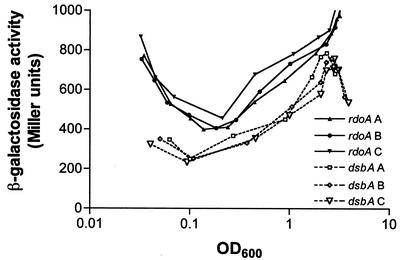
It has been previously shown that a dsbA transcriptional fusion (pMEG2) is growth phase dependent with maximal expression in stationary phase (19). The activity of the rdoA promoter fusion was compared to that of the dsbA::lacZ promoter fusion vector to identify similarities and variations in the expression patterns of these two growth-phase-dependent promoters. The magnitude of rdoA promoter expression was higher than that of the dsbA promoter at all stages of growth (Fig. 2). For both promoters, β-galactosidase activity decreases initially, indicating that β-galactosidase turnover exceeds synthesis. At an OD600 of 0.1, the dsbA promoter drives a net increase in activity while the rdoA promoter activity does not surpass the β-galactosidase turnover rate until the OD600 = 0.25. The β-galactosidase levels then increase at a slightly higher rate with the rdoA promoter than the dsbA promoter until stationary phase, where the dsbA promoter-driven activity falls off somewhat. This variation in terms of the growth-phase-dependent activity of these promoters suggests that different regulatory mechanisms influence the transcription of the rdoA and dsbA promoters.
FIG. 2.
A comparison of rdoA::lacZ (pPrdoA) and dsbA::lacZ (pMEG2) expression in the wild-type serovar Typhimurium SL1344 strain throughout the growth curve in rich medium. The β-galactosidase activities of pPrdoA (rdoA) and pMEG2 (dsbA) from three separate assays are plotted (A, B, and C). Each point in a curve represents the average activity of two samples.
E. coli NlpE overexpression stimulates serovar Typhimurium rdoA promoter activity.
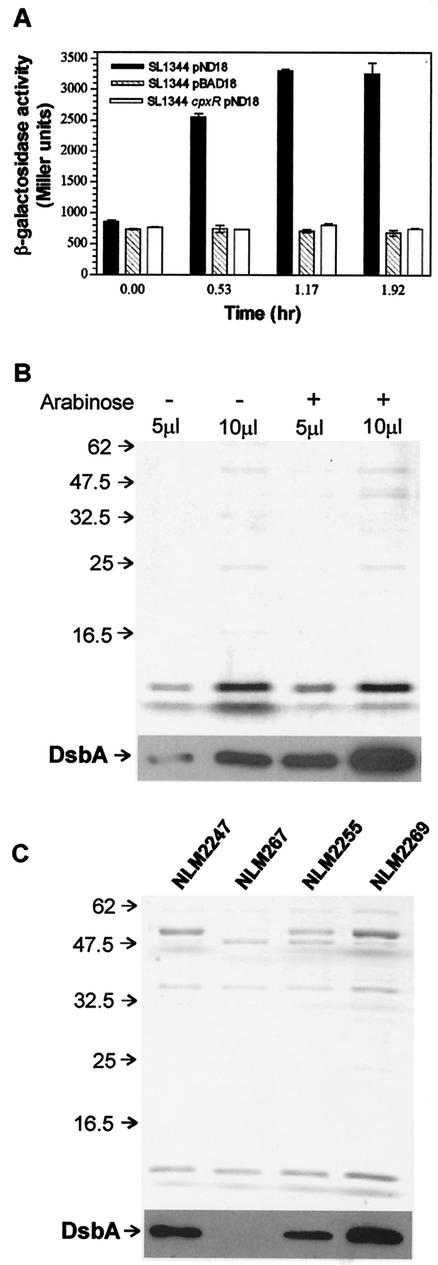
The sequence of the serovar Typhimurium rdoA ORF and that of the corresponding E. coli yihE ORF are very similar (78% nucleotide sequence identity, and the deduced amino acid sequences are 86% identical for both ORFs). Comparing the promoter insert region of pPrdoA to the same region in E. coli revealed high sequence identity, indicating that the most likely promoter in the insert contains a putative CpxR-P-responsive promoter box as characterized for yihE (51). Overexpression of E. coli outer membrane lipoprotein, NlpE, has been shown to stimulate the homologous yihE promoter fivefold by activating the Cpx pathway (12, 51, 55). To ascertain if the rdoA promoter is also inducible by the Cpx pathway in wild-type serovar Typhimurium, SL1344 cells containing pPrdoA reporter plasmid were transformed with an inducible E. coli NlpE overproducing plasmid (pND18) or control vector (pBAD18). β-galactosidase assay results indicated that rdoA promoter activity increased approximately threefold over that of the control vector after induction of NlpE expression (Fig. 3A). This correlated with increased levels of DsbA protein when NlpE was overexpressed (Fig. 3B) and did not occur when rdoA promoter activity was measured in a cpxR null background (Fig. 3A). These findings are similar to the yihE promoter expression pattern, as measured by a nuclease protection assay, and increased DsbA protein levels, measured by pulse-chase immunoprecipitation, reported by Pogliano et al. (51) during the activation of the Cpx pathway in E. coli. Henceforth, the E. coli NlpE-overproducing plasmid, pND18, was utilized as a tool to potently stimulate serovar Typhimurium rdoA promoter expression.
FIG. 3.
Effects of inducing NlpE on the rdoA promoter and DsbA protein levels. (A) A comparison of promoter activity of rdoA with or without induction of the Cpx response via expression of NlpE by comparing rdoA::lacZ (pPrdoA) in cells containing an arabinose-inducible nlpE (pND18) or control (pBAD18) vector. The cpxR::Ω strain is included to demonstrate that the effect on the rdoA promoter is mediated by the Cpx pathway. Cells were grown to an OD600 of 0.3 in the presence of 0.2% glucose. The cultures were then split, and 0.4% arabinose was added to one. Cultures were assayed for β-galactosidase activity at the time of adding arabinose (t = 0) and several times later. (B) Upper panel, protein gel of periplasmic extracts from cells grown under NlpE-uninduced (−) and induced (+) conditions at two loading concentrations (5 and 10 μl) stained with Coomassie blue to indicate total protein levels (DsbA is not visible on this gel); lower panel, Western immunoblot developed with anti-DsbA antiserum. DsbA is more abundant when NlpE is overexpressed. (C) Upper panel, protein gel of periplasmic extracts from the wild-type strain (NLM2247), dsbA null strain (NLM267), rdoA null strain (NLM2255), and rdoA null strain containing a copy of rdoA in trans (NLM2269) grown under NlpE-inducing conditions (all strains contain pDN18 and were grown in the presence of arabinose) stained with Coomassie blue to indicate total protein levels (DsbA is not visible); lower panel, Western immunoblot developed with anti-DsbA antiserum. Inducing NlpE does not increase the DsbA levels to that of the wild type (NLM2247) in the rdoA null (NLM2255) strain. The wild-type phenotype is regained when rdoA is provided in trans (NLM2269).
E. coli NlpE overexpression induces serovar Typhimurium rdoA and dsbA transcript levels but does not significantly alter the levels of rdoA-dsbA cotranscript.
Reports from studies with E. coli have established that the yihE-dsbA cotranscript is necessary for dsbA expression (7, 51). We evaluated the DsbA protein in a rdoA null background and found that DsbA levels decreased under conditions of NlpE overexpression compared to the wild type and that normal DsbA levels could be restored by providing a copy of rdoA in trans (Fig. 3C). DsbA was also quantitated from 2D gels from at least three separately prepared samples for each growth condition and found to be twofold reduced in the rdoA null strain compared with the wild type under NlpE-induced conditions. These protein expression results suggested that the contribution of the rdoA transcript to dsbA expression needed to be clarified, and this question was evaluated with serovar Typhimurium and with E. coli.
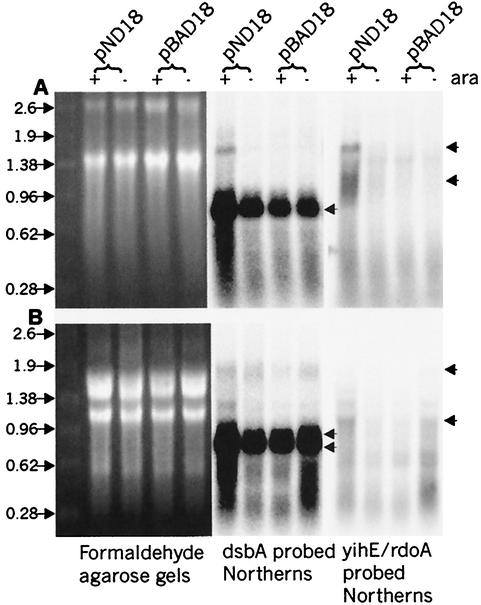
To identify the transcripts originating from the serovar Typhimurium rdoA-dsbA region and clarify their possible effect on dsbA regulation, Northern hybridization using serovar Typhimurium dsbA and rdoA internal probes was employed on total RNA harvested from wild-type SL1344 strains containing either the NlpE-overexpressing plasmid (pND18) or control vector (pBAD18) (Fig. 4B). Northern blot analysis of membranes probed with a dsbA-specific fragment detected two distinct dsbA transcripts in wild-type serovar Typhimurium (of approximately 0.8 and 0.7 kb), with a 2.5-fold increase in the level of the smaller transcript under NlpE-overproducing conditions compared to the level for “induced” cells containing the control vector. The two dsbA-encoding transcripts described here and by Goecke et al. (19) have been mapped to two distinct dsbA promoters located in the 3′ end of the RdoA coding region.
FIG. 4.
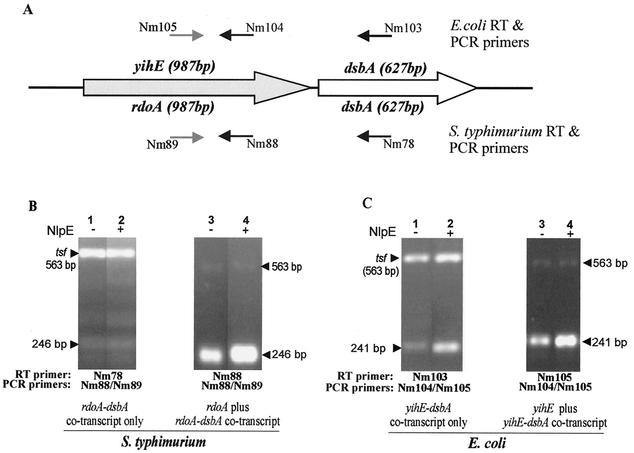
Transcript analysis of yihE, rdoA, and dsbA in serovar Typhimurium and E. coli. RNA formaldehyde agarose gels show total RNA amounts loaded in each lane. E. coli (A) and S. typhimurium (B) cells containing pBAD18 (control) vector or pND18 (NlpE-overexpressing plasmid) induced (+) with 0.4% arabinose or uninduced/repressed (−) with 0.2% glucose are analyzed. The left panels show the agarose gels, while the right panels show the corresponding Northern blots of a single membrane probed with either a dsbA probe or a yihE/rdoA probe. Cotranscript (approximately 1.8 kb) was detected using an E. coli dsbA probe or a yihE probe (upper arrow, panel A) and a serovar Typhimurium dsbA probe (upper arrow, panel B). The most abundant transcript in both organisms is dsbA, with one transcript of approximately 0.8 kb in E. coli and two transcripts of approximately 0.7 and 0.8 in serovar Typhimurium (double arrows, panel B). The yihE or rdoA transcripts of approximately 1.0 kb are indicated with arrows for both organisms. In serovar Typhimurium, fragmentation of the 23S rRNA occurs, which results in a different rRNA banding pattern than is seen with E. coli rRNA (27).
A putative rdoA-dsbA cotranscript (size, approximately 1.8 kb) was rarely clearly identified using a serovar Typhimurium dsbA probe (Fig. 4B) despite numerous attempts. This faint 1.8-kb signal matched the expected length of a cotranscript and could also be detected using an rdoA probe and long exposure times (data not shown). Additional transcripts detected of approximately 1.38 kb are too small to encode functional cotranscripts. The most prominent 1-kb transcript detected with the rdoA probe matches the expected length of rdoA mRNA. Densitometric quantitation revealed an approximately 1.5-fold increase of the 1.0-kb rdoA signal from wild-type cells overexpressing NlpE versus control vector. This result agrees with the rdoA transcriptional fusion reporter assays, in which an increase in rdoA promoter expression were observed under the same conditions. In a dsbA null strain overexpressing NlpE, considerably higher levels of the 1-kb transcript (an additional threefold increase) was observed than was found with wild-type cells overexpressing NlpE (data not shown).
To discern transcriptional similarities or differences that may exist in the same region in E. coli, Northern blot analysis was also performed with E. coli MG1655 strains that carried NlpE-overproducing or control plasmid (Fig. 4A). A striking difference between the two organisms was clearly demonstrated in which a yihE-dsbA cotranscript (of approximately 1.8 kb) was identified by using both E. coli dsbA and yihE probes (Fig. 4A). In contrast to serovar Typhimurium, a distinct increase of this transcript was distinguished between pND18-induced RNA samples and uninduced RNA samples, although the signal still appeared to be faint (approximately a 3-day exposure to phosphorimager screen) compared to the more abundant 0.7-kb transcript (an approximately 4-h exposure to phosphorimager screen). The E. coli RNA membrane probed for yihE displayed two separate bands of approximately 1.8 and 1 kb (Fig. 4A). The signals matched predicted sizes of cotranscript and monocistronic yihE mRNA. Distinct increases of both bands in cells overexpressing NlpE versus the control vector were also observed. Furthermore, strains containing the control vector and the glucose-repressed NlpE plasmid did not reveal detectable levels of either of these transcript signals.
Since the rdoA transcript was not easily detected using Northern analysis, RT-PCR was also performed to compare rdoA monocistronic and rdoA-dsbA cotranscript amounts in pND18 (nlpE)- and pBAD18 (control vector)-overexpressing conditions (see Fig. 5A for primers and Materials and Methods for strategy). Figure 5B shows RT-PCR amplicons of rdoA-dsbA cotranscript or cotranscript plus rdoA monocistronic transcript from SL1344 cells overexpressing NlpE and controls. The RT-PCR products were compared to measure differential cotranscript or rdoA monocistronic transcript expression, based upon an internal control transcript, tsf, encoding the elongation factor EF-Tsf (26). The expression of the internal control is constant in serovar Typhimurium at the growth phase studied and appears in similar amounts in SL1344 cells overexpressing NlpE or pBAD18 control vector cells for the specific RT-PCR amplicon examined. Figure 5B shows that there is very little difference in the amount of cotranscript in serovar Typhimurium cells under NlpE-overproducing conditions compared to the control vector. However, judged against the internal control, a definite increase in the quantity of the rdoA monocistronic mRNA plus cotranscript was observed in NlpE-overexpressing cells versus results for control vector-containing cells. Since we know from the RT-PCR analysis of the cotranscript alone that its expression level is comparatively equal between strains overexpressing NlpE and the control vector, this must mean there is a significant increase in the quantity of the rdoA monocistronic transcript in NlpE-overexpressing cells. This result confirms Northern blotting data on the increase in the amounts of rdoA monocistronic transcripts under the same conditions.
FIG. 5.
Transcript analysis of yihE, rdoA, rdoA-dsbA, and yihE-dsbA by RT-PCR. The primers utilized in the RT-PCR are illustrated in panel A, which shows the organization of the rdoA (yihE)-dsbA region in serovar Typhimurium and E. coli. Serovar Typhimurium primer Nm78 binds 400 bp downstream of the dsbA start codon, while primers Nm88 and Nm89 bind 855 and 612 bp downstream of the rdoA start codon. E. coli primer Nm103 binds 404 bp downstream of the dsbA start codon, while primers Nm105 and Nm104 bind 855 and 614 bp downstream of the yihE start codon. In panels B and C the lower amplicons are the target amplicons, while the upper bands are the tsf (encoding the elongation factor EF-Tsf) internal control amplicon used as a standard in the qualitative assessment of pND18 (NlpE)-induced (+) or pBAD18 (control vector)-induced (−) samples of serovar Typhimurium (B) and E. coli (C) RNA preparations. The size of the EF-tsf amplicon is 563 bp, while the serovar Typhimurium target amplicon for the rdoA-dsbA- and rdoA-specific transcript is 246 bp. The E. coli target amplicon for the yihE-dsbA- or yihE-specific transcript is 241 bp.
Comparative RT-PCR analysis was also extended to E. coli MG1655 strains that carried NlpE-overproducing or control plasmid. Figure 5C illustrates an increase in the yihE-dsbA cotranscript in strains overproducing NlpE versus results with the control vector. This result agrees with Northern blot data using dsbA and yihE probes. Moreover, an increase in the amount of cotranscript plus yihE monocistronic RT-PCR amplicon was detected in the E. coli strain overproducing NlpE versus results with the control vector. This is due only in part to an increase in cotranscript and also involves an increase in the level of yihE monocistronic mRNA.
Phenotypes associated with RdoA.
Although the data presented here point to different transcripts contributing to rdoA and dsbA expression levels, still nothing is known about the function of RdoA. It was thought that RdoA could play a role as an intermediate in the signaling pathways initiated by activating the Cpx system. The effect of rdoA expression on dsbA transcription was examined (Fig. 6), and it was determined that an increase in dsbA promoter activity occurred in response to NlpE expression. Although this change was modest (20% above uninduced level), this increase was not seen in the rdoA null strain. There was also a notable difference between wild-type and rdoA null strains in mid-log-phase cells without NlpE expression (Fig. 6, t = 0), indicating that the absence of rdoA increases dsbA transcription. This difference was not seen in stationary phase cells. It may be that lack of rdoA indirectly triggers dsbA transcription during exponential phase growth through other, as yet undefined, pathways.
FIG. 6.
Overexpression of NlpE increases dsbA promoter activity. A comparison of dsbA::lacZ expression using strains containing pMEG2 and either the vector control (pBAD18) or the NlpE expressing vector (pND18) at t = 0 and 2 h after induction of NlpE expression with 0.2% arabinose. Cells were grown to an OD600 of 0.6 prior to induction. The asterisks indicate that the mean is statistically significantly different from the mean with SL1344 pBAD18 at either time point (P < 0.05).
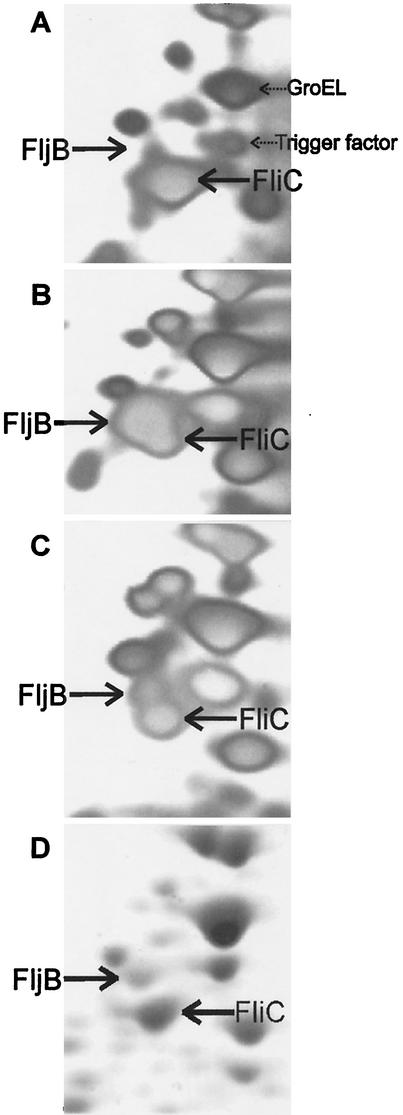
To further define the role of RdoA in serovar Typhimurium, 2D gel electrophoresis analysis was carried out in an effort to identify proteins affected by the presence or absence of RdoA. If RdoA is part of a signaling pathway, it would be likely to have an effect on more than one target protein. The 2D gel analysis was carried out by comparing the protein profiles of serovar Typhimurium SL1344 pBAD18, SL1344 pND18, SL1344 rdoA pBAD18, and SL1344 rdoA pND18 all grown in the presence of arabinose. DsbA levels were observed to decrease by twofold in the rdoA null strain in an RdoA/NlpE overexpression-dependent manner via 2D gel electrophoresis (data not shown). Many other proteins were noted that also changed in abundance under NlpE overexpression conditions (N. Martin, unpublished observations). One of the more intriguing changes seen was an NlpE/RdoA-dependent shift in flagellin production. Figure 7 shows a portion of the 2D gels containing four different proteins that were identified by peptide mass fingerprinting via mass spectrometry, FliC, FljB, GroEL, and Trigger factor. In control cells (Fig. 7A), the flagellar protein type predominantly expressed is FliC. There is also a predominant spot identified as GroEL, with a much less abundant spot corresponding to Trigger factor. The overexpression of NlpE results in a switch to the predominant expression of FljB and also results in enhanced production of Trigger factor (Fig. 7B). When NlpE is overexpressed in the rdoA null strain, flagellar protein expression switches back to predominantly FliC and, in this case, Trigger factor levels stay approximately the same as when NlpE is overexpressed in wild-type cells. The protein profile (ratio of FljB to FliC) in the rdoA null strain with pBAD18 is very similar to that for the wild type without NlpE expression (Fig. 7D). The results show that the absence of RdoA alters the response to NlpE overproduction, presumably via the Cpx pathway, suggesting a role for RdoA in the transduction of Cpx-related signals to a subset of the Cpx-responsive targets.
FIG. 7.
Inducing the Cpx pathway alters flagellin expression (phase variation) in a Cpx/RdoA-dependent fashion. 2D gel analysis of the flagellins expressed in serovar Typhimurium under NlpE-inducing and noninducing conditions shows that the primary flagellin produced in wild-type cells containing pBAD18 is FliC (A). In wild-type cells containing pND18 (expressing NlpE), the flagellin switches to primarily FljB (B). In the absence of rdoA, cells containing pND18 revert to producing much less FljB (C) while cells containing pBAD18 also express predominately FliC (D). All cultures were grown in the presence of 0.2% arabinose for 2 h after addition at an OD600 of 0.6. Two additional protein spots, GroEL and Trigger factor, identified by mass spectrometry, are also indicated for comparison. Because small variations in total protein loading and staining can affect the overall spot intensity, it is important to compare the ratios of FljB to FliC in each gel rather than spot intensity.
DISCUSSION
This work sought to clarify the nature of transcripts originating from the rdoA promoter, namely the rdoA monocistronic and rdoA-dsbA cotranscripts, in the regulation of dsbA expression and also to examine the role of RdoA in serovar Typhimurium. The results clearly establish that rdoA promoter activity is growth phase dependent. This expression pattern occurred in wild-type and dsbA null background strains as well as in wild-type cells grown in rich LB media and minimal media at neutral pH. The growth-phase-dependent expression of the rdoA promoter was found to be independent of RpoS (data not shown). RpoS or σs is the stationary phase sigma factor that regulates the expression of a number of genes that are involved in the adaptive changes required for stress management during stationary phase (36, 39). Other promoters in E. coli, such as the P1 promoter of osmC gene (8), the promoter of the glgCAP operon (25), and the trxA thioredoxin gene promoter (38), have been reported to be growth phase regulated, also independently of RpoS.
Expression of the rdoA promoter is sensitive to the media conditions. For instance, minimal medium growth conditions increased promoter activity (approximately 300 to 400 Miller units) throughout growth. The dsbA promoter is also more highly active in minimal medium, (19) although the reason is not clear. In the case of acidic minimal medium, the rdoA promoter was turned off during stationary phase. While the factors responsible for such a decrease are unknown, the results point out that rdoA expression is not essential for viability of serovar Typhimurium during stationary phase under acid-stressed conditions. Studies of gene expression under low-pH conditions have focused mainly on inducible gene expression, although clearly some uncharacterized genes are turned off (15). Two major low-pH-inducible systems or acid tolerance responses (ATR) have been described for serovar Typhimurium, based on the growth phase at which each becomes induced (14). The log phase ATR system, induced when exponentially growing cells are adapted to pH 5.8 and the pH is lowered further to pH 4.5, results in the induction of 50 acid shock proteins, a subset of them members of the RpoS, PhoPQ, or fur regulons (5, 14). The second, a distinct RpoS- and fur-independent, stationary-phase ATR, induces more than 10 stationary phase acid shock proteins (2, 14). Few genes have been identified that are repressed by low-pH conditions, two examples being serovar Typhimurium OmpF, a major outer membrane porin (15), and Shigella sonnei VirF, a transcriptional activator of the IpaBCD host cell invasion apparatus (43).
Although the rdoA (this work) and dsbA (19) promoters both show strong growth phase dependence, different properties were revealed in terms of growth-stage-dependent increases in promoter activity. In the case of the dsbA promoter, the global regulator H-NS, a “histone-like” protein, has been demonstrated to modulate dsbA promoter expression during mid-exponential phase and plays a role in the repression of dsbA transcription in early-exponential-phase growth (C. V. Gallant, T. V. Ponnampalam, H. Spencer, and N. L. Martin, unpublished results). Although the exact regulatory mechanism behind the induction in rdoA expression remains to be elucidated, the continued high level of expression of the rdoA promoter during stationary phase suggests that the rdoA gene product may be important for stationary phase processes for serovar Typhimurium.
For a serovar Typhimurium dsbA null strain, an elevated response of the rdoA promoter (an approximately 200-Miller-unit increase) is observed compared with results for a wild-type strain at every stage of growth. These results indicate that a lack of disulfide bond-forming ability (provided by DsbA) in the periplasm has a positive feedback effect on rdoA transcription for serovar Typhimurium. This implies that a signal transduction mechanism exists linking the presence of DsbA (in the periplasm) to rdoA transcription (in the cytoplasm). Such a system is most likely the Cpx pathway, responding to a higher incidence of misfolded proteins in the periplasm as described for E. coli (11, 51). Although the cpxRA genes have been sequenced for serovar Typhimurium, this two-component signal transduction pathway has not been characterized for this organism. However, support for the Cpx pathway being involved in stimulating rdoA transcription comes from the following: (i) the information available for E. coli on the Cpx up-regulation of the homologous yihE gene (12, 51); (ii) the presence of a CpxR-P recognition sequence in the promoter of rdoA and the increase of rdoA promoter activity under conditions of E. coli NlpE overproduction, which is not seen in a cpxR null background; and (iii) the increase in the levels of the 1-kb rdoA monocistronic transcript for both wild-type and dsbA null strains with NlpE overexpression.
The increase in serovar Typhimurium rdoA promoter activity under NlpE-overproducing conditions, coupled to the parallel increases of the 1-kb rdoA transcript and the 0.7-kb dsbA transcript under similar conditions, suggests that Cpx-mediated signals affect dsbA expression, either directly at the dsbA promoter or indirectly through an rdoA-related mechanism. The presence of a conserved CpxR binding site in the rdoA promoter suggests that the Cpx response is mediated through rdoA; however, Northern blot analysis and RT-PCR of NlpE induced and uninduced serovar Typhimurium RNA preparations showed very minor variations in rdoA-dsbA cotranscript production, far less than expected if the cotranscript serves as a source for functional rdoA and dsbA transcripts. Homologues of yihE/rdoA identified in Pseudomonas (P. stutzeri, 45% protein identity; P. aeruginosa, 43% protein identity; P. fluorescens, 44% protein identity) and Shewanella putrefaciens (52% protein identity), as well as the Bacillus subtilis YerI protein (25% protein identity), are not found immediately upstream of these organisms' dsbA genes. In these bacteria, yihE and dsbA may be coregulated via a regulon, but in each of these cases the contribution of a yihE-dsbA cotranscript to DsbA expression levels is impossible. In serovar Typhimurium, if a cotranscript is synthesized, its processing must be extremely rapid and complete, such that no detectable difference in the levels of the bicistronic mRNA is observed between NlpE-overexpressed RNA preparations and uninduced RNA preparations, even in the sensitive RT-PCR based assay system used here. Evidence from other studies suggests that such rapid processing is unlikely. For instance, the well-characterized RNase E processing of the E. coli Pap operon, papBA transcript, clearly shows differential amounts of the polycistronic papBA mRNA under different promoter-inducing conditions (45, 46). In another example, substitution of the native promoter of the E. coli rph-pyrE operon with an inducible promoter and analysis of the transcripts synthesized under induced and uninduced conditions showed obvious variations in the levels of the dicistronic rph-pyrE mRNA (1). Rapid processing of an rdoA-dsbA cotranscript could be up-regulated by the Cpx pathway as an alternative explanation for the lack of significant difference in the levels of the bicistronic mRNA between NlpE-overexpressed and uninduced RNA preparations. However, such a mechanism that links the activation of the Cpx pathway and increased RNase activity or mRNA turnover has not been described to date in the literature. Preliminary RNase protection assays have failed to detect any additional evidence for rdoA-dsbA cotranscription products (data not shown). Transcriptional regulation of this rdoA/dsbA region is therefore unique in serovar Typhimurium, since Pogliano et al. (51) and Danese and Silhavy (11) established that the cotranscript induced via the Cpx pathway in E. coli plays a significant role in the regulation of dsbA expression. In our hands, a clear increase in the yihE-dsbA cotranscript and the yihE-specific transcript was detected by both Northern hybridization and RT-PCR, confirming the view that the cotranscript contributes to dsbA expression in E. coli. The presence of extremely limited amounts of larger-than-1-kb rdoA-containing transcripts in serovar Typhimurium therefore seems more likely to be due to leaky transcriptional termination between the rdoA and dsbA coding regions.
Although it is clear that rdoA has an effect on dsbA levels, the precise mechanism of rdoA influence remains to be elucidated for serovar Typhimurium. One potential posttranscriptional mechanism would involve rdoA mediating an increase in stability of the 0.7-kb dsbA transcript, such that this transcript accumulates under Cpx-induced conditions. An analogous mode of regulation occurs in the posttranscriptional control of polynucleotide phosphorylase (pnp) during cold acclimation for E. coli (68). The increase of pnp transcript during cold shock is mainly due to the increased stability of the pnp mRNA brought about by a protein-RNA interaction and less, if at all, to the activation of the two upstream promoters (68).
Alternatively, RdoA could be a transcriptional regulator involved in relaying or modulating Cpx-initiated signals. There is an NlpE/RdoA-dependent effect at the level of transcription of the dsbA promoter; however, it is still unknown if RdoA is directly mediating this effect or if RdoA acts via another component. By using 2D gel electrophoresis, it was also established that RdoA is involved in flagellar phase variation under NlpE-overproducing conditions. Flagellar phase variation has been under investigation for a number of years, but the environmental signals initiating phase variation are poorly understood. It is known that phase variation is mediated by the reversible inversion of a 996-bp portion of the chromosome that either positions a promoter allowing expression of FljB along with a repressor of FliC (FljA) or displaces this promoter so that FljB and FljA are not expressed and the FliC gene, which is at a separate chromosomal location, is expressed (54, 69, 70). This DNA inversion is mediated by the Hin recombinase (44). At another level of transcriptional control, the abundance of functional FliA, a flagellar gene-specific sigma factor, influences flagellar filament gene expression (32), but since both the fliC and fljB promoters are responsive to FliA, it is not likely to be involved with Cpx-mediated phase variation. The nature of the actual signaling pathways influencing phase variation will be the subject of further study.
It is also interesting to speculate on the reason that the Cpx pathway is involved in phase variation. Otto and Silhavy (47) recently presented convincing evidence that the E. coli Cpx pathway and NlpE are involved in sensing and responding to bacterial cell adhesion to environmental surfaces. They hypothesize that NlpE may be involved in relaying an adhesion-specific signal to the Cpx system, as they showed that NlpE is required for normal adhesion to occur and NlpE is unlikely to be able to act directly as an adhesin. A change in flagellar type may also be important in an initial interaction with a surface or some additional virulence aspect in serovar Typhimurium. Using “phase-locked” mutants of serovar Typhimurium, it has been demonstrated that FliC-expressing strains have a selective advantage over FljB-expressing strains in a murine model for typhoid fever, although no difference in the ability to invade epithelial cells or in the induction of enteropathogenesis was found between FliC- and FljB-expressing strains (28).
Although the E. coli yihE-dsbA and serovar Typhimurium rdoA-dsbA regions share more than 79% homology at the DNA level and both organisms are closely related, they differ significantly in terms of their pathogenicity and the types of environment encountered. Therefore, differences in the transcriptional regulation of dsbA and rdoA as a response to the need for these proteins, important for growth and survival in specific conditions, is not unexpected. One poorly understood issue is the mechanism of rdoA influence over dsbA expression and the ultimate role of RdoA in the bacterial cell. A DNA array study of yihE and dsbA null strains of Shigella flexneri showed significant alterations in transcription levels of more than 100 different genes involved in diverse processes such as glycolysis, central intermediary metabolism, structural cell proteins, and transport proteins (37), lending little clarity to the role of yihE. Curiously, in the same study dsbA mutants caused a slight decrease in cpxA/R transcription, contrary to previous studies showing induction of the Cpx pathway in the absence of disulfide oxidoreductase activity (51). The work described here has identified two proteins, DsbA and FljB, whose expression is at least partially dependent upon RdoA. Further studies aimed at elucidating the function of RdoA and its relationship to DsbA, flagellar phase variation, and the Cpx pathway for serovar Typhimurium are currently under way.
Acknowledgments
We are grateful to Tom Silhavy for providing the E. coli NlpE-overexpressing plasmid (pND18) and Tracy Raivio for providing MC4100 cpxR::Ω. We also acknowledge the Canadian Foundation for Innovation for providing funding to establish the Protein Function Discovery Facility at Queen's University, where the mass spectrometry work was carried out. We thank D. Hyndman of the Protein Function Discovery Facility for his assistance.
This work was supported by a Canadian Institutes of Health Research (CIHR) grant to N.L.M.
REFERENCES
- 1.Andersen, J. T., P. Poulsen, and K. F. Jensen. 1992. Attenuation in the rph-pyrE operon of Escherichia coli and processing of the dicistronic mRNA. Eur. J. Biochem. 206:381-390. [DOI] [PubMed] [Google Scholar]
- 2.Bang, I. S., B. H. Kim, J. W. Foster, and Y. K. Park. 2000. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:2245-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, J. C. 1994. Building bridges: disulphide bond formation in the cell. Mol. Microbiol. 14:199-205. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell, J. C., and J. Beckwith. 1993. The bonds that tie: catalyzed disulfide bond formation. Cell 74:769-771. [DOI] [PubMed] [Google Scholar]
- 5.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 7.Belin, P., and P. L. Boquet. 1994. The Escherichia coli dsbA gene is partly transcribed from the promoter of a weakly expressed upstream gene. Microbiology 140(Pt 12):3337-3348. [DOI] [PubMed] [Google Scholar]
- 8.Bouvier, J., S. Gordia, G. Kampmann, R. Lange, R. Hengge-Aronis, and C. Gutierrez. 1998. Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol. Microbiol. 28:971-980. [DOI] [PubMed] [Google Scholar]
- 9.Cosma, C. L., P. N. Danese, J. H. Carlson, T. J. Silhavy, and W. B. Snyder. 1995. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol. Microbiol. 18:491-505. [DOI] [PubMed] [Google Scholar]
- 10.Dailey, F. E., and H. C. Berg. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danese, P. N., and T. J. Silhavy. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 12.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 13.Fabianek, R. A., H. Hennecke, and L. Thony-Meyer. 2000. Periplasmic protein thiol:disulfide oxidoreductases of Escherichia coli. FEMS Microbiol. Rev. 24:303-316. [DOI] [PubMed] [Google Scholar]
- 14.Foster, J. W. 1999. When protons attack: microbial strategies of acid adaptation. Curr. Opin. Microbiol. 2:170-174. [DOI] [PubMed] [Google Scholar]
- 15.Foster, J. W., and H. K. Hall. 1990. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fourney, R. M., J. Miyakoshi, R. S. Day III, and M. C. Patterson. 1988. Northern blotting: efficient RNA staining and transfer. Focus 10:5-6. [Google Scholar]
- 17.Frech, C., M. Wunderlich, R. Glockshuber, and F. X. Schmid. 1996. Preferential binding of an unfolded protein to DsbA. EMBO J. 15:392-398. [PMC free article] [PubMed] [Google Scholar]
- 18.Freedman, R. B. 1995. The formation of protein disulphide bonds. Curr. Opin. Struct. Biol. 5:85-91. [DOI] [PubMed] [Google Scholar]
- 19.Goecke, M., C. Gallant, P. Suntharalingam, and N. L. Martin. 2002. Salmonella typhimurium DsbA is growth-phase regulated. FEMS Microbiol. Lett. 206:229-234. [DOI] [PubMed] [Google Scholar]
- 20.Grauschopf, U., J. R. Winther, P. Korber, T. Zander, P. Dallinger, and J. C. Bardwell. 1995. Why is DsbA such an oxidizing disulfide catalyst? Cell 83:947-955. [DOI] [PubMed] [Google Scholar]
- 21.Gross, C. A., C. L. Chan, and M. A. Lonetto. 1996. A structure/function analysis of Escherichia coli RNA polymerase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351:475-482. [DOI] [PubMed] [Google Scholar]
- 22.Guddat, L. W., J. C. Bardwell, and J. L. Martin. 1998. Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization. Structure 6:757-767. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, S., M. Abe, M. Kimoto, S. Furukawa, and T. Nakazawa. 2000. The dsbA-dsbB disulfide bond formation system of Burkholderia cepacia is involved in the production of protease and alkaline phosphatase, motility, metal resistance, and multi-drug resistance. Microbiol. Immunol. 44:41-50. [DOI] [PubMed] [Google Scholar]
- 25.Hengge-Aronis, R., and D. Fischer. 1992. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol. Microbiol. 6:1877-1886. [DOI] [PubMed] [Google Scholar]
- 26.Holmstrom, K., T. Tolker-Nielsen, and S. Molin. 1999. Physiological states of individual Salmonella typhimurium cells monitored by in situ reverse transcription-PCR. J. Bacteriol. 181:1733-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu, D., L. M. Shih, and Y. C. Zee. 1994. Degradation of rRNA in Salmonella strains: a novel mechanism to regulate the concentrations of rRNA and ribosomes. J. Bacteriol. 176:4761-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda, J. S., C. K. Schmitt, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, P. Adams, C. D. O'Connor, and A. D. O'Brien. 2001. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect. Immun. 69:3021-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishihara, T., H. Tomita, Y. Hasegawa, N. Tsukagoshi, H. Yamagata, and S. Udaka. 1995. Cloning and characterization of the gene for a protein thiol-disulfide oxidoreductase in Bacillus brevis. J. Bacteriol. 177:745-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joly, J. C., and J. R. Swartz. 1997. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry 36:10067-10072. [DOI] [PubMed] [Google Scholar]
- 31.Kamitani, S., Y. Akiyama, and K. Ito. 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 11:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlinsey, J. E., S. Tanaka, V. Bettenworth, S. Yamaguchi, W. Boos, S. I. Aizawa, and K. T. Hughes. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220-1231. [DOI] [PubMed] [Google Scholar]
- 33.Kingston, R. E. 1996. Analysis of RNA by Northern and slot blot hybridization. John Wiley & Sons Inc., New York, N.Y.
- 34.Klapper, A., B. MacKay, and M. D. Resh. 1992. Rapid high resolution western blotting: from gel to image in a single day. BioTechniques 12:650-654. [PubMed] [Google Scholar]
- 35.Kwon, A. R., T. G. Oh, D. H. Kim, and E. C. Choi. 1999. Molecular cloning of the arylsulfate sulfotransferase gene and characterization of its product from Enterobacter amnigenus AR-37. Protein Expr. Purif. 17:366-372. [DOI] [PubMed] [Google Scholar]
- 36.Lee, I. S., J. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155-167. [DOI] [PubMed] [Google Scholar]
- 37.Li, M. S., J. S. Kroll, and J. Yu. 2001. Influence of the yihE gene of Shigella flexneri on global gene expression: an analysis using DNA arrays. Biochem. Biophys. Res. Commun. 288:91-100. [DOI] [PubMed] [Google Scholar]
- 38.Lim, C. J., T. Daws, M. Gerami-Nejad, and J. A. Fuchs. 2000. Growth-phase regulation of the Escherichia coli thioredoxin gene. Biochim. Biophys. Acta 1491:1-6. [DOI] [PubMed] [Google Scholar]
- 39.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 40.Mileykovskaya, E., and W. Dowhan. 1997. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J. Bacteriol. 179:1029-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 42.Missiakas, D., and S. Raina. 1997. Protein folding in the bacterial periplasm. J. Bacteriol. 179:2465-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama, S., and H. Watanabe. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nanassy, O. Z., and K. T. Hughes. 1998. In vivo identification of intermediate stages of the DNA inversion reaction catalyzed by the Salmonella Hin recombinase. Genetics 149:1649-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilsson, P., S. Naureckiene, and B. E. Uhlin. 1996. Mutations affecting mRNA processing and fimbrial biogenesis in the Escherichia coli pap operon. J. Bacteriol. 178:683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsson, P., and B. E. Uhlin. 1991. Differential decay of a polycistronic Escherichia coli transcript is initiated by RNaseE-dependent endonucleolytic processing. Mol. Microbiol. 5:1791-1799. [DOI] [PubMed] [Google Scholar]
- 47.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pahl, H. L., and P. A. Baeuerle. 1994. Oxygen and the control of gene expression. Bioessays 16:497-502. [DOI] [PubMed] [Google Scholar]
- 49.Peek, J. A., and R. K. Taylor. 1992. Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 89:6210-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 51.Pogliano, J., A. S. Lynch, D. Belin, E. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 52.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 53.Rietsch, A., and J. Beckwith. 1998. The genetics of disulfide bond metabolism. Annu. Rev. Genet. 32:163-184. [DOI] [PubMed] [Google Scholar]
- 54.Silverman, M., and M. Simon. 1980. Phase variation: genetic analysis of switching mutants. Cell 19:845-854. [DOI] [PubMed] [Google Scholar]
- 55.Snyder, W. B., L. J. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum symbiotic plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 57.Spector, M. P. 1998. The starvation-stress response (SSR) of Salmonella. Adv. Microb. Physiol. 40:233-279. [DOI] [PubMed] [Google Scholar]
- 58.Tomb, J. F. 1992. A periplasmic protein disulfide oxidoreductase is required for transformation of Haemophilus influenzae Rd. Proc. Natl. Acad. Sci. USA 89:10252-10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turcot, I., T. V. Ponnampalam, C. W. Bouwman, and N. L. Martin. 2001. Isolation and characterization of a chromosomally encoded disulphide oxidoreductase from Salmonella enterica serovar Typhimurium. Can. J. Microbiol. 47:711-721. [PubMed] [Google Scholar]
- 60.Urban, A., M. Leipelt, T. Eggert, and K. E. Jaeger. 2001. DsbA and DsbC affect extracellular enzyme formation in Pseudomonas aeruginosa. J. Bacteriol. 183:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogel, T. J., and P. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 62.Warwicker, J. 1998. Modeling charge interactions and redox properties in DsbA. J. Biol. Chem. 273:2501-2504. [DOI] [PubMed] [Google Scholar]
- 63.Watarai, M., T. Tobe, M. Yoshikawa, and C. Sasakawa. 1995. Disulfide oxidoreductase activity of Shigella flexneri is required for release of Ipa proteins and invasion of epithelial cells. Proc. Natl. Acad. Sci. USA 92:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 65.Wunderlich, M., and R. Glockshuber. 1993. Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 2:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan, J. X., R. Wait, T. Berkelman, R. A. Harry, J. A. Westbrook, C. H. Wheeler, and M. J. Dunn. 2000. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 21:3666-3672. [DOI] [PubMed] [Google Scholar]
- 67.Yu, J. 1998. Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect. Immun. 66:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zangrossi, S., F. Briani, D. Ghisotti, M. E. Regonesi, P. Tortora, and G. Deho. 2000. Transcriptional and post-transcriptional control of polynucleotide phosphorylase during cold acclimation in Escherichia coli. Mol. Microbiol. 36:1470-1480. [DOI] [PubMed] [Google Scholar]
- 69.Zieg, J., M. Hilmen, and M. Simon. 1978. Regulation of gene expression by site-specific inversion. Cell 15:237-244. [DOI] [PubMed] [Google Scholar]
- 70.Zieg, J., and M. Simon. 1980. Analysis of the nucleotide sequence of an invertible controlling element. Proc. Natl. Acad. Sci. USA 77:4196-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]