Abstract
Sulfolobus solfataricus secretes an acid-resistant α-amylase (amyA) during growth on starch as the sole carbon and energy source. Synthesis of this activity is subject to catabolite repression. To better understand α-amylase function and regulation, the structural gene was identified and disrupted and the resulting mutant was characterized. Internal α-amylase peptide sequences obtained by tandem mass spectroscopy were used to identify the amyA coding sequence. Anti-α-amylase antibodies raised against the purified protein immunoprecipitated secreted α-amylase activity and verified the enzymatic identity of the sequenced protein. A new gene replacement method was used to disrupt the amyA coding sequence by insertion of a modified allele of the S. solfataricus lacS gene. PCR and DNA sequence analysis were used to characterize the altered amyA locus in the recombinant strain. The amyA::lacS mutant lost the ability to grow on starch, glycogen, or pullulan as sole carbon and energy sources. During growth on a non-catabolite-repressing carbon source with added starch, the mutant produced no detectable secreted amylase activity as determined by enzyme assay, plate assay, or Western blot analysis. These results clarify the biological role of the α-amylase and provide additional methods for the directed genetic manipulation of the S. solfataricus genome.
Sulfolobus solfataricus is a hyperthermophilic member of the archaea which inhabits acidic geothermal environments. It exhibits diverse modes of metabolism at high temperatures (70 to 90°C), including lithoautotrophy with sulfur and carbon dioxide (5, 23, 37) and chemoheterotrophy with sugars or amino acids (7, 15). Despite this metabolic flexibility, mechanisms regulating carbohydrate consumption by this organism are poorly understood. On occasion, plant-derived carbohydrates such as starch and cellulose contribute to the geothermal pool carbon cycle. In the absence of plant life, however, the endogenous microbial community itself may be an important source of reduced carbon compounds. For example, since hyperthermophilic archaea accumulate glycogen as an intracellular storage polymer (24), lysis could make glycogen available for consumption by surviving cells.
In hot acid environments polysaccharide degradation occurs rapidly, reflecting high rates of chemical hydrolysis and oxidation. Consequently, successful competition for these carbohydrates necessitates glycosyl hydrolase secretion. Secreted α-amylases promote the consumption of exogenous starch by releasing linear maltodextrins for subsequent assimilation. These enzymes have been classified into two sequence families (13 and 57) based on the presence of distinctive conserved domains (19). A growing number of glycosyl hydrolases, including α-amylases and pullulanases, have been characterized from hyperthermophilic archaea (8, 21, 32, 35), including members of family 57 (10, 22). However, the lack of suitable genetic methods for hyperthermophilic archaea have precluded studies on the biological significance of these enzymes.
S. solfataricus also produces an α-amylase which is secreted into the culture supernatant during growth on starch as the sole carbon and energy source (16). The purified enzyme is a homodimer with a subunit mass of 120 kDa and catalyzes the hydrolysis of starch, dextrin, and α-cyclodextrin. Assimilation of starch-derived carbon in this organism is coupled to production of a cell-associated α-glucosidase which converts maltodextrins into glucose (30, 31). S. solfataricus employs a catabolite repression (CR) system to regulate production of glycosyl hydrolases, including synthesis of the secreted α-amylase (3). Catabolite effects on α-amylase production affect induced and basal levels of enzyme synthesis. CR-I refers to the ability of added glucose to repress α-amylase synthesis in the presence of the inducer, starch (16). CR-II refers to a hierarchal relationship between α-amylase abundance and the type of sole carbon and energy source supporting growth, which is independent of inducer. (18). Aspartate was identified as the most repressing sole carbon source studied, while glutamate was the least repressing. A genetic component to the CR regulatory system governing α-amylase synthesis has also been described (17). Mutations in a locus called car decrease levels of the α-amylase. Efforts to understand how the CR system exerts its effects on α-amylase synthesis have been stymied by the lack of identity of the α-amylase structural gene. The studies described here were undertaken to remedy this problem.
MATERIALS AND METHODS
Archaeal strains and cultivation.
S. solfataricus strains were grown at 80°C at pH 3.0 in Brock's salts basal salts medium as described previously (30). Escherichia coli strain DH5α (Gibco-BRL) was used as described elsewhere (18) for propagating recombinant plasmids. Strain PBL2002 (lacS::IS1217) is a spontaneous derivative of wild-type S. solfataricus (PBL2000) with an insertion of IS1217 at position 1242 in lacS. Strain PBL2003 contains a modified but functional allele of lacS* lacking an EcoRV site produced by gene replacement at the lacS locus. Strain PBL2004 (amyA::lacS*) was derived from strain PBL2003 by targeted disruption. Tryptone, lactose, glucose, pullulan, and hydrolyzed starch from potatoes were added at a final concentration of 0.2% (wt/vol). l-Glutamic acid was used at a 5 mM concentration. Growth in liquid culture was monitored spectrophotometrically at a wavelength of 540 nm. Unless otherwise indicated, strains were subcultured from a glucose minimal medium. Starch plate assays employed a solid medium prepared using 0.6% (wt/vol) Gelrite (Kelco) and basal salts containing 5 mM glutamate and 0.2% (wt/vol) hydrolyzed starch from potatoes adjusted to pH 3.0 with sulfuric acid. Magnesium chloride was added at a final concentration of 8.0 mM to solidify the medium. Plates were incubated at 80°C in plastic containers with sufficient hydration to prevent dessication. A solution of 2% (wt/vol) potassium iodide and 0.6125% (wt/vol) iodine was added to the plates to visualize zones of starch hydrolysis. The procedure for preparation and use of competent S. solfataricus was as described previously (33). Prior to use, plasmids were treated with HaeIII methylase (New England Biolabs). Electrocompetent cells (2 × 109) were mixed with plasmid DNA, electroporated using a Gene Pulser (Bio-Rad), and then transferred to tryptone liquid medium and incubated for 8 h to allow recovery. Cells were then transferred to minimal lactose medium and incubated until growth was observed (typically 6 days). Lactose-utilizing cells were then plated on minimal lactose plates to obtain single colonies.
Recombinant DNA procedures.
All recombinant DNA procedures were as described previously (18, 29). Forward PCR primers for amyA were as follows: 1172F2, 5′ TGAAGGATCCATGAGCGTAAACTTTACTGTGTGAAGC 3′; 1172FA, 5′ TATACATATGATAAAAATTGCAATTTTAGCCATGGG 3′. The reverse primer was 1172R1, 5′ GTCAGGATCCGTAAGTTCCCATGGAACTTGTTGATTTGTT 3′. 1172F2 starts at position +124 relative to the amyA start codon, and 1172R1 starts 101 bp 5′ to the amyA stop codon. Both 1172FA and 1172R1 encode added BamHI sites. 1172FA starts at position +1 relative to the amyA start codon and includes an added NdeI site. PCR with 1172F2 and 1172R1 will produce a truncated amyA amplicon lacking sequences encoding 41 amino acids from the N-terminal end and 23 amino acids from the C-terminal end. Primers for lacS were as follows: forward primer LacSF, 5′ ATGCAATCCGGATACTAGGAGGAGTAGCATATAATTACG 3′; reverse primer LacSR, 5′ CAATGATCCGGAATCTAAACGACTTTCCAATTAGGCTAA 3′. Both LacSF and LacSR encode added BspEI sites. LacSF starts 169 bp 5′ to the lacS start codon, while LacSR starts 157 bp 3′ to the lacS stop codon. The lacS* allele was created using forward primer LacSE1 (5′ TTCAGTAACGTACATATAGAGATGATGATACCTATTCCAGTATTTCG 3′) and reverse primer LacSE2 (5′ CGAAATACTGGAATAGGTATCATCTCTATATG). Plasmid pAmy1 was constructed by insertion of a BamHI-digested PCR amyA amplicon produced with 1172F2 and 1172R1 into the BamHI site of pNEB193 (New England Biolabs). Plasmid pAmy2 was constructed by insertion of a BspEI-digested PCR lacS* amplicon into the BspEI site of pAmy1. The lacS* insertion in pAmy2 is positioned in a reverse orientation relative to amyA.
Protein and enzyme assays.
Protein concentrations were measured using the BCA protein assay reagent kit (Pierce). α-Amylase levels were determined using a dextrinization assay (27) as described previously (16) with the following modifications. A reaction mixture containing either culture supernatant clarified by centrifugation at 3,000 × g in an SS34 rotor and Sorval RC2B centrifuge at +4°C or purified enzyme was combined with 100 μg of starch in 10 mM sodium acetate (pH 3.5) and incubated at 80°C for 120 min. The reaction was terminated by cooling at 4°C for 5 min and then equilibrated to room temperature. Color was developed by addition of 0.005 ml of an iodine solution (4% [wt/vol] potassium iodide, 1.25% [wt/vol] iodine). The sample absorbance was determined at a wavelength of 600 nm and was corrected for an unincubated but otherwise identical sample. One unit of amylase activity was equivalent to the amount of protein which hydrolyzed 1 μg of starch in 1 min. All samples were assayed in duplicate, and the averages of the sample results are reported.
Protein electrophoresis, sequencing, Western blotting, and glycosylation analysis.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions using SeeBlue prestained molecular mass standards (Invitrogen). Prior to electrophoresis, samples were adjusted to 2% (wt/vol) SDS and 3 mM β-mercaptoethanol and boiled for 10 min. SDS-polyacrylamide gels were stained with Coomassie blue R250 to visualize protein. Peptide sequences were determined by tandem mass spectrometry electrospray analysis using material excised from SDS-PAGE gels. Gel slices were infused with trypsin to digest the protein, and the resulting peptides were recovered and separated by capillary electrophoresis prior to injection. Chemiluminescent Western blot analysis was performed with the ECL system (Amersham-Pharmacia Biotech) as described elsewhere (29). S. solfataricus culture supernatants were clarified by centrifugation at 3,000 × g for 30 min from cells grown as indicated. The supernatant was concentrated 100-fold by lyophilization prior to processing for SDS-PAGE. Purified α-amylase representing 10 ml of culture supernatant was used as a standard for Western blot analysis. Protein glycosylation was analyzed using purified α-amylase preparations and a glycan detection kit (Roche) with glycosylation control proteins, as described by the manufacturer.
Antibody production and immunoprecipitation assays.
Polyclonal antibodies for the α-amylase were prepared using BALB/c mice as described previously (4). Polyclonal antibodies for the β-glycosidase (LacS) were as described elsewhere (18). Immunoprecipitation assays for the presence of α-amylase employed active samples of clarified culture supernatant fractionated by High S Macro Prep ion-exchange chromatography (Bio-Rad) at room temperature. A 5-ml resin bed equilibrated with pH 3.5 10 mM sodium acetate buffer (SA) was loaded with culture supernatant at 1.5 ml/min using a peristaltic pump. The column was washed with three to five bed volumes of SA at a reduced flow rate of 0.3 ml/min, and bound protein was eluted with a 100-ml linear gradient of 0 to 1 M sodium chloride in SA at the same flow rate. Fractions were collected in 3-ml volumes and assayed directly. α-Amylase activity typically eluted at 0.1 M sodium chloride. A 200-μl aliquot of an active fraction was combined with 50 μl of anti-α-amylase antibodies or anti-β-glycosidase antibodies and incubated for 2 h at 4°C with gentle agitation. An equivalent of Staphylococcus aureus protein A (Sigma) suspension constituting 350 μg was added and the mixture was incubated for 3 h at 4°C with gentle agitation. The sample was clarified by centrifugation and the supernatant and protein A pellet were assayed for α-amylase activity.
Bioinformatic analysis.
Sequences used for phylogenetic analysis were derived by BLAST queries against the NCBI database by using the SSO1172 domain spanning positions 451 to 643. CLUSTAL W (36) and a 192-residue segment of the retrieved files were used to create multiple sequence alignments. PHYLIP 3.57c (11) and the distance method of analysis were used for phylogenetic studies. SEQBOOT was used to generate 100 bootstrapped data sets, distance matrices were determined using PROTDIST and the Dayhoff PAM matrix option, unrooted trees were inferred by neighbor-joining analysis using NEIGHBOR, CONSENSE was used to identify the most likely tree, and FITCH was used to create branch lengths proportional to distance values.
RESULTS
Identification of the α-amylase gene.
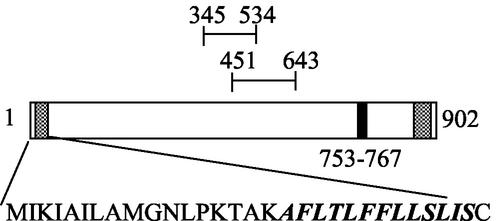
Tandem mass spectrometry electrospray analysis of α-amylase trypsinized fragments provided two internal peptide sequences of ALYTSTLLSEVF and EQALLYSTII. Since isoleucine and leucine have identical masses, the sequencing technique could not discriminate them. When adjustments were made for this constraint, a consensus peptide sequence of EQAL(L)YTST(I/L)(I/L)SEVF was derived. Queries of this sequence by FastaP against the S. solfataricus genome (reference 34 and http://www-archbac.u-psud.fr/projects/sulfolobus/) closely matched a region (13 of 14 residues) at positions 753 to 767 in the C-terminal third of a conserved hypothetical open reading frame (ORF) labeled SSO1172. SSO1172 could encode a protein with a predicted mass of 101 kDa and an isoelectric point of 5.05.
The purified S. solfataricus α-amylase has an apparent mass of 120 kDa as determined by SDS-PAGE and gel filtration chromatography (16). This is significantly larger than the predicted mass of the protein encoded by SSO1172. Consequently, additional efforts were conducted to confirm the relationship between the purified protein used to derive the peptide sequences and the α-amylase activity. Polyclonal antibodies prepared against the purified protein were used to test if the enzyme activity could be immunoprecipitated from cation exchange fractions of culture supernatant containing α-amylase activity (Table 1). α-Amylase activity precipitation was dependent on the type of antibodies used as well as on the addition of protein A and active chromatographic samples. α-Amylase activity remained in the supernatant despite addition of protein A and active sample fractions when control antibodies specific for an unrelated protein (LacS) were used instead. These results verified the relationship between the 120-kDa protein and α-amylase activity. Disparity between predicted and apparent protein masses could reflect posttranslational modification of the native protein, such as protein glycosylation. However, using the purified protein and a method involving sugar hydroxyl group derivatization with the hapten digoxigenin followed by Western blot analysis, no evidence of protein glycosylation was apparent.
TABLE 1.
Immunoprecipitation of α-amylase activity
| Reactant additions | Activitya (U/ml)
|
||||
|---|---|---|---|---|---|
| Protein A | AmyA | Anti-AmyA | Anti-LacS | Supernatant | Pellet |
| − | + | − | − | 34.11 | |
| + | + | − | − | 28.39 | <0.04 |
| + | + | + | − | <0.04 | 19.71 |
| + | + | − | + | 61.56 | <0.04 |
One unit of activity equals 1 μg of starch hydrolyzed per min per ml of sample.
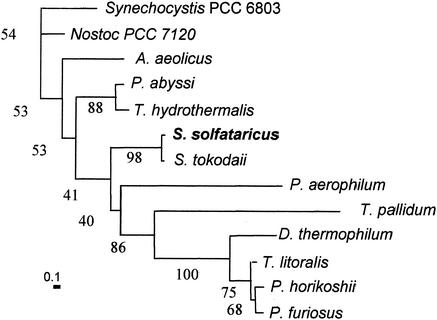
SSO1172 encodes a gene of 2,709 bp and 902 amino acids (Fig. 1). Regions of elevated hydrophobicity are apparent at both ends of the protein. Comparison between the N-terminal region of SSO1172 and that of other membrane-associated or secreted S. solfataricus proteins (1, 9) suggested that this region encoded a signal peptide. Comparison with other functionally related hyperthermophilic archaeal enzymes revealed conservation of a C-terminal hydrophobic region (10). An internal sequence spanning positions 345 to 534 exhibited homology to the glycosyl hydrolase family 57 (GHF 57) domain, providing further support for the identity of the gene. Based on these findings, a protein phylogeny for SSO1172 was prepared (Fig. 2). Sequences for comparison were recovered by BLAST using the amino acid sequences spanning the GHF 57 domain. However, a consensus neighbor-joining distance tree with a majority of bootstrap values greater than 50% was apparent only when using a C-terminal portion of the GHF 57 domain comprising 87 residues, combined with an additional C-terminal 105 residues. The S. solfataricus sequence clustered most closely with that of the related organism Sulfolobus tokodaii, followed by the crenarchaeote, Pyrobaculum aerophilum. Euryarchaeal sequences were also apparent but exhibited more divergent relatedness. Protein members of the glycosyl hydrolase family 13 contain four short conserved amino acid sequences (21), while a similar set of distinctive residues have not been reported for members of GHF 57. Multiple sequence alignments guided by the phylogenetic grouping evident in the nearest-neighbor distance tree were prepared to examine the possible occurrence of such residues. A large number of conserved residues are apparent from alignments of the crenarchaeal sequences (data not shown); however, inclusion of the most closely related euryarchaeal sequences greatly reduces the number of conserved positions. The conserved crenarchaeal residues present in the S. solfataricus α-amylase amino acid sequence are PFYHxxxPL (position 462 to 470), DxxxxIxxG (479 to 487), GxWxPEMAxD (501 to 510), IxxTxLxQ (522 to 529), PfxVxxxxGxxIxVxxRxxxLSNxxxFxxFxxxxxL (547 to 583), and LxxxY (625 to 629). The conserved archaeael residues are: PxxHxxxPxxxxxxxxxD (462 to 479), GxWxxExA (504 to 511), and IxxxxRxxxLSxxxxF (562 to 577).
FIG. 1.
Schematic of AmyA. Hatched regions designate hydrophobic sequences. The filled region indicates the location of the internal sequenced peptides. The overline labeled 345-534 indicates the location of the GHF 57 domain PFAM3065. The overline labeled 451-643 indicates the location of the sequences used for phylogenetic comparison. The putative N-terminal signal peptide is shown underneath, with the italicized and bolded portion showing a hydrophobic-rich region.
FIG. 2.
Phylogenetic analysis of α-amylase sequences. A neighbor-joining distance tree is shown, based on comparison of 192 residues. Distances are indicated by the bar in the lower left corner, which represents 10 substitutions per 100 residues. Percent occurrence among 100 trees is given for all nodes. Sequences were as follows: Synechocystis strain PCC 6803 (unknown protein), gi 1001420; Nostoc strain PCC 7120 (hypothetical protein), gi 17228805; Aquifex aeolicus (hypothetical protein), gi 2983309; Pyrococcus abyssi (putative amylopullulanase), gi 14520398; Thermococcus hydrothermalis (type II pullulanase), gi 4731920; S. solfataricus, gi 15898025; S. tokodaii (hypothetical protein), gi 15921357; Pyrobaculum aerophilum (hypothetical protein), gi 18314078; Treponema pallidum (putative α-amylase), gi 3322410; Dictyoglomus thermophilum (1,4-α-d-glucan glucanohydrolase), gi 113763; Thermococcus litoralis (4-α-glucanotransferase), gi 2351202; Pyrococcus horikoshii (putative α-amylase), gi 6647403; and Pyrococcus furiosus (α-amylase), gi 1351936.
Disruption of the α-amylase gene.
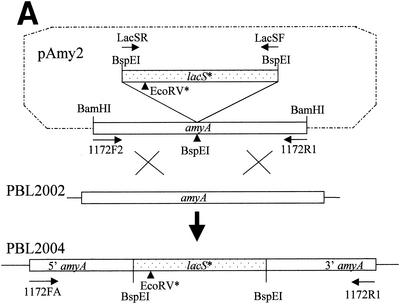
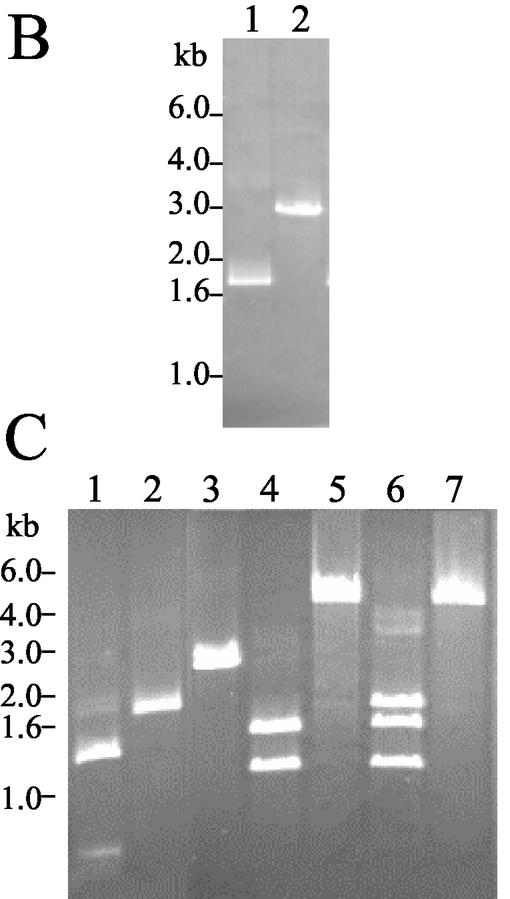
To test whether SSO1172 encoded an α-amylase, a loss-of-function mutation was constructed in this gene. The strategy for this construction was to interrupt the SSO1172 coding sequence by insertion of a functional marker gene (Fig. 3A). The resulting chromosomal recombinant could then be tested for altered phenotypic properties associated with the gene disruption. A modified but otherwise functional allele of lacS was used as the marker gene. The lacS modification was made by overlap extension PCR (20) using a primer encoding a single base difference (A to G) from the natural sequence. This change resulted in a silent substitution mutation at position 1132 relative to the lacS start codon and the loss of a unique EcoRV site. The modified lacS gene (lacS*) along with 169 and 157 bp of 5′ and 3′ flanking DNA sequence, respectively, was amplified by PCR using primers to add BspEI sites at either end and then cloned into pNEB193. The resulting plasmid was used to transform the Lac− strain, PBL2002, to lactose utilization in liquid medium. The lacS gene in PBL2002 was disrupted by spontaneous transposition of IS1217 (Fig. 3B). Lactose-utilizing recombinants were then purified and analyzed by PCR and restriction analysis, and one of these was named PBL2003. PCR amplification of lacS followed by restriction digestion was used to distinguish between the natural and modified lacS alleles present in strains PBL2000 and PBL2003 (Fig. 3C). A 1.8-kb PCR product was produced from the wild-type strain, PBL2000, which could be cut by EcoRV and formed two fragments of 1.3 and 0.5 kb (Fig. 3C, lane 1). The same size product was also produced from strain PBL2003, but this product was resistant to EcoRV digestion (Fig. 3C, lane 2). DNA sequence analysis demonstrated the presence of the A-to-G substitution mutation at position 1132.
FIG. 3.
Disruption of the amyA locus. (A) Schematic of the disruption strategy. The locations of PCR primers are indicated by arrows. Formation of the disrupted amyA allele requires two recombination events between plasmid pAmy2 and amyA which are indicated by the crossed lines. (B) PCR analysis of lacS in PBL2000 and PBL 2002 using primers LacSF and LacSR. Lane 1, PBL2000; lane 2, PBL2002. (C) PCR analysis of disrupted locus. Lane 1, lacS from PBL2000 amplified using LacSF and LacSR and digested by EcoRV; lane 2, lacS from PBL2003 amplified using LacSF and LacSR and digested by EcoRV; lane 3, amyA from strain PBL2002 amplified using 1172FA and 1172R1; lane 4, amyA amplified from strain PBL2003 and digested by BspEI; lane 5, amyA amplified from disrupted strain PBL2004 using 1172FA and 1172R1; lane 6, amyA amplified from disrupted strain PBL2004 using 1172FA and 1172R1 and digested by BspEI; lane 7, amyA amplified from disrupted strain PBL2004 using 1172FA and 1172 R1 and digested by EcoRV.
The modified lacS* allele present in strain PBL2003 was then amplified by PCR and used to disrupt a cloned and truncated copy of amyA, creating plasmid pAmy2 at a unique BspEI site (Fig. 3A). This plasmid construct was used to transform strain PBL2002 to lactose utilization. Lactose-utilizing recombinants were purified and characterized by PCR and restriction analysis. Two classes of recombinants were recovered at nearly equal frequencies: those which had undergone recombination at lacS and those which had undergone recombination at amyA. One of the latter group was named PBL2004. PCR amplification of amyA from strain PBL2002 using primers 1172FA and 1172R1 produced a fragment of nearly 3 kb (Fig. 3C, lane 3). Digestion of this product at a unique BspEI site located at position 1511 in the ORF produced two fragments of 1.5 and 1.3 kb, respectively (Fig. 3C, lane 4). PCR amplification of the amyA locus from strain PBL2004 produced a single fragment of 4.8 kb, nearly 1.8 kb larger than that observed with strain PBL2002 (Fig. 3C, lane 5). The change in amplicon size was consistent with the insertion of the lacS* allele. BspEI digestion of this product from PBL2004 produced three fragments of 1.3, 1.5, and 1.8 kb (Fig. 3C, lane 6). The two smaller fragments comigrate with those from BspEI digestion of the amyA amplicon produced from strain PBL2002. The identity of the lacS allele present in the disrupted amyA locus in strain PBL2004 was evident by the apparent loss of sensitivity of this PCR product to EcoRV digestion (Fig. 3C, lane 7).
Characterization of the amyA::lacS* disruption mutant.
The consequence of amyA disruption initially was assessed using a solid medium plate assay for α-amylase activity (Fig. 4). Cells of the wild-type and disruption mutant strains, PBL2000 and PBL2004, respectively, were grown in a minimal glucose medium and 2 × 108 cells were washed and applied to the surface of the starch-containing plate. The plating medium also contained glutamate as an alternative carbon and energy source. Glutamate is a nonrepressing carbon source for α-amylase synthesis (16). After 2 days of incubation at 80°C, the plate was flooded with iodine. A large zone of clearing was readily apparent from the wild-type strain, resulting from the action of secreted α-amylase (Fig. 4A). In contrast, no zone of clearing was evident for the amyA disruption strain (Fig. 4B), indicating an absence of secreted enzyme.
FIG. 4.
Starch plate assay. Strains were grown in liquid medium, washed, and applied to the plate. (A) Wild-type strain (PBL2000); (B) amyA-disrupted strain (PBL2004). After 2 days of incubation at 80°C, the plate was flooded with an iodine solution and photographed.
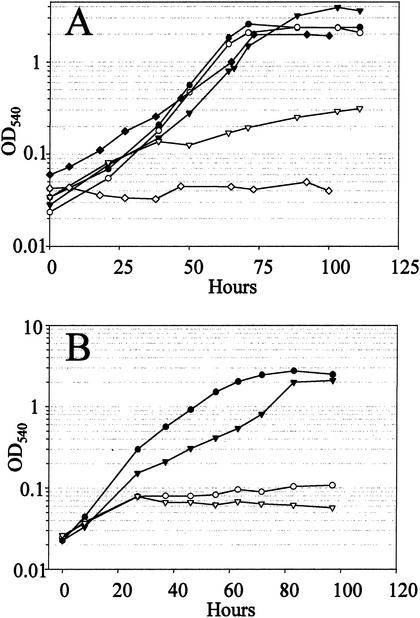
To assess the importance of the α-amylase in carbohydrate metabolism, growth of the wild-type and amyA mutant strains was compared in batch cultures using selected carbon sources (Fig. 5). Both strains grew well on glucose as the sole carbon and energy source (Fig. 5A). In contrast, at a starch concentration of 0.2% (wt/vol) the wild-type strain doubled about 5 times, achieving a final cell density of 2 × 109 cells/ml, while the disruption mutant exhibited no significant growth over the same period of incubation. Supplementation of the starch medium with the nonrepressing amino acid, glutamate, at a growth-limiting concentration allowed the mutant to grow for a limited period, while the wild-type strain achieved high cell density. This result suggested that the inability of the mutant to grow in the starch medium was not merely the result of energy deprivation precluding sufficient α-amylase synthesis to support transition to the new carbon and energy source. The mutant also failed to grow on either glycogen or pullulan (Fig. 5B). These results indicate that the α-amylase is essential for growth on starch, glycogen, and pullulan.
FIG.5.
Growth phenotypes. Growth of the wild-type strain, PBL2000 (filled symbols), and the amyA disruption strain, PBL2004 (open symbols), on selected carbon sources. (A) Glucose (circles), starch (diamonds), and starch plus glutamate (inverted triangles); (B) glycogen (circles) and pullulan (inverted triangles).
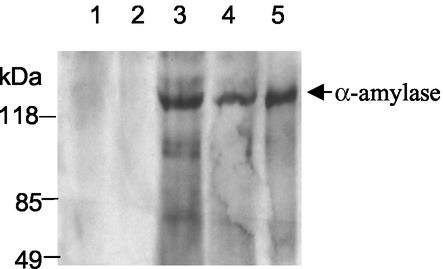
Levels of secreted α-amylase activity were measured for the wild-type strain and the amyA disruption mutant during growth on several carbon sources (Table 2). Clarified culture supernatants for the wild-type strain exhibited three- to fourfold-more secreted activity during growth on starch relative to that on glucose, reflecting the catabolite-repressing activity of glucose on α-amylase synthesis (16, 18). α-Amylase production also was evaluated during growth on glucose and on starch with added glutamate. Glutamate was added to provide an alternative carbon and energy source in addition to starch. Enzyme levels of the wild-type strain during growth in this medium were less than those observed on starch alone because the culture was sampled prior to glutamate exhaustion and before maximum levels of α-amylase secretion were achieved. The amyA disruption mutant produced no detectable secreted α-amylase activity during growth in glucose or glutamate with added starch medium, despite its ability to grow on these carbon sources. Cell yields in the latter medium were limited by the amount of added glutamate. To address the possibility that α-amylase was still produced by the mutant strain but in an inactive form, Western blot analysis of concentrated culture supernatants was conducted using cells grown on a medium containing starch and glutamate. Secreted α-amylase was detected in samples from the wild-type strain, PBL2000 (Fig. 6, lanes 3 and 4), which comigrated with a purified α-amylase standard (Fig. 6, lane 5). In contrast, no cross-reacting material was evident in concentrated culture supernatant samples from the amyA disruption mutant, PBL2004 (Fig. 6, lanes 1 and 2).
TABLE 2.
α-Amylase synthesis
| Strain/genotype | α-Amylase activity (U/ml)a
|
||||
|---|---|---|---|---|---|
| Starch | Glucose | Glucose and starchb | Pullulan | Glycogen | |
| PBL2000/amyA+ | 0.43 | 0.14 | 0.16 | 0.36 | 0.42 |
| PBL2004/amyA::lacS∗ | NGc | <0.04 | <0.04 | NG | NG |
One unit of activity equals 1 μg of starch hydrolyzed per min per ml of sample. Samples were analyzed at cell densities of 109/ml.
Samples were analyzed at cell densities of 108/ml.
NG, no growth.
FIG. 6.
Western blot analysis of culture supernatants. Western blotting samples were amyA-disrupted strain PBL2004 (lanes 1 and 2), wild-type strain PBL2000 (lanes 3 and 4), and purified α-amylase (lane 5). Lanes 2 and 4 contain 1.0-ml equivalent of supernatant; lanes 1 and 3 contain 3.0-ml equivalent of supernatant. Cross-reacting material was visualized by chemiluminescence.
DISCUSSION
The results presented here identify the gene (amyA) encoding the major secreted α-amylase (AmyA) of S. solfataricus. AmyA is essential for growth on starch but dispensable for growth on other carbon sources such as glucose. AmyA also is required for growth on glycogen and pullulan. Starch and glycogen are related glucose polymers differing primarily in chain length, while pullulan is comprised instead of α-1,6-linked maltotriose subunits. The finding that AmyA is required for glycogen utilization therefore is not unexpected; however, the failure of the mutant to utilize pullulan suggests AmyA also has significant activity towards this polymer and that the resulting hydrolytic products are readily assimilated and metabolized. Despite the existence of other ORFs in the annotated S. solfataricus genome with sequence homology to amylases and related glycosyl hydrolases (34), none is sufficient to replace the role of amyA in mediating exogenous starch utilization. The role of such genes in carbohydrate metabolism therefore remains to be determined. Such genes may be involved in the intracellular utilization of storage carbohydrates such as glycogen. The secreted α-amylase also is unrelated to another activity previously characterized from this organism which has also been referred to as an α-amylase (28). This latter activity is actually a cytoplasmic glycosyltrehalose trehalohydrolase whose function appears instead to be synthesis of the osmolyte trehalose.
Phylogenetic analysis of the S. solfataricus α-amylase protein sequence demonstrates that it is most closely related to archaeal proteins of similar function. These included enzymes with characterized α-amylase and pullulanase activities (10, 22, 26) as well as a single bacterial example from the anaerobe Dictyoglomus sp. (13). Interestingly, the archaeal orthologs occur in genomic locations flanked by putative carbohydrate transporters, suggesting that this group of genes may have a common evolutionary origin. In S. solfataricus, the α-amylase occurs adjacent to ORFs SSO1168 to 1171, which encode a cluster of sugar ABC transporters. Since all of the closest matching sequences were derived from thermophilic or hyperthermophilic organisms, certain residues present within the GHF 57 domain may contribute to thermostability or high-temperature enzyme optima. The conserved positions apparent in multiple sequence alignments of the crenarchaeal sequences may constitute such residues. Most of these positions in related euryarchaeal genes are not conserved, indicating that significant divergence has occurred across the archaeal subdivisions. The enzyme from S. solfataricus, however, must overcome the additional constraint of low pH. The conserved GHF 57 domain constitutes only a portion of the α-amylase sequence. Attempts to locate sequences with homology to the N-terminal half of the α-amylase yielded only matches to enzymes from hyperthermophilic acidophiles and not neutrophiles, suggesting that this region may play a role in mediating acid resistance.
The discrepancy between the apparent mass of the purified natural S. solfataricus α-amylase and the predicted mass of the amyA ORF remains to be explained. It is possible that some degree of secondary structure is maintained during sample processing and separation on SDS-PAGE, or that some form of posttranslational modification occurs, such as addition of a lipid or rare sugar. Studies on recombinant forms of the protein will facilitate studies on this question. Use of an alternative nonrepressing carbon source to support growth of the amyA mutant strain excluded the trivial possibility that lack of α-amylase synthesis resulted from an inability to make protein resulting from energy limitation. Availability of a loss-of-function amyA mutant provides a direction selection for gain-of-function derivatives. This phenotype may be useful in engineering alternative forms of this or other α-amylases to realize improved functional parameters.
Disruption of the S. solfataricus amyA locus is the first demonstration of directed chromosomal recombination in this organism or any member of the crenarchaeal subdivision of the archaea and the first example in a hyperthermophile. Directed recombination methods have been described for methanogenic and halophilic archaea belonging to the euryarchaeal subdivision (2, 6, 12, 14, 25, 38). The ability to selectively manipulate the S. solfataricus chromosome by site-directed recombination coupled with the availability of its genome sequence should allow studies which distinguish essential from nonessential loci as well as investigations into the role of selected genes in basic metabolic and regulatory processes. For example, as noted here, the repositioned copy of the S. solfataricus lacS gene maintained expression at sufficient levels to allow growth on lactose as the sole carbon and energy source. Thus, direct manipulations of the lacS sequence could be used to identify regions under control of the catabolite repression system. Current efforts concern the directed alteration of other S. solfataricus genes and refinement of the gene replacement method.
Acknowledgments
This research was supported by National Science Foundation grants MCB-9974453 and MCB-0085216.
REFERENCES
- 1.Albers, S. V., and A. M. Driessen. 2002. Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: a genomic survey. Arch. Microbiol. 177:209-216. [DOI] [PubMed] [Google Scholar]
- 2.Beneke, S., H. Bestgen, and A. Klein. 1995. Use of the Escherichia coli uidA gene as a reporter in Methanococcus voltae for the analysis of the regulatory function of the intergenic region between the operons encoding selenium-free hydrogenases. Mol. Gen. Genet. 248:225-228. [DOI] [PubMed] [Google Scholar]
- 3.Bini, E., and P. Blum. 2001. Archaeal catabolite repression: a gene regulatory paradigm. Adv. Appl. Microbiol. 50:339-366. [DOI] [PubMed] [Google Scholar]
- 4.Blum, P., J. Bauernfiend, J. Ory, and J. Krska. 1992. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J. Bacteriol. 174:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brock, T. D., K. M. Brock, R. T. Belly, and R. L. Weiss. 1972. Sulfolobus: a genus of sulfur oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84:54-68. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Kupiec, R., C. Blank, and J. A. Leigh. 1997. Transcriptional regulation in archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc. Natl. Acad. Sci. USA 94:1316-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rosa, M., A. Gambacorta, and J. D. Bu'lock. 1975. Extremely thermophilic acidophilic bacteria convergent with Sulfolobus acidocaldarius. J. Gen. Microbiol. 86:156-164. [DOI] [PubMed] [Google Scholar]
- 8.Dong, G., C. Vieille, A. Savchenko, and J. G. Zeikus. 1997. Cloning, sequencing, and expression of the gene encoding extracellular α-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl. Environ. Microbiol. 63:3569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elferink, M. G., S. V. Albers, W. N. Konings, and A. J. Driessen. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39:1494-1503. [DOI] [PubMed] [Google Scholar]
- 10.Erra-Pujada, M., P. Debeire, F. Duchiron, and M. J. O'Donohue. 1999. The type II pullulanase of Thermococcus hydrothermalis: molecular characterization of the gene and expression of the catalytic domain. J. Bacteriol. 181:3284-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—phylogeny inference package, version 3.2. Cladistics 5:164-166. [Google Scholar]
- 12.Ferrando-May, E., B. Brustmann, and D. Oesterhelt. 1993. A C-terminal truncation results in high-level expression of the functional photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. Mol. Microbiol. 9:943-953. [DOI] [PubMed] [Google Scholar]
- 13.Fukusumi, S., A. Kamizono, S. Horinouchi, and T. Beppu. 1988. Cloning and nucleotide sequence of a heat-stable amylase gene from an anaerobic thermophile Dictyoglomus thermophilum. Eur. J. Biochem. 174:15-21. [DOI] [PubMed] [Google Scholar]
- 14.Gernhardt, P., O. Possot, M. Foglino, L. Sibold, and A. Klein. 1990. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol. Gen. Genet. 221:273-279. [DOI] [PubMed] [Google Scholar]
- 15.Grogan, D. W. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haseltine, C., M. Rolfsmeier, and P. Blum. 1996. The glucose effect and regulation of α-amylase production in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 178:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haseltine, C., R. Montalvo-Rodriguez, A. Carl, E. Bini, and P. Blum. 1999. Extragenic pleiotropic mutations that repress glycosyl hydrolase expression in the hyperthermophilic archaeon Sulfolobus solfataricus. Genetics 152:1353-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haseltine, C., R. Montalvo-Rodriguez, E. Bini, A. Carl, and P. Blum. 1999. Coordinate transcriptional control in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 181:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janecek, S., E. Leveque, A. Belarbi, and B. Haye. 1999. Close evolutionary relatedness of α-amylases from archaea and plants. J. Mol. Evol. 48:421-426. [DOI] [PubMed] [Google Scholar]
- 22.Jeon, B. S., H. Taguchi, H. Sakai, T. Ohshima, T. Wakagi, and H. Matsuzawa. 1997. 4-α-Glucanotransferase from the hyperthermophilic archaeon Thermococcus litoralis: enzyme purification and characterization, and gene cloning, sequencing and expression in Escherichia coli. Eur. J. Biochem. 248:171-178. [DOI] [PubMed] [Google Scholar]
- 23.Kandler, O., and K. O. Stetter. 1981. Evidence for autotrophic CO2 assimilation in Sulfolobus brierleyi via a reductive carboxylic acid pathway. Zentralbl. Bakteriol. Hyg. Abt. 1 Orig. C 2:111-121. [Google Scholar]
- 24.Konig, J., R. Skorko, W. Zillig, and W.-D. Reiter. 1982. Glycogen in thermoacidophilic archaebacteria of the genera Sulfolobus, Thermoproteus, Desulfurococcus and Thermococcus. Arch. Microbiol. 132:297-303. [Google Scholar]
- 25.Krebs, M. P., E. N. Spudich, H. G. Khorana, and J. L. Spudich. 1993. Synthesis of a gene for sensory rhodopsin I and its functional expression in Halobacterium halobium. Proc. Natl. Acad. Sci. USA 90:3486-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laderman, K. A., K. Asada, T. Uemori, H. Mukai, Y. Taguchi, I. Kato, and C. B. Anfinsen. 1993. Alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus, cloning and sequencing of the gene and expression in Escherichia coli. J. Biol. Chem. 268:24402-24407. [PubMed] [Google Scholar]
- 27.Manning, G. G., and L. L. Campbell. 1961. Thermostable α-amylase of Bacillus stearothermophilus. J. Biol. Chem. 236:2952-2957. [PubMed] [Google Scholar]
- 28.Miura, Y., M. Kettoku, M. Kato, K. Kobayashi, and K. Kondo. 1999. High level production of thermostable α-amylase from Sulfolobus solfataricus in high-cell density culture of the food yeast Candida utilis. J. Mol. Microbiol. Biotechnol. 1:129-134. [PubMed] [Google Scholar]
- 29.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolfsmeier, M., and P. Blum. 1995. Purification and characterization of a maltase from the extremely thermophilic crenarchaeote Sulfolobus solfataricus. J. Bacteriol. 177:482-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolfsmeier, M., C. Haseltine, E. Bini, A. Clark, and P. Blum. 1998. Molecular characterization of the α-glucosidase gene (malA) from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 180:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savchenko, A., C. Vieille, and J. G. Zeikus. 2001. Alpha-amylases and amylopullulanase from Pyrococcus furiosus. Methods Enzymol. 330:354-363. [DOI] [PubMed] [Google Scholar]
- 33.Schleper, C., K. Kubo, and W. Zillig. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. USA 89:7645-7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeote Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunna, A., M. Moracci, M. Rossi, and G. Antranikian. 1997. Glycosyl hydrolases from hyperthermophiles. Extremophiles 1:2-13. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood, A. P., D. P. Kelly, and P. R. Norris. 1987. Autotrophic growth of four Sulfolobus strains on tetrathionate and the effect of organic nutrients. Arch. Microbiol. 146:382-389. [Google Scholar]
- 38.Zhang, J. K., A. K. White, H. C. Kuettner, P. Boccazzi, and W. W. Metcalf. 2002. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J. Bacteriol. 184:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]