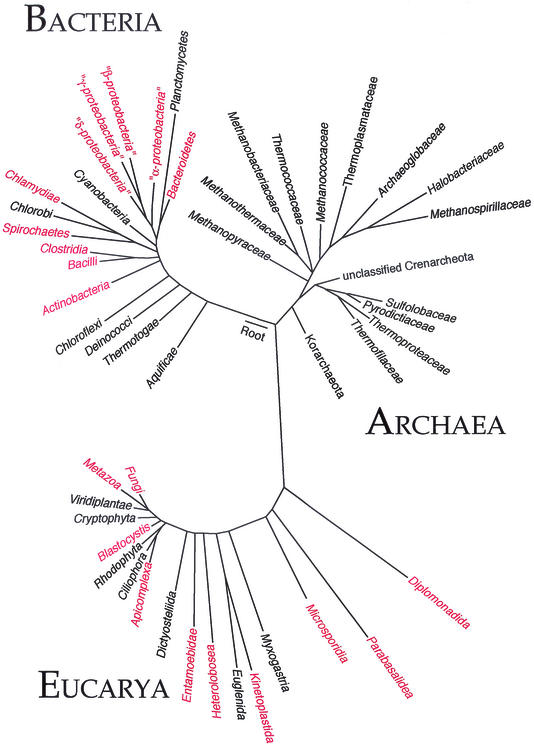
Over the past 50 years, the search for microorganisms in seemingly uninhabitable environments, such as at the extremes of pH, temperature, nutrient concentration, and pressure, has yielded a great deal of information about the diversity and origins of microbial life. Archaea, one of the three domains of life, is a highly diverse and abundant group of prokaryotes, and includes a number of “extremophiles” that thrive in such environments as hot springs, salt lakes, and submarine volcanic habitats (33). Recent molecular studies have also revealed that archaea, like bacteria, are commonly mesophilic (10). It is puzzling that despite being one of the most numerous and ubiquitous life forms on earth, no member of the domain Archaea has been described as a human pathogen (47) (Fig. 1).
FIG. 1.
Universal phylogenetic tree based on comparison of small subunit rRNA sequences. Phyla or divisions of Eucarya and Bacteria in which known human pathogens exist are shown in red. Archaea branches denote families rather than phyla, except for the Korarchaeota. Note that pathogens exist within the Fusobacteria, Acanthamoebidae, Stramenophiles, and Mesomycetozoa; however, these divisions are not pictured. (Modified from reference 44 with permission of the publisher.)
Archaea differ from eukarya and bacteria in terms of genetic, biochemical, and structural features. For example, archaea possess unique flagellins and ether-linked lipids (59) and lack murein in their cell walls (24). Multiple archaeal genomes have been sequenced to completion, contributing to a better understanding of the unique cellular processes of archaea and their role in adaptation to extreme environments. Despite the information emerging about archaeal genomes, structure, and function, much remains unknown. Over half of archaeal genes encode unique proteins with unknown functions (18). Difficulties in the isolation and cultivation of archaea also contribute to a relative lack of knowledge. No definitive virulence genes or factors have been described in archaea to date. Nevertheless, archaea may have the means, and they certainly have the opportunity, to cause disease.
Archaea share some characteristics with known pathogens that may reflect the potential to cause disease. Such characteristics include ample access to a host (i.e., opportunity) and capabilities for long-term colonization and coexistence with endogenous flora in a host. The detection of anaerobic archaea in the human colonic (36), vaginal (2), and oral microbial flora (1) demonstrates their ability to colonize the human host. Details regarding their survival in such human niches, including human immune system evasion and competition with normal human flora, however, are largely unavailable. Whether or not members of the Archaea possess virulence factors as commonly defined (i.e., the means for causing disease) is unclear. Of course, generalizations about a group of organisms as diverse as archaea are problematic. With this in mind, one might ask whether there are any fundamental features shared by archaea that should preclude them from acting as pathogens. In this paper, we discuss human diseases in which archaea may play a role as well as potential virulence characteristics of archaea, possible explanations for the current absence of information about archaea as pathogens, and molecular methods that might be utilized in the search for such pathogens.
POTENTIAL PATHOGENICITY OF ARCHAEA
Might archaea be capable of causing disease? Current data suggest that archaea are able to colonize and survive in humans. However, no concerted efforts have been undertaken to implicate archaea in human disease. The full spectrum of outcomes from these archaea-human interactions, whether it includes altered host physiology, tissue damage, or clinical disease, remains a mystery. In general, pathogens are distinguished from commensals by their reliance on a strategy for survival and replication in or transmission from a host that regularly leads to cellular, tissue, or organismal damage. Strategies can vary enormously, but they often define a signature for a family of pathogens. These strategies involve gaining access to the host in sufficient numbers, adhering to and colonizing a niche, evading host defenses, and multiplying in the host (50). If some archaea behave as pathogens they probably follow this basic scheme, but they may use fundamentally different mechanisms at some or all of these steps.
(i) Access and colonization.
The ubiquity of archaea in nature provides them ample opportunity for access to and colonization of susceptible hosts. In fact, we know of multiple niches for archaea in the human host. Methanogens, for example, have been identified among the human colonic (36), subgingival (1, 6, 29), and vaginal flora (2). Given their requirement for strictly anaerobic microenvironments, methanogens are likely to exist in human anatomic sites where well-characterized anaerobic bacteria flourish, possibly as coinhabitants.
Specific surface interactions between host cells and archaea have not been characterized, nor have specific mechanisms involved in cell or tissue tropism. However, potential means for movement and attachment have been described. Unique flagella have been identified for many genera of archaea, and such flagella have more structural similarities to the type IV pili of bacteria than they do to bacterial flagella (55). Multiple archaeal genomes contain partial tad loci, which may encode Tad-like proteins involved in fibril formation and surface adherence (23). Pyrodictium species, which are disk-shaped hyperthermophilic crenarchaeotes, grow in culture as mycelium-like cell masses (33). A network of hollow flagellum-like filaments connects the Pyrodictium cells in a web-like arrangement, the purpose of which is unclear. It is possible such structures may be involved in host-microbe adherence or microbe-microbe interactions within a potential host.
Once an archaeal pathogen reaches its specific host niche, it should be able to compete successfully with other endogenous flora for survival and persistence, in a manner similar to that of the known bacterial pathogens. Clues to the nature of archaeal interactions within the endogenous flora may be gleaned from recent studies of methanogen survival in arthropods. Methanogens have been identified in the hindguts of many terrestrial arthropods, including millipedes, cockroaches, termites, and scarab beetles (20). Three types of symbiosis have been described for such arthropods as follows: colonization of the hindgut lumen by free methanogens, association of methanogens with chitinous structures in the hindgut (chitinous bristles in the cockroach hindgut are covered with a mucous layer containing a complex biofilm), and the internalization of methanogens within intestinal anaerobic protists. Such protists include amoebomastigotes, flagellates, trichomonads, and ciliates (21). For example, methanogens resembling Methanobrevibacter species have been demonstrated in the cytoplasm of the ciliate Nyctotherus ovalis within the cockroach hindgut by electron microscopy (17). Sequence-based methods have also been used to identify methanogens in the termite hindgut (41). Methanogen-protozoa symbioses may be a common feature of many anaerobic ecosystems. The existence of such anaerobic protists containing archaeal species in the human intestinal flora has not been described to date.
In addition to the strict anaerobic conditions required for methanogen survival in the human colon, adaptation to the specific local nutrient environment must take place if methanogens are to compete with other members of the microflora. Syntrophy between archaea and sulfate-reducing bacteria has been described at the sulfate-methane interface in the sea floor and in methane-rich sediments (4, 11, 42). Syntrophy is a situation in which two or more organisms cooperate in the consumption of a substance (e.g., methane) not capable of being catabolized by either alone (33). It has been postulated that syntrophy occurs in the anaerobic microbial community of deep periodontal pockets, where methanogens consume H2 produced by secondary fermentors, and contributes to periodontal disease (7). It is possible that this energy-efficient metabolic process exists in other anaerobic communities within the human host as well. In a different setting, a relative lack of methane oxidizers compared with the number of sulfate-reducing bacteria in human intestinal microflora, with a resultant increase in local hydrogen sulfide, may contribute to human disease. For example, the presence of intestinal sulfate-reducing bacteria and an increase in potentially toxic levels of hydrogen sulfide have been postulated to play a role in the pathogenesis of ulcerative colitis, a form of inflammatory bowel disease in humans (31). Could a relative paucity or overabundance of archaea contribute to this and other human diseases? At the same time, perhaps the presence of archaea is necessary and required in certain anaerobic niches in order to maintain a desirable balanced biochemical microenvironment in the host.
(ii) Recognition by host defenses.
Pathology due to microbial infection usually results either from direct damage to the host by the microorganism or from a host inflammatory response. The latter arises from a variety of interrelated mechanisms, including innate recognition of pathogen-associated molecular patterns, active induction of host response pathways by pathogen virulence factors, and acquired capabilities for antigen recognition. The first of these mechanisms may be the most primitive and highly conserved across a wide range of known microbial virulence scenarios.
In thinking about the absence of known archaeal pathogens and the possibility that archaeal pathogens do in fact exist, one is drawn to the issue of archaeal cell wall structure, patterned molecules, and conserved features of the members of this domain. As noted above, archaea lack murein in their cell walls and their lipids are composed of branched phytonyl chains attached to glycerol backbones via ether bonds, in contrast to the ester-linked fatty-acyl chains common to bacteria and eucarya. In addition, lipopolysaccharide has not been demonstrated in archaea. When incorporated into liposomes (forming “archaeosomes”), the unique polar lipids of archaea have been shown to serve as potent immune adjuvants both in vitro and in vivo (28). Such archaeal lipid structures demonstrate more potent adjuvant activity than conventional adjuvants such as liposomes or alum in vivo, regardless of the source archaeal species (including numerous methanogens, thermophiles, and halobacteria) (27). For example, vaccination of mice with archaeosome-entrapped listeriolysin leads to rapid and prolonged specific immunity against Listeria monocytogenes infection and is superior to several other antigen-delivery vaccine strategies (9). Archaeosomes recruit and activate antigen-presenting cells by enhancing expression of major histocompatibility complex class II and costimulatory molecules and evoke strong antigen-specific responses to entrapped antigens when injected into the peritoneal cavities of mice, in a manner similar to that by lipopolysaccharide (28). These findings suggest specific recognition of archaeal polar lipids by cellular components of the mammalian human system. It remains unknown whether human Toll-like receptors are involved in such a process (34). Perhaps further investigation into the mechanisms of archaeosome adjuvanticity will provide insight into the specific interactions of archaea with the host immune system.
(iii) Evasion of host defenses.
Pathogens often rely on specific mechanisms to evade the host immune response. Such mechanisms include the elaboration of toxins or enzymes, the use of antiphagocytic capsules, modification of the microbial outer surface, and induced or deliberate internalization into host cells. It is unknown whether archaea have specific means to evade human immune responses. The paracrystalline cell surface S-layer of many archaea may play a role in evading an immune response, in a similar manner by which the S-layers of Campylobacter and Aeromonas species (16, 19) are postulated to play a role in virulence. Studies of some endosymbiotic methanogens in arthropods and ruminants based on transmission electron microscopy reveal these archaea inside membrane-bound vacuoles (15). However, it is not clear whether these organisms persist or multiply in this niche. Pathogens usually evolve a distinct set of biochemical strategies to achieve the goal of multiplication within a host cell. While archaea utilize a number of unique biochemical pathways to survive in nonhuman environments, it is not known whether such pathways are involved in archaeal survival in the human host or in other mesophilic environments. One expects that continued genome sequencing of archaea, functional studies of gene products, and the development of relevant models of host interaction will shed light on biochemical pathways and potential virulence factors involved in evasion of host defenses.
(iv) Genetic potential of archaea as pathogens.
Virulence genes are often associated with transmissible genetic elements (40). It is plausible that archaea, in the context of a complex population of human bacterial flora, have the potential for virulence gene acquisition by such mechanisms. Lateral genetic transfers between archaea and bacteria over evolutionary time periods have already been proposed, based in part on the genetic analysis of the recently sequenced bacterium Thermotoga maritima (37) and the discovery of archaeal genes that encode C1-transfer and methanogenic coenzymes in the aerobic bacterium Methylobacterium extorquens (8). Evidence of rapid, real-time exchange of genetic material among or with the archaea is not available. Some halobacteria contain extremely large plasmids (45), but it is presumed that these plasmids are not exchanged frequently in nature.
Full genome sequencing offers a more complete picture of virulence-associated genetic organization. At least 16 archaeal genome sequences have been completed (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html). Over 50% of the predicted genes have no obvious counterparts among known genes in bacteria or eukaryotes. In particular, none of these genes encode known toxins or virulence-associated products. The completed genomes of archaea, however, may still provide clues to the presence of possible virulence factors. For example, a survey of genes that encode putative transcriptional regulators in four archaeal genomes revealed possible members of the bacterial LysR and sensor transduction regulator families (30). Some of the members of these families are known to be associated with regulation of virulence factors in bacterial pathogens. Confirmation of virulence in archaea is fraught with difficulty, as there are few tractable genetic systems among these organisms, they have no clear virulence phenotype, and there are no obvious animal model systems in which to evaluate virulence.
POTENTIAL ASSOCIATIONS OF ARCHAEA WITH HUMAN DISEASE
Methanogens are the only archaea that have been identified in humans, despite human contact with other archaeal types, such as extreme halophiles (commonly found on such high-salt foods as sausages, salt pork, and fish (26). Methanogens were also once thought to be the exclusive archaeal component in the rumen, until Thermoplasma-associated sequences were discovered in this ecosystem using 16S rRNA sequence-based approaches (54). The failure to identify nonmethanogens in humans may be in large part due to the lack of any concerted efforts to define the abundance or diversity of archaea in human microenvironments. One might expect that if methanogens were found in diseased human tissue, they would coexist with common anaerobic bacterial species at the same site. Methanogens may follow virulence strategies in humans similar to those of the known anaerobic bacteria. Anaerobic infectious diseases are common in humans, especially at or near sites of colonization by the endogenous microflora, and they are usually associated with malignancy, gastrointestinal or genitourinary surgery, bite wounds, and aspiration (14). Such diseases are often polymicrobial, with routine cultivation techniques revealing the presence of multiple, endogenous anaerobic and aerobic bacteria. Anaerobes usually act as opportunists and require a breakdown in a normal anatomic barrier or delivery to a privileged anatomical site (e.g., surgery or trauma), as well as the concomitant presence of aerobes. Given their colonization of the alimentary tract in most humans, one might implicate methanogens in the causation of known anaerobic microbial processes usually attributed to members of the oral and gastrointestinal endogenous flora, e.g., brain, liver, and intra-abdominal abscesses, periodontal disease, and aspiration pneumonia.
Selective host genetic susceptibility to archaeal pathology is a possibility that is currently difficult to assess. This scenario presupposes specific archaeal recognition and response mechanisms. It could, however, explain how common members of the endogenous flora might contribute to disease in only a subset of hosts. The pathogenesis of inflammatory bowel disease in humans, for example, remains a mystery but may involve an inappropriate activation of the intestinal mucosal immune system in predisposed hosts (32). In theory, this immune response may be elicited in response to specific intestinal archaea, or conversely, immune tolerance to these organisms may be disrupted (13).
Methanobrevibacter smithii is the dominant methanogen isolated from the human colon (35, 36). Isolation of M. smithii involves the plating of fecal samples on antibiotic-containing medium and analyzing the incubating cultures for methane production (39). Cultures demonstrating the presence of methane production are selected for fluorescence microscopy, taking advantage of the natural fluorescence of methanogens (12). Interestingly, the isolation of M. smithii from fecal specimens has been shown to be more common among patients with diverticulosis than among normal subjects (57). Potential problems with the use of these isolation methods may contribute to the finding of only a single predominant archaeal species in humans. First, the use of antibiotics may inhibit the growth of some archaea. Archaeal antimicrobial susceptibilities have been poorly characterized. Second, reliance on methane production clearly excludes nonmethanogens. Third, the specificity and sensitivity of fluorescence microscopy for methanogen detection are not known but are likely to be suboptimal (33).
Measurement of breath methane excretion is a method that has been used to determine gut methane production (5) and may be an indirect means of determining the contribution of active methanogenic archaea to disease. Higher levels of breath methane have been detected in patients with precancerous conditions (ulcerative colitis and colonic polyposis) (46) and cancer of the colon (22, 46) than in healthy patients. Other studies have failed to confirm these findings and have noted that breath methane concentrations can be drastically altered by confounding factors such as the use of laxatives and enemas (25). There have been no prospective clinical trials using breath methane as a screening tool in the early identification of precancerous or malignant diseases of the colon. Thus, it is unclear whether methane excretion increases after such diseases develop or methanogen proliferation contributes to the pathophysiology of colonic diseases.
Several studies suggest susceptibility of archaeal species to currently available antimicrobial agents. For example, “defaunation” of cockroaches with low concentrations of metronidazole results in a rapid drop in methane production, presumably due to methanogen eradication from the hindgut (17). It is unknown whether such antibiotics directly kill methanogens in the lumen of the gut, kill their ciliate protozoal hosts, or effect the local anaerobic bacterial population indirectly by altering the concentrations of coexistent methanogens. Since archaeal cell wall polymers differ from bacterial peptidoglycans, antibacterial agents like vancomycin and β-lactams are expected to have no activity against archaea (24). In a study from Spain (6), a search for antibiotic resistance within the genus Halobacterium revealed that most extreme halophiles were resistant to β-lactams and aminoglycosides but were sensitive to many other antimicrobials, including macrolides, chloramphenicol, novobiocin, rifampin, bacitracin, and fluoroquinolones. Interestingly, certain archaea (Archaeoglobus spp., Pyrococcus, and Methanococcus jannaschii) have recently been discovered to contain efflux pumps of the resistance nodulation division superfamily (56). These efflux permeases transport hydrophobic drugs out of certain classes of bacteria, and they may serve similar functions in archaea. Antimicrobial therapy used in the routine clinical practice of medicine may have unrecognized effects on archaeal populations within the human body and, in some cases, may have led to beneficial effects due to unsuspected antiarchaeal activity.
CONCLUSION AND FUTURE STUDIES
Despite the rapid accumulation of information about archaeal biochemistry, phylogenetic diversity, and genetics, little is known about their diversity and abundance within the human body or the types of interactions in which archaea engage with human cells, components of the human immune system, or other members of the endogenous flora. No clear association between archaea and human disease has been described to date, in part due to limitations in our ability to detect, identify, and isolate these microorganisms. If archaea are involved in human disease, it is likely that such involvement will be elucidated using molecular methods, given the relative difficulty of their cultivation (48, 49, 58).
Among the non-cultivation-based approaches, several may prove useful for the identification of potential archaeal pathogens, including nucleic acid-based methods, gas chromatography, mass spectroscopy, and microscopy. Over the past decade, PCR amplification, cloning, and sequencing of phylogenetically informative molecules, such as small subunit ribosomal DNA, have become standard tools for identifying both known and unknown microorganisms in complex microbial communities (10, 43, 51). The large number of unique genes evident within archaeal genomes should provide additional opportunities for the selection of specific archaeal targets (3, 52). Although the use of DNA microarrays for high-throughput microbial classification and identification is still in its early developmental phases, this approach offers the advantage of massively parallel screening of nucleic acid molecules from complex populations. Quantification will be crucial in situations where the disease process is dependent on the relative abundance of the pathogen.
Gas chromatography and mass spectroscopy may be used to identify and detect unique archaeal lipid profiles in disease-associated sites (38, 42, 53). Reliability of detection and discrimination between different archaeal types must await development of more robust reference data sets. Microscopy can play a supporting role in connecting archaea to human disease. Fluorescent in situ hybridization is limited in sensitivity but allows physical associations to be made between phylogenetically reliable sequences and pathology. The potential utility of in situ immunofluorescence and immunohistochemistry for archaeal detection is suggested by the work of Doddema and Vogels (12). The limited availability of well-characterized and specific archaeal antigens poses problems for the near-future application of antibody-based methods.
When Archaea was first identified as a separate domain of life, its members were viewed as extremophiles and thus foreign to the human habitat and microbial ecosystem. This moniker was not a true reflection of archaeal physiology but rather a result of limited data and a narrow scientific perspective. Molecular approaches have now revealed archaea in decidedly nonextreme environments. It is not unreasonable that the application of similar techniques to the human “environment” may expand our perspective yet again.
Acknowledgments
We thank Norman R. Pace (University of Colorado) for his assistance with Fig. 1.
Editor: D. A. Portnoy
REFERENCES
- 1.Belay, N., R. Johnson, B. S. Rajagopal, E. C. de Macario, and L. Daniels. 1988. Methanogenic bacteria from human dental plaque. Appl. Environ. Microbiol. 54:600-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belay, N., B. Mukhopadhyay, E. Conway de Macario, R. Galask, and L. Daniels. 1990. Methanogenic bacteria in human vaginal samples. J. Clin. Microbiol. 28:1666-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhuiya, M. W., H. Sakuraba, C. Kujo, N. Nunoura-Kominato, Y. Kawarabayasi, H. Kikuchi, and T. Ohshima. 2000. Glutamate dehydrogenase from the aerobic hyperthermophilic archaeon Aeropyrum pernix K1: enzymatic characterization, identification of the encoding gene, and phylogenetic implications. Extremophiles 4:333-341. [DOI] [PubMed] [Google Scholar]
- 4.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 5.Bond, J. H., Jr., R. R. Engel, and M. D. Levitt. 1971. Factors influencing pulmonary methane excretion in man. An indirect method of studying the in situ metabolism of the methane-producing colonic bacteria. J. Exp. Med. 133:572-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonelo, G., A. Ventosa, M. Megias, and F. Ruiz-Berraquero. 1984. The sensitivity of halobacteria to antibiotics. FEMS Microbiol. Lett. 21:341-345. [Google Scholar]
- 7.Carlsson, J. 2000. Growth and nutrition as ecological factors, p. 68-130. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology: the molecular basis. Horizon Scientific Press, Wymongham, United Kingdom.
- 8.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 9.Conlan, J. W., L. Krishnan, G. E. Willick, G. B. Patel, and G. D. Sprott. 2001. Immunization of mice with lipopeptide antigens encapsulated in novel liposomes prepared from the polar lipids of various Archaeobacteria elicits rapid and prolonged specific protective immunity against infection with the facultative intracellular pathogen Listeria monocytogenes. Vaccine 19:3509-3517. [DOI] [PubMed] [Google Scholar]
- 10.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. F. 2000. Resolving a methane mystery. Nature 407:577-579. [DOI] [PubMed] [Google Scholar]
- 12.Doddema, H. J., and G. D. Vogels. 1978. Improved identification of methanogenic bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 36:752-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duchmann, R., I. Kaiser, E. Hermann, W. Mayet, K. Ewe, and K. H. Meyer zum Buschenfelde. 1995. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease (IBD). Clin. Exp. Immunol. 102:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finegold, S. M. 2000. Anaerobic bacteria: general concepts, p. 2519-2537. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.
- 15.Finlay, B. J., G. Esteban, K. J. Clarke, A. G. Williams, T. M. Embley, and R. P. Hirt. 1994. Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol. Lett. 117:157-161. [DOI] [PubMed] [Google Scholar]
- 16.Garduno, R. A., A. R. Moore, G. Olivier, A. L. Lizama, E. Garduno, and W. W. Kay. 2000. Host cell invasion and intracellular residence by Aeromonas salmonicida: role of the S-layer. Can. J. Microbiol. 46:660-668. [DOI] [PubMed] [Google Scholar]
- 17.Gijzen, H. J., C. A. Broers, M. Barughare, and C. K. Stumm. 1991. Methanogenic bacteria as endosymbionts of the ciliate Nyctotherus ovalis in the cockroach hindgut. Appl. Environ. Microbiol. 57:1630-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham, D. E., R. Overbeek, G. J. Olsen, and C. R. Woese. 2000. An archaeal genomic signature. Proc. Natl. Acad. Sci. USA 97:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grogono-Thomas, R., J. Dworkin, M. J. Blaser, and D. G. Newell. 2000. Roles of the surface layer proteins of Campylobacter fetus subsp. fetus in ovine abortion. Infect. Immun. 68:1687-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackstein, J. H., and C. K. Stumm. 1994. Methane production in terrestrial arthropods. Proc. Natl. Acad. Sci. USA 91:5441-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstein, J. H., and G. D. Vogels. 1997. Endosymbiotic interactions in anaerobic protozoa. Antonie Leeuwenhoek 71:151-158. [DOI] [PubMed] [Google Scholar]
- 22.Haines, A., G. Metz, J. Dilawari, L. Blendis, and H. Wiggins. 1977. Breath-methane in patients with cancer of the large bowel. Lancet ii:481-483. [DOI] [PubMed]
- 23.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandler, O., and H. Konig. 1998. Cell wall polymers in Archaea (Archaebacteria). Cell. Mol. Life Sci. 54:305-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlin, D. A., R. D. Jones, J. R. Stroehlein, A. J. Mastromarino, and G. D. Potter. 1982. Breath methane excretion in patients with unresected colorectal cancer. JNCI 69:573-576. [PubMed] [Google Scholar]
- 26.Kobayashi, T., B. Kimura, and T. Fujii. 2000. Haloanaerobium fermentans sp. nov., a strictly anaerobic, fermentative halophile isolated from fermented puffer fish ovaries. Int. J. Syst. E vol. Microbiol. 50(Pt. 4):1621-1627. [DOI] [PubMed] [Google Scholar]
- 27.Krishnan, L., C. J. Dicaire, G. B. Patel, and G. D. Sprott. 2000. Archaeosome vaccine adjuvants induce strong humoral, cell-mediated, and memory responses: comparison to conventional liposomes and alum. Infect. Immun. 68:54-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan, L., S. Sad, G. B. Patel, and G. D. Sprott. 2001. The potent adjuvant activity of archaeosomes correlates to the recruitment and activation of macrophages and dendritic cells in vivo. J. Immunol. 166:1885-1893. [DOI] [PubMed] [Google Scholar]
- 29.Kulik, E. M., H. Sandmeier, K. Hinni, and J. Meyer. 2001. Identification of archaeal rDNA from subgingival dental plaque by PCR amplification and sequence analysis. FEMS Microbiol. Lett. 196:129-133. [DOI] [PubMed] [Google Scholar]
- 30.Kyrpides, N. C., and C. A. Ouzounis. 1999. Transcription in archaea. Proc. Natl. Acad. Sci. USA 96:8545-8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine, J., J. K. Furne, and M. D. Levitt. 1996. Ashkenazi Jews, sulfur gases, and ulcerative colitis. J. Clin. Gastroenterol. 22:288-291. [DOI] [PubMed] [Google Scholar]
- 32.Macpherson, A., U. Y. Khoo, I. Forgacs, J. Philpott-Howard, and I. Bjarnason. 1996. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut 38:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Prokaryotic diversity: the Archaea, p. 546-572. In M. T. Madigan, J. M. Martinko, and J. Parker (ed.), Brock biology of microorganisms. Prentice-Hall, Inc., Upper Saddle River, N.J.
- 34.Means, T. K., D. T. Golenbock, and M. J. Fenton. 2000. The biology of Toll-like receptors. Cytokine Growth Factor Rev. 11:219-232. [DOI] [PubMed] [Google Scholar]
- 35.Miller, T. L., and M. J. Wolin. 1983. Stability of Methanobrevibacter smithii populations in the microbial flora excreted from the human large bowel. Appl. Environ. Microbiol. 45:317-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, T. L., M. J. Wolin, E. C. de Macario, and A. J. Macario. 1982. Isolation of Methanobrevibacter smithii from human feces. Appl. Environ. Microbiol. 43:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nesbo, C. L., S. L'Haridon, K. O. Stetter, and W. F. Doolittle. 2001. Phylogenetic analyses of two “archaeal” genes in Thermotoga maritima reveal multiple transfers between archaea and bacteria. Mol. Biol. Evol. 18:362-375. [DOI] [PubMed] [Google Scholar]
- 38.Nishihara, M., S. Nagahama, M. Ohga, and Y. Koga. 2000. Straight-chain fatty alcohols in the hyperthermophilic archaeon Pyrococcus furiosus. Extremophiles 4:275-277. [DOI] [PubMed] [Google Scholar]
- 39.Nottingham, P. M., and R. E. Hungate. 1968. Isolation of methanogenic bacteria from feces of humans. J. Bacteriol. 96:2178-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1099. [DOI] [PubMed] [Google Scholar]
- 41.Ohkuma, M., S. Noda, K. Horikoshi, and T. Kudo. 1995. Phylogeny of symbiotic methanogens in the gut of the termite Reticulitermes speratus. FEMS Microbiol. Lett. 134:45-50. [DOI] [PubMed] [Google Scholar]
- 42.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 43.Ovreas, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 45.Pfeifer, F., G. Weidinger, and W. Goebel. 1981. Characterization of plasmids in halobacteria. J. Bacteriol. 145:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pique, J. M., M. Pallares, E. Cuso, J. Vilar-Bonet, and M. A. Gassull. 1984. Methane production and colon cancer. Gastroenterology 87:601-605. [PubMed] [Google Scholar]
- 47.Reeve, J. N. 1999. Archaebacteria then…Archaes now (are there really no archaeal pathogens?). J. Bacteriol. 181:3613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 49.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 50.Relman, D. A., and S. Falkow. 2000. A molecular perspective of microbial pathogenicity, p. 2-13. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.
- 51.Sandaa, R. A., O. Enger, and V. Torsvik. 1999. Abundance and diversity of Archaea in heavy-metal-contaminated soils. Appl. Environ. Microbiol. 65:3293-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siebers, B., H. Brinkmann, C. Dorr, B. Tjaden, H. Lilie, J. van der Oost, and C. H. Verhees. 2001. Archaeal fructose-1,6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase. J. Biol. Chem. 276:28710-28718. [DOI] [PubMed] [Google Scholar]
- 53.Silvey, P., P. C. Pullammanappallil, L. Blackall, and P. Nichols. 2000. Microbial ecology of the leach bed anaerobic digestion of unsorted municipal solid waste. Water Sci. Technol. 41:9-16. [PubMed] [Google Scholar]
- 54.Tajima, K., T. Nagamine, H. Matsui, M. Nakamura, and R. I. Aminov. 2001. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol. Lett. 200:67-72. [DOI] [PubMed] [Google Scholar]
- 55.Thomas, N. A., S. L. Bardy, and K. F. Jarrell. 2001. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25:147-174. [DOI] [PubMed] [Google Scholar]
- 56.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 57.Weaver, G. A., J. A. Krause, T. L. Miller, and M. J. Wolin. 1986. Incidence of methanogenic bacteria in a sigmoidoscopy population: an association of methanogenic bacteria and diverticulosis. Gut 27:698-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson, K. H., R. Blitchington, R. Frothingham, and J. A. Wilson. 1991. Phylogeny of the Whipple's-disease-associated bacterium. Lancet 338:474-475. [DOI] [PubMed] [Google Scholar]
- 59.Zillig, W. 1991. Comparative biochemistry of Archaea and Bacteria. Curr. Opin. Genet. Dev. 1:544-551. [DOI] [PubMed] [Google Scholar]