Abstract
MpaA amidase was identified in Escherichia coli by its amino acid sequence homology with the ENP1 endopeptidase from Bacillus sphaericus. The enzymatic activity of MpaA, i.e., hydrolysis of the γ-d-glutamyl-diaminopimelic acid bond in the murein tripeptide l-alanyl-γ-d-glutamyl-meso-diaminopimelic acid, was demonstrated in the cell extract of a strain expressing mpaA from a multicopy plasmid. An mpaA mpl (murein peptide ligase) double mutant accumulated large amounts of murein tripeptide in its cytoplasm, consistent with the premise that MpaA degrades the tripeptide if its recycling via the peptidoglycan biosynthetic pathway is blocked.
About 40% of Escherichia coli cell wall murein is degraded each generation by lytic transglycosylases and endopeptidases (7, 11, 18). The resulting anhydro-muropeptides are efficiently recycled (11). One such example, N-acetylglucosamine-anhydro-N-acetylmuramyl-l-alanyl-γ-d-glutamyl-meso-diaminopimelyl-d-alanine (GlcNAc-anhMurNAc-l-Ala-γ-d-Glu-Dap-d-Ala),is transported into the cytoplasm via AmpG, a permease specific for muropeptides containing GlcNAc-anhMurNAc (3a, 11). It is rapidly broken down to yield GlcNAc, anhMurNAc, d-Ala, and murein tripeptide (l-Ala-γ-d-Glu-Dap), by NagZ, a β-N-acetylglucosamidase (3, 20), AmpD, an anhMurNAc-l-Ala amidase (9, 12), and LdcA, an l,d-carboxypeptidase (19). Murein tripeptide is then ligated to UDP-MurNAc by Mpl, a murein peptide ligase (13), and thus free tripeptide reenters the biosynthetic pathway for murein synthesis. The two amino sugars are also recycled for murein and lipopolysaccharide synthesis (15).
Over the course of our studies on cell wall recycling, two observations together strongly suggested that significant hydrolysis of murein tripeptide by an amidase must be taking place in the cytoplasm: (i) an mpl mutant accumulates an amount of murein tripeptide that is only a little more than 2-fold that of UDP-MurNAc-pentapeptide (13), while in an ampD deletion mutant, the amount of anhMurNAc-tripeptide present in the cytoplasm is about 40-fold more than the amount of UDP-MurNAc-pentapeptide (11); (ii) the amount of the murein dipeptide l-Ala-d-Glu present in the cytoplasm of the mpl mutant is much greater than in the wild type (13). Another indication of the presence of a γ-d-Glu-Dap amidase activity was the earlier demonstration of small amounts of Dap-d-Ala in the cytoplasm of E. coli (8). We therefore searched for the putative amidase, and here we report the identification of a gene, mpaA, and demonstrate that MpaA cleaves the γ-d-Glu-Dap amide bond in murein peptides.
Homology-based identification of a γ-d-Glu-Dap amidase in E. coli.
A sporulation-related enzyme, ENP1 from Bacillus sphaericus NCTC 9602, that cleaves the γ-d-Glu-Dap amide bond has been reported (1, 6, 10). It has two domains: a 100-amino-acid N-terminal domain consisting of two tandem LysM motif sequences involved in binding to peptidoglycan, and a 296-amino-acid C-terminal catalytic domain with a zinc binding site. In a search of the E. coli database, regions of the YcjI amino acid sequence displayed significant homology with the catalytic domain of ENP1 although the overall identity between the two sequences was low (13%). In particular, the putative zinc binding triad His162-Glu165-His307 of ENP1 corresponded to His69-Glu72-His177 of YcjI (Fig. 1). YcjI is encoded by an unknown open reading frame located at 29.9 min on the E. coli genome (2). YcjI appears to lack a signal sequence, and therefore YcjI presumably resides in the E. coli cytoplasm. We speculated that YcjI would be the putative amidase hydrolyzing the γ-d-Glu-Dap bond. Based on its biochemical activity reported below, YcjI has been named MpaA for murein peptide amidase A.
FIG. 1.
Alignment of the amino acid sequences of the ENP1 endopeptidase from B. sphaericus and YcjI (MpaA) from E. coli by T-COFFEE program (http://www.ch.embnet.org/software/TCoffee.html). Identical amino acids and conserved amino acids are indicated by stars and colons, respectively. Amino acid numbers for both proteins are indicated on the right. ENP1 and YcjI sequences are referred to as Q03415 and P51983 in the Swiss-Prot library. The putative zinc binding triad His162-Glu165-His307, catalytic dyad Tyr347-Glu366, and substrate binding residue Asp255 of ENP1 are underlined.
Accumulation of murein tripeptide in an mpl mpaA double mutant.
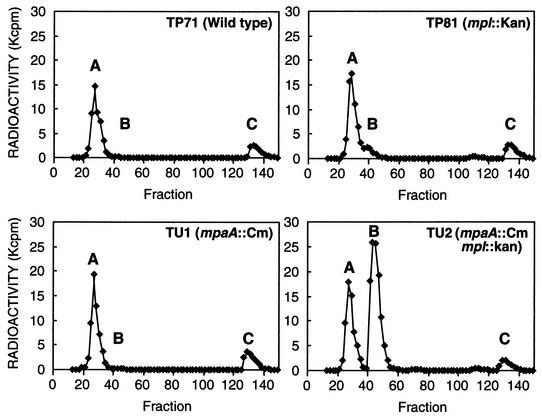
A mpaA null strain (mpaA::Cm) was constructed by one-step inactivation of the chromosomal gene in E. coli by using PCR and the phage λ Red recombinase (4). To generate mpaA::Cm, strain TP81 carrying pKD46 (Para/Red, Ampr) was transformed with a DNA fragment amplified by using the plasmid pKD3 (Cmr) as a template and primers 5′-GATACAAGATCGATGACATAGCGTTGAAGAAAAGGATTGTAGGCTGGAGCTGCTTCG and 5′-GATGCCAGCGCAGCAAGTTTGCCATCGCAAAGAGATATTATGAATATCCTCCTTAGT(the italicized sequence is homologous to the mpaA-flanking sequence, and the underlined sequence is homologous to the Cmr gene). mpaA::Cm was transduced into the wild-type strain, TP71 (11) and the isogenic mpl::Kan mutant strain, TP81 (this work), by transduction with T4gt7 phage (14) to form the mpaA::Cm mutant, TU1, and the mpaA::Cm mpl::Kan double mutant, TU2. Growth characteristics of TU1 and TU2 were similar to those of the parent TP71 in L broth at 37°C. If MpaA hydrolyzes murein tripeptide, much more tripeptide should accumulate in the mpl mpaA double mutant than in the mpl mutant. To compare the amounts of murein tripeptide present in the cytoplasm of the various strains, TP71 (wild type), TP81 (mpl::Kan), TU1 (mpaA::Cm), and TU2 (mpl::Kan, mpaA::Cm) were grown at 37°C with vigorous agitation in M9 minimal medium containing 1 μCi of [3H]Dap/ml (20 μCi/mmol; Moravek Biochemicals, Inc., Brea, Calif.) and supplemented with 0.2% glucose, 0.1% Casamino Acids, 1 mM MgCl2, 4 μg of thiamine/ml, and 100 μg each of lysine, threonine, and methionine/ml. When the turbidity of the culture reached 50 to 70 Klett units, cells were harvested, chilled rapidly, washed with cold water, resuspended in water, and boiled for 5 min. Boiled samples were centrifuged to remove cell debris. The hot-water extracts (supernatants) were analyzed by high-pressure liquid chromatography (HPLC) as described below. Samples were adjusted to a pH of ∼2.5 with trifluoroacetic acid (TFA), absorbed on a C18 Hypersil reverse-phase column (250 by 4.6 mm; 3-μm particle size) from Bischoff (Leonberg, Germany), and eluted at 0.5 ml/min with 0.05% TFA for 30 min, followed by a gradient from 0.05% TFA to 10% of acetonitrile containing 0.035% TFA over 40 min. Fractions of 0.25 ml were collected, and the radioactivity in each fraction was determined by liquid scintillation counting. As shown in Fig. 2, no tripeptide (peak B, identified as the murein tripeptide, l-Ala-γ-d-Glu-Dap, by mass spectrometry) was detectable in the mpl+ strains TP71 and TU1. As expected, the mpl mutant, TP81, had a readily detectable amount of tripeptide about equal in quantity to that of UDP-MurNAc-pentapeptide (peak C). In contrast, the mpl mpaA double mutant, TU2, accumulated a significantly larger amount (over 10-fold) of tripeptide. These results indicate that in the absence of Mpl, most of the murein tripeptide was hydrolyzed by MpaA.
FIG. 2.
HPLC analysis of hot-water extracts of E. coli wild type, the mpl::Kan mutant, the mpaA::Cm mutant, and the double mutant. As described in the text, cells were labeled with [3H]Dap at 37°C and harvested at mid-log phase. All values are corrected by the cell turbidity (5 ml of culture, 100 Klett units). Fraction A, Dap; fraction B, l-Ala-γ-d-Glu-Dap; fraction C, UDP-MurNAc-pentapeptide.
Direct demonstration of the γ-d-Glu-Dap amidase activity of MpaA.
To demonstrate the enzymatic activity of MpaA, we incubated whole-cell extracts with purified murein tripeptide (16) as a substrate. Enzymatic digests were analyzed by HPLC as described above, a procedure which separates Dap and murein tripeptide. Amounts of 0.1 mg of protein from extracts of TP71 (wild-type) cells or TU1 (mpaA::Cm) cells did not cleave any murein tripeptide when incubated at 37°C for 60 min in 0.1 ml of 0.1 M potassium phosphate buffer (pH 7.0). To demonstrate the hydrolysis of murein tripeptide by MpaA in vitro, mpaA was cloned (17) into the multicopy pGEM-T Easy vector (Promega, Madison, Wis.) by using a DNA fragment containing the mpaA gene and its putative σ70-regulated promoter sequence. The mpaA DNA fragment was amplified from the E. coli chromosome by PCR using 5′-TGCAAACAACTTCCGGC and 5′-TTTCGCCGATCTTGACG as primers, generating the plasmid pTmpaA, with the T7 promoter upstream of mpaA.
A strain carrying pTmpaA grew slowly in rich or minimal medium at 37°C, although it had normal morphology. A protein of the expected molecular weight of MpaA was not detectable by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in the extract of cells carrying pTmpaA even when MpaA was overexpressed from the T7 promoter in strain BL21(DE3). The low level of MpaA is probably due to the presence of the rare start codon, TTG. However, 0.1 mg of protein of whole-cell extract of uninduced TU2 carrying pTmpaA released 41% of the Dap from Dap-labeled murein tripeptide in 60 min at 37°C.
Specificity of MpaA.
Though UDP-MurNAc-pentapeptide contains a γ-d-Glu-Dap bond, an equal amount of extract from TU2/pTmpaA did not cleave this substrate. Hence, MpaA would not be expected to interfere with murein synthesis. Since the amount of anhMurNAc-tripeptide present in the ampD mutant is huge (11), one would predict that anhMurNAc-tripeptide must be a poor substrate for MpaA. This proved to be the case. A cell extract from TP73 (ΔampDE mutant) carrying pTmpaA did not hydrolyze anhMurNAc-tripeptide in vitro. Note that AmpD amidase efficiently cleaves the anhMurNAc-l-Ala bond of this substrate to release tripeptide (9, 12). These results indicate that MpaA is specific for cleavage of the γ-d-Glu-Dap bond of free murein tripeptide but not of peptides linked to muramic acid. The fact that Dap-d-Ala has been found in extracts of E. coli (8) indicates that MpaA cleaves murein tetrapeptide as well as murein tripeptide, but we have not tested this possibility directly. Unlike MpaA, ENP1 hydrolyzes the γ-d-Glu-DAP bond of MurNAc-tripeptide and MurNAc-tetrapeptide, as well as the amide bond of free tripeptide and tetrapeptide (1). The different substrate specificities of MpaA and ENP1 might be related to their cellular roles: ENP1 works on peptidoglycan of the forespore during sporulation, and MpaA works on free murein peptide in the recycling pathway.
Recently, Dawn et al. have shown that YcjG, divergently coded downstream of mpaA, is l-Ala-d/l-Glu epimerase, resulting in the conversion of l-Ala-d-Glu to l-Ala-l-Glu (5). These authors also cloned mpaA (ycjI), showed that MpaA does not cleave the γ-d-Glu-l-Lys bond of l-Ala-γ-d-Glu-l-Lys, and speculated that it may require the γ-d-Glu-Dap bond (5). Under nutrient-limiting conditions, MpaA together with the epimerase could function to metabolize murein tripeptide.
Acknowledgments
This work was supported in part by Public Health Service grant GM51610 from the National Institute of General Medical Sciences.
We thank Debabrata RayChaudhuri for his critical reading of the manuscript.
REFERENCES
- 1.Arminjon, F., M. Guinand, M.-J. Vacheron, and G. Michel. 1977. Specificity profiles of the membrane-bound γ-d-glutamyl-(l)meso-diaminopimelate endopeptidase and LD-carboxypeptidase from Bacillus sphaericus 9602. Eur. J. Biochem. 73:557-565. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, Q., H. Li, K. Merdek, and J. T. Park. 2000. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 182:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawn, M., Z. Schmidt, B. K. Hubbard, and J. A. Gerlt. 2001. Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as l-Ala-d/l-Glu epimerases. Biochemistry 40:15707-15715. [DOI] [PubMed] [Google Scholar]
- 6.Garnier, M., M.-J. Vacheron. M. Guinand, and G. Michel. 1985. Purification and partial characterization of the extracellular γ-d-glutamyl-(l)meso-diaminopimelate endopeptidase I, from Bacillus sphaericus NCTC 9602. Eur. J. Biochem. 148:539-543. [DOI] [PubMed] [Google Scholar]
- 7.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodell, E. W., and U. Schwarz. 1985. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J. Bacteriol. 162:391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtje, J.-V., U. Kopp, A. Ursinus, and B. Wiedemann. 1994. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol. Lett. 122:159-164. [DOI] [PubMed] [Google Scholar]
- 10.Hourdou, M.-L., M. Guinand, M.-J. Vacheron, G. Michel, L. Denoroy, C. Duez, S. Englebert, B. Joris, G. Weber, and J.-M. Ghuysen. 1993. Characterization of the sporulation-related γ-d-glutamyl-(l)meso-diaminopimelic-acid-hydrolysing peptidase I of Bacillus sphaericus NCTC 9602 as a member of the metallo(zinc) carboxypeptidase A family. Modular design of the protein. Biochem. J. 292:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs, C., L.-J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J.-M. Frere. 1995. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-L-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 13.Mengin-Lecreulx, D., J. van Heijenoort, and J. T. Park. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate: l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1992. A short course in bacterial genetics, a laboratory manual. p. 268-274. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park, J. T., D. RayChaudhuri, H. Li, S. Normark, and D. Mengin-Lecreulx. 1998. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate. J. Bacteriol. 180:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Shockman, G. D., and J.-V. Holtje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. New comprehensive biochemistry, vol. 27. Elsevier Science Publishers B. V., Amsterdam, The Netherlands.
- 19.Templin, M. F., A. Ursinus, and J.-V. Holtje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 18:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Votsch, W., and M. F. Templin. 2000. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J. Biol. Chem. 275:39032-39038. [DOI] [PubMed] [Google Scholar]