Abstract
The shdA gene is carried on a 25-kb genetic island at centisome 54 (CS54 island) of the Salmonella enterica serotype Typhimurium chromosome. In addition to shdA, the CS54 island of Salmonella serotype Typhimurium strain LT2 contains four open reading frames designated ratA, ratB, sivI, and sivH. DNA hybridization analysis revealed that the CS54 island is comprised of two regions with distinct phylogenetic distribution within the genus Salmonella. Homologues of shdA and ratB were detected only in serotypes of Salmonella enterica subsp. I. In contrast, sequences hybridizing with ratA, sivI, and sivH were present in S. enterica subsp. II and S. bongori in addition to S. enterica subsp. I. Deletion of the ratA and sivI genes did not alter the ability of Salmonella serotype Typhimurium to colonize the organs of mice. Insertional inactivation of the sivH gene resulted in defective colonization of the Peyer's patches of the terminal ileum but normal colonization of the cecum, mesenteric lymph nodes, and spleen. Deletion of the shdA gene resulted in decreased colonization of the cecum and Peyer's patches of the terminal ileum and colonization to a lesser degree in the mesenteric lymph nodes and spleen 5 days post-oral inoculation of mice. A strain containing a deletion in the ratB gene exhibited a defect for the colonization of the cecum but not of the Peyer's patches, mesenteric lymph nodes, and spleen. The shdA and ratB deletion strains exhibited a shedding defect in mice, whereas the sivH deletion strain was shed at numbers similar to the wild type. These data suggest that colonization of the murine cecum is required for efficient fecal shedding in mice.
Salmonella-induced enterocolitis is currently the leading food-borne illness with a lethal outcome in the United States (33). The causative agents, nontyphoidal serotypes of Salmonella enterica subsp. I, are introduced into the human food supply primarily because these pathogens persist within populations of livestock and domestic fowl (11, 13, 15, 17-19, 44). Fecal contamination of the environment is the factor most important for the transmission of S. enterica subsp. I serotypes among animals on the farm and during transport (14, 16, 32, 45, 46). Slaughter of infected animals and the subsequent contamination of food products which may occur during processing explains the high prevalence of S. enterica subsp. I serotypes in meat and meat products in the United States (41).
Salmonella bongori and Salmonella enterica subsp. II, IIIa, IIIb, IV, VI, and VII can cause intestinal and extraintestinal infections in humans with symptoms that are indistinguishable from those resulting from infection with nontyphoidal serotypes of S. enterica subsp. I (1). However, human cases of infection with S. bongori or S. enterica subsp. II, IIIa, IIIb, IV, VI, and VII are rare (1) because these pathogens are mainly associated with cold-blooded vertebrates and are infrequently isolated from livestock, domestic fowl, or derived food products (38). These data suggest that serotypes of S. enterica subsp. I possess genetic determinants that enable them to persist in the intestines of livestock and domestic fowl but which are absent from serotypes of S. bongori and S. enterica subsp. II, IIIa, IIIb, IV, VI, and VII. Candidates for such genes have been identified by genomic comparison of Salmonella serotypes. Hybridization of genomic DNA from S. bongori, S. enterica subsp. IIIa, and S. enterica subsp. I serotypes Paratyphi A and B with a Salmonella enterica serotype Typhimurium LT2 microarray and comparison of the complete genome sequences identified a subset of 216 LT2 genes which have close homologues in one or several S. enterica subsp. I serotypes but are absent from Escherichia coli K-12, Klebsiella pneumoniae, S. enterica subsp. IIIa, and S. bongori (31, 39). Porwollik et al. point out that only 88 of these genes, including bigA, envF, sifAB, srfJ, srgAB, saf, stb, stc, std, stf, sti, and shdA, are named, which indicates that this group of genes has remained largely unstudied (39).
It was recently shown that one of these genes, shdA, is required for persistent shedding of Salmonella serotype Typhimurium with the feces from orally inoculated mice (29). The ShdA protein of Salmonella serotype Typhimurium, a member of the autotransporter family, is an outer membrane protein that binds fibronectin (28). The shdA gene is located on an approximately 25-kb island in the xseA-yfgK intergenic region of Salmonella serotype Typhimurium (31) at centisome 54 (CS54 island). The phylogenetic distribution of shdA and its role in the ability of Salmonella serotype Typhimurium to be shed with feces raises the question as to whether other genes carried on the CS54 island are required for intestinal persistence.
Here we describe the molecular characterization of the CS54 island of Salmonella serotype Typhimurium strain ATCC 14028. We investigate the extent of the subsp. I-specific DNA region and the contribution of the carried genes to the colonization of the mouse and shedding with feces.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The Salmonella reference collections B and C have been described previously (9, 10). Salmonella serotype Typhimurium strain IR715 is a virulent, nalidixic acid-resistant derivative of strain ATCC 14028 and has been described previously (43). E. coli strains S17-1 λpir and DH5α have been described previously (22, 42). Strains were routinely cultured aerobically at 37°C in Luria-Bertani (LB) broth supplemented with antibiotics as appropriate at the following concentrations: carbenicillin, 100 mg/liter; tetracycline, 20 mg/liter; chloramphenicol, 30 mg/liter; kanamycin, 100 mg/liter; and nalidixic acid, 50 mg/liter. For the detection of phoA expression, 20 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (XP)/liter was added to LB agar plates. For counter selection of the sacRB marker, bacteria were grown in sucrose broth as described previously (27). The bacteriophage P22 HT105/1 int mutant was used for generalized transduction of antibiotic resistance markers between Salmonella serotype Typhimurium strains as previously described (3).
Cloning and sequence determination of the S. enterica serotype Typhimurium xseA-yfgK intergenic region.
The cloning of the 3′ terminus of the xseA gene and the shdA gene on the recombinant cosmid pRK824 has been previously described (29). Recombinant clones pRA59, pRA64, pRA71, and pRA73 were derived from pRK824 by subcloning restriction fragments into vectors pUC18 or pBluescriptKS(−) in E. coli strain DH5α. The complete nucleotide sequence in the CS54 island was determined by using an AutoRead sequencing kit (Pharmacia) and an ALF automated sequenator. The nucleotide sequences were analyzed with the MacVector, version 6.0.1, software package (Oxford Molecular Group).
Southern hybridization.
Isolation of genomic DNA, digestion with restriction enzymes, and Southern transfer of DNA onto a nylon membrane were performed as previously described (2). Hybridization was performed at 65°C in solutions without formamide. Two 15-min washes were performed under nonstringent conditions at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate. Labeling of DNA probes with fluorescein-11-dUTP and detection with anti-fluorescein antiserum were performed with the Gene Images labeling and detection kit (Amersham-Pharmacia).
Construction of Salmonella serotype Typhimurium strains with deletions of shdA, ratA, ratB, sivI, and sivH.
To construct a strain carrying a deletion of the shdA gene (bp−5 to +6090) (Fig. 1) DNA regions flanking the shdA open reading frame (ORF) were amplified with the primers 5′ GCGGCGTAGATGAGAATACC 3′, 5′ GAAGATCTCGCACGGCGCTCCAGAC 3′, 5′ GAAGATCTCAATCTGCGCTATAACTGG 3′, and 5′ GGCCCGTCAGCAAACCGC 3′. Reaction products of the predicted size were digested with BglII and ligated by using the rapid DNA ligation kit (Roche) and cloned into the pCR2.1 vector (TA-cloning kit; Invitrogen) in E. coli strain DH5α. The resulting plasmid was designated pAH20. The XbaI/HindIII insert of pAH20 was subcloned into vector pUC18, yielding pAH24, and a BamHI-restricted kanamycin resistance cassette (KSAC; Pharmacia) was cloned onto the BglII site to give rise to plasmid pAH32. The entire insert of pAH32, consisting of the flanking regions of shdA with a kanamycin resistance cassette, was excised by using NotI and SpeI and subcloned into the XbaI/NotI-digested pEP185.2 vector (26) to give rise to plasmid pAH34. Plasmid pAH34 maintained in E. coli strain S17-1 λpir was introduced into Salmonella serotype Typhimurium strain IR715 by conjugal transfer, and exconjugants were selected on LB plates supplemented with nalidixic acid and kanamycin. An exconjugant that was resistant to kanamycin but sensitive to chloramphenicol was identified and designated strain AH9 (Fig. 1). Deletion of the shdA gene in AH9 was confirmed by Southern hybridization by using a shdA-specific probe derived from the pRA38 insert (29) (data not shown).
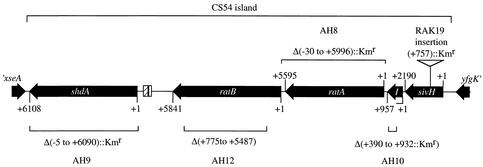
FIG. 1.
ORFs of the CS54 island of Salmonella serotype Typhimurium strain ATCC 14028 and deletion or insertion mutations constructed for phenotypic analysis. ORFs are indicated by arrows. The position of a dispersed repeat (cross-hatched bar) and remnant of an IS1 element (open bar) are indicated. The lengths (in base pairs) of the ORFs are indicated from the ATG codon (+1) to the stop codon. The position of the deletion or insertion in shdA, ratB, ratA, sivI, or sivH in strains AH9, AH12, AH8, AH10, and RAK19 are indicated.
The construction of a Salmonella serotype Typhimurium strain in which the wild-type shdA gene was reintroduced into strain AH9 for complementation (strain RAK60) was as follows. A DNA fragment at the 3′ end of the shdA ORF was amplified by PCR with primers 5′ GCTCTAGAGCGGCGTAGATGAGAATACC 3′ and 5′ CCGATATCACATATTTCGCACGGCGCTC 3′. The PCR product was digested with XbaI and SmaI, ligated into pFUSE (4) previously restricted with XbaI and SmaI, and transformed into E. coli S17-1 λpir to yield plasmid pRA133. Plasmid pRA133 was introduced into Salmonella serotype Typhimurium strain IR715 by conjugal transfer. Exconjugants that had pRA133 recombined into the chromosome were selected on LB plus nalidixic acid and chloramphenicol, and one nalidixic acid-resistant and chloramphenicol-resistant exconjugant was designated strain RAK23. The chromosomal construct was confirmed by Southern hybridization of EcoRV-digested RAK23 genomic DNA by using the pRA133 insert as a probe (data not shown). A bacteriophage P22 HT105/1 int mutant lysate propagated on strain RAK23 was used to transduce the chloramphenicol resistance marker of pRA133 present in the chromosome adjacent to the shdA stop codon into strain AH9. Transductants were selected for by plating on LB-plus-chloramphenicol plates containing 5 mM EGTA. Cotransduction of the wild-type shdA gene into strain AH9 for complementation was tested for by replica plating transductants on LB-plus-chloramphenicol plates and LB-plus-kanamycin plates. A phage P22-sensitive, chloramphenicol-resistant, kanamycin-sensitive transductant was designated RAK60 and purified from contaminating P22 phage by streaking to single colonies twice on Evans blue uracil (EBU) plates (7).
To construct a strain carrying a deletion of the ratA gene (−30 to +5996) (Fig. 1), DNA regions flanking the ratA ORF were amplified with the primers 5′ GGTGAGGGTGGTAAAAATCACGTC 3′, 5′ GAAGATCTTTCTCAGGTACGTCAGTGAAATCG 3′, 5′GAAGATCTTCGTAATTCAACCCACTTTGC 3′, and 5′ GGCCGCTGATGCGGATCG 3′. Reaction products of the predicted size were digested with BglII, ligated by using the rapid DNA ligation kit (Roche), and cloned into the pCR2.1 TOPO vector (TA-cloning kit; Invitrogen) in E. coli TOP10 cells. The resulting plasmid was designated pAH10. The XbaI/HindIII insert from pAH10 was subcloned into vector pUC18, yielding pAH18. A BamHI-restricted kanamycin resistance cassette (KSAC; Pharmacia) was cloned into the BglII site to give rise to plasmid pAH31. The SpeI/NotI insert from pAH31 was subcloned into the suicide vector pEP185.2, yielding plasmid pAH33. Plasmid pAH33 maintained in E. coli strain S17λpir was introduced into Salmonella serotype Typhimurium strain IR715 by conjugal transfer, and exconjugants were selected on LB plus nalidixic acid and kanamycin. A kanamycin-resistant, chloramphenicol-sensitive exconjugant was designated strain AH8. Deletion of the ratA gene in AH8 was confirmed by Southern hybridization with a ratA-specific probe derived from pRA64 (data not shown).
The construction of a strain carrying a nonpolar deletion in the ratB (+775 to +5487) (Fig. 1) gene was as follows. DNA regions flanking the ratB gene were amplified with primers 5′ TTAAGCATGAGATCGTCTGCTCC 3′ and 5′ GAAGATCTTCAATACCGCTATCCGTTTGGG 3′ and primers 5′ GAAGATCTCATTGTTTACCACGCCCTGG 3′ and 5′ GGAATGAACTGCAACGAAATGGAC 3′. Both PCR products were digested with BglII and ligated together by using the rapid DNA ligation kit (Roche) and subcloned into the pCR2.1 TOPO vector (TA-cloning kit; Invitrogen) to yield plasmid pAH8-2. The XbaI/HindIII insert from pAH8-2 was subcloned into pUC18, yielding pAH17. The BamHI fragment from pAH17 was subcloned into the suicide vector pRDH10 (27) that had been digested with BamHI to give rise to plasmid pAH40. Plasmid pAH40 maintained in E. coli strain S17λpir was introduced into Salmonella serotype Typhimurium strain IR715 by conjugal transfer, and exconjugants were selected on LB plus nalidixic acid and chloramphenicol. Plasmid pRDH10 carries the sacRB locus, which may be used as a counter selectable marker during growth in the presence of 6% sucrose and in the absence of NaCl at 30°C. In order to select for the loss of pRDH10 from the chromosome due to a recombination between homologous DNA derived from the chromosome and pAH40, exconjugants were grown in sucrose broth for 18 h at 30°C with shaking. Serial 10-fold dilutions were plated on sucrose plates and incubated at 30°C for 18 h. Colonies growing on these plates were replica plated on LB plus nalidixic acid and LB chloramphenicol, and several chloramphenicol-sensitive variants were selected. To distinguish between variants that had lost the pRDH10 plasmid from the chromosome by homologous recombination resulting in regeneration of the wild-type ratB locus from those resulting in the introduction of a deletion in the ratB locus, primers 5′ TTAAGCATGAGATCGTCTGCTCC 3′ and 5′ GGAATGAACTGCAACGAAATGGAC 3′ were used to amplify the sequence across the deletion. One resolved exconjugant was designated AH12. The deletion of ratB in strain AH12 was confirmed by Southern hybridization by using a probe constructed from plasmid pAH17 (data not shown).
The construction of a Salmonella serotype Typhimurium strain in which the wild-type ratB gene was reintroduced into strain AH12 for complementation (strain RAK58) was as follows. A DNA fragment at the 3′ end of the ratB ORF was amplified by PCR with primers 5′ GAAGATCTCGCACGGCGCTCCAGAC 3′ and 5′ GCGGCGTAGATGAGAATACC 3′. The PCR product was digested with XbaI and SacI ligated into pGP704 (26) previously restricted with XbaI and SacI and transformed into E. coli S17-1 λpir to yield plasmid pMAR4. Plasmid pMAR4 was introduced into Salmonella serotype Typhimurium strain IR715 by conjugal transfer. Exconjugants that had pMAR4 recombined into the chromosome were selected on LB plus nalidixic acid and carbenicillin, and one nalidixic acid-resistant and carbenicillin-resistant exconjugant was designated strain MRZ4. The chromosomal construct was confirmed by Southern hybridization (data not shown). A bacteriophage P22 HT105/1 int mutant lysate propagated on strain MRZ4 was used to transduce the carbenicillin resistance marker of pMAR4 present in the chromosome adjacent to the ratB stop codon in strain AH12. Transductants were selected for by plating on LB-plus-carbenicillin plates containing 5 mM EGTA. Colony PCR with primers 5′ GAATTCCTCAACGCCGCGAAGGTC 3′ and 5′ GTCGACCTAACCGGATGTCAGCCCTAC 3′ was used to test for cotransduction of the wild-type ratB gene into strain AH12. A phage P22-sensitive transductant that was positive in the PCR screen was designated RAK58 and purified from contaminating P22 phage by streaking to single colonies twice on EBU plates (7).
To construct a strain carrying a deletion of the sivI gene (+390 to +932) (Fig. 1), DNA regions flanking the sivI ORF were amplified with primers 5′ CCCCACCTTCACTTTTATGC 3′, 5′ GAAGATCTTGGTCTGTCCATCGACTACAAACG 3′, 5′GAAGATCTGGTCGCTGCCATTCTGATC 3′, and 5′ GCGTCAGCGTTGTGGCTG 3′. Reaction products of the predicted size were digested with BglII, ligated with the rapid DNA ligation kit (Roche) into the pCR2.1 TOPO vector, and transformed into E. coli TOP10 cells (TA-cloning kit; Invitrogen). The resulting plasmid was designated pAH9. The XbaI/HindIII insert from pAH9 was subcloned into the vector pUC18, yielding pAH19. A BamHI-restricted kanamycin resistance cassette (KSAC; Pharmacia) was cloned into the BglII site to give rise to plasmid pAH22. The SpeI/NotI insert from pAH31 was subcloned into the suicide vector pEP185.2, yielding plasmid pAH35. Plasmid pAH35 was conjugated from E. coli strain S17λpir into Salmonella serotype Typhimurium strain IR715, and exconjugants selected on LB plus nalidixic acid and kanamycin. A kanamycin-resistant, chloramphenicol-sensitive exconjugant was designated strain AH10. The deletion of the sivI gene in AH10 was confirmed by Southern hybridization with a sivI-specific probe (data not shown).
Strain RAK19, which contains an insertion of a kanamycin resistance cassette 757 nucleotides from the ATG start codon of the sivH gene, was constructed as follows. A 1,230-bp fragment was amplified with primers 5′ TCTCTAGATTTACCACCGGCATCACCA 3′ and 5′ TATTAGTGTCTGGAGTATC 3′ and cloned into pBR322 (8). An ScaI-restricted kanamycin resistance cassette was ligated into the unique ScaI site in the cloned fragment. The resulting fragment was subcloned into pGP704 with the DraI and XbaI restriction enzymes, and the resulting recombinant plasmid was designated pGPK1. This plasmid was introduced into Salmonella serotype Typhimurium strain C5 (40) by conjugal transfer from E. coli S17-1 λpir. Exconjugants were selected on LB plus nalidixic acid and kanamycin. Exconjugants which had undergone two recombination events resulting in allelic exchange of the plasmid-borne sivH::Kmr allele were identified by replica plating on LB-plus-kanamycin plate and LB-plus-ampicillin plates. One exconjugant that was kanamycin resistant and ampicillin sensitive was selected and designated ST59. Phage P22 propagated on strain ST59 was used to transduce the sivH::Kmr allele from strain ST59 into strain IR715. A phage P22-sensitive transductant, designated RAK19, was purified from contaminating P22 phage by streaking to single colonies twice on EBU plates (7).
The construction of a Salmonella serotype Typhimurium strain in which the wild-type sivH gene was reintroduced into strain RAK19 for complementation (strain RAK59) was as follows. A DNA fragment at the 5′ end of the sivH ORF was amplified by PCR with primers 5′ GAAGATCTTCAGAATGCGAATCCTTCGCAC 3′ and 5′ GTATGCGAACAAGCGTAACAC 3′. The PCR product was digested with XbaI and SacI, ligated into pGP704 previously restricted with XbaI and SacI, and transformed into E. coli S17-1 λpir to yield plasmid pMAR3. Plasmid pMAR3 was introduced into Salmonella serotype Typhimurium strain IR715 by conjugal transfer. Exconjugants that had pMAR3 recombined into the chromosome were selected on LB plus nalidixic acid and carbenicillin, and one nalidixic acid-resistant and carbenicillin-resistant exconjugant was designated strain MRZ3. The chromosomal construct was confirmed by Southern hybridization (data not shown). A bacteriophage P22 HT105/1 int mutant lysate propagated on strain MRZ3 was used to transduce the chloramphenicol resistance marker of pMAR3 present in the chromosome adjacent to the sivH into strain RAK19. Transductants were selected for by plating on LB plus carbenicillin containing 5 mM EGTA. Cotransduction of the wild-type sivH gene into strain RAK19 for complementation was tested for by replica plating transductants on LB plus carbenicillin and LB plus kanamycin. A phage P22-sensitive, carbenicillin-resistant, kanamycin-sensitive transductant was designated RAK60 and purified from contaminating P22 phage by streaking to single colonies twice on EBU plates (7).
A Salmonella serotype Typhimurium strain containing an insertion of a kanamycin resistance cassette in the phoN gene was constructed as follows. The DNA region containing the phoN gene was PCR amplified with primers 5′ GACTCTAGAATAACCGTCCGGGAAATG 3′ and 5′ TAACCCGGGATTTGGTGGAGAGTG 3′. The PCR product was digested with XbaI and SmaI and cloned into suicide plasmid pGP704 digested with XbaI and SmaI, yielding plasmid pTN102. A SacI-digested kanamycin resistance cassette (KSAC; Pharmacia) was cloned into a unique SacI site in the phoN ORF in pTN102. This construct was transformed in E. coli strain S17-1 λpir and transferred to Salmonella serotype Typhimurium strain IR715 by conjugation. Exconjugants were selected on LB plus nalidixic acid and kanamycin plates and patched on LB plus carbenicillin. A kanamycin-resistant, carbenicillin-sensitive colony was selected, and the strain was designated AJB715. AJB715 formed white colonies when grown on LB plates supplemented with XP. The insertion of the kanamycin resistance cassette in the phoN gene in strain AJB715 was confirmed by Southern hybridization (data not shown).
Animal experiments.
Throughout this study, 6- to 8-week-old female BALB/c (ByJ; Jackson Lab) and 8- to 12-week-old CBA/J (Jackson Lab) mice were used. Bacteria were routinely cultured as standing overnight cultures prior to infection. In all experiments, the bacterial titer of the inoculum was determined by spreading serial 10-fold dilutions on agar plates containing the appropriate antibiotics and determining the number of CFU.
For competitive infection experiments, groups of 5 mice were infected by oral gavage with an approximately 1:1 mixture of mutant and isogenic parents at a dose of approximately 109 CFU/mouse. Fecal pellets were homogenized in 1 ml of phosphate-buffered saline. The limit of detection was approximately 0.08 CFU/mg of feces. The cecum, 3 Peyer's patches of the terminal ileum adjacent to the cecum, the mesenteric lymph nodes, and the spleen were harvested aseptically and homogenized in 5 ml of phosphate-buffered saline, pH 7.4. Dilutions of fecal pellets and homogenized organs were plated on LB plates containing the appropriate antibiotics. For competitive infection experiments with strains AJB715 and AH12, LB-plus-nalidixic-acid agar plates were supplemented with XP to distinguished between colonies expressing PhoN (AH12) and colonies that were PhoN negative (AJB715). Data were normalized by dividing the output ratio (CFU of the mutant/CFU of the wild type) by the input ratio (CFU of the mutant/CFU of the wild type). In case only one bacterial strain was recovered from fecal pellets, the limit of detection was determined for the missing strain and used to calculate the minimum mutant-to-wild type ratio. All data were converted logarithmically prior to the calculation of averages and statistical analysis. A Student t test was used to determine whether the log value of the mutant-to-wild type ratio recovered from infected organs or fecal pellets was significantly different from the log value of the mutant-to-wild type ratio present in the inoculum.
Nucleotide sequence accession number.
The complete nucleotide sequence of the insert of pRK824 was determined and deposited in the GenBank database under accession no. AF140550.
RESULTS
Nucleotide sequence analysis of the CS54 island.
The shdA gene is carried at one end of a genetic island present in serotypes of S. enterica subsp. I but absent from serotypes of S. enterica subsp. II to VII and the closely related species S. bongori and E. coli (29). In order to investigate the extent of the subsp. I-specific genetic island, the complete nucleotide sequence of the insert of pRK824, a cosmid from a gene bank of Salmonella serotype Typhimurium strain ATCC 14028, was determined and deposited in the GenBank database (accession no. AF140550). From the stop codon of xseA, which is downstream of shdA and defines one border of the genetic island, a DNA sequence with no significant homology to the E. coli K-12 MG1655 genome nucleotide sequence extended for 24,030 bp. Following this was an ORF, carried on the reverse strand, whose deduced amino acid sequence exhibited 97.5% identity over the terminal 40 amino acids of YfgK of E. coli K-12 (Fig. 1). The ORFs of the CS54 island of Salmonella serotype Typhimurium strain ATCC 14028 were identical to those of Salmonella serotype Typhimurium strain LT2 (31), except that ratB in ATCC 14028 was truncated by a point mutation. Four ORFs were carried on the CS54 island in addition to shdA (Fig. 1A). The properties of these potential coding sequences are summarized in Table 1. The GC content of the island varied between 28%, in the shdA-ratB intergenic region, and 59.9%, in the shdA, ratA, and ratB ORFs. The shdA-ratB intergenic region contained a 134-bp sequence with homology to dispersed repeat sequences (93 to 69% identity) that are repeated 15 times in the E. coli K-12 MG1655 genome (6). No function has been assigned to these E. coli repeats. The shdA-ratB intergenic region also contained a partial ORF with homology (84% over 26 amino acids) to the terminal 26 residues of the IS1 element from Shigella dysenteriae (accession no. P03832) (35).
TABLE 1.
ORFs of the xseA-yfgK intergenic region of Salmonella serotype Typhimurium strain ATCC 14028
| Gene designation | % GC content | Peptide lengtha | Peptide mass (kDa) | Signal peptideb | Protein description |
|---|---|---|---|---|---|
| shdA | 58.0 | 2,035 | 207 | 1-60 | Similar to MisL (35%), AIDA-I (34%), IcsA (30%) (carboxy terminus) |
| ratB | 59.9 | 1,947 | 204 | 1-27 | Similar to RatA |
| ratA | 59.0 | 1,865 | 200 | 1-30 | Similar to RatB |
| sivI | 53.3 | 319 | 33.9 | 1-28 | Similar to SivH (34.5%) (over terminal 110 residues) |
| sivH | 52.3 | 730 | 81.2 | 1-19 | Similar to invasin (49.5%), intimin (48%) (amino terminus) |
Given as the number of amino acid residues.
Amino acid residue positions are given.
A region of approximately 13 kb in the middle portion of the island was comprised of 13 imperfect direct repeats which were carried within the ratA and ratB ORFs. Sequence analysis of the deduced amino acid sequence of RatA and RatB indicated the presence of six direct repeats in each protein (designated RatA I to VI and RatB I to VI) (Fig. 2). The repeats ranged in size from 198 (in RatB I) to 227 (in RatA VI) amino acids, and they ranged in identity from 62% (RatA I and RatB I) to 29% (RatA III and RatA VI). The repeats did not exhibit similarity with any sequence in the available databases. Compared to strain LT2, ratB in strain ATCC 14028 had a frameshift mutation, which reduced the molecular mass of the predicted protein from 257 kDa (RatB in LT2) to 206 kDa (RatB in ATCC 14028).
FIG. 2.
Position and alignment of the imperfect repeats of RatA and RatB. In the Clustal alignment of the imperfect repeats (top), dark shaded boxes indicate identical residues, light shaded boxes indicate residues with similar biochemical properties, and dashes indicate gaps in alignment. The positions of the repeats in the RatA and RatB proteins (arrows) are indicated (filled bars) (bottom).
Adjacent to yfgK, an ORF, designated sivH (Salmonella invasin homologue), was identified whose deduced amino acid sequence exhibited homology in its N-terminal 350 residues with invasin of Yersinia pseudotuberculosis (49.5% identity) and intimin of E. coli O111 (enteropathogenic E. coli) (48% identity). The amino termini of invasin and intimin serve as membrane-spanning anchors in the bacterial outer membrane. Invasin binds β1 integrin via a domain located in the 192 C-terminal amino acid residues (30). Intimin binds the bacterium-encoded Tir receptor via a domain in the 280 C-terminal amino acid residues (20). Since the C-termini of invasin and intimin did not share homology with SivH, sequence comparison did not provide clues about a possible binding specificity of SivH. However, homology with the N-termini of invasin and intimin suggested that SivH may be an integral outer membrane protein.
Downstream of sivH, an ORF of 957 bp, designated sivI, was identified whose deduced amino acid sequence exhibited 34% identity to SivH over the 110 C-terminal amino acids. The remaining sequence of SivI had no homology with sequences in the available databases.
Distribution of the CS54 island within the genus Salmonella.
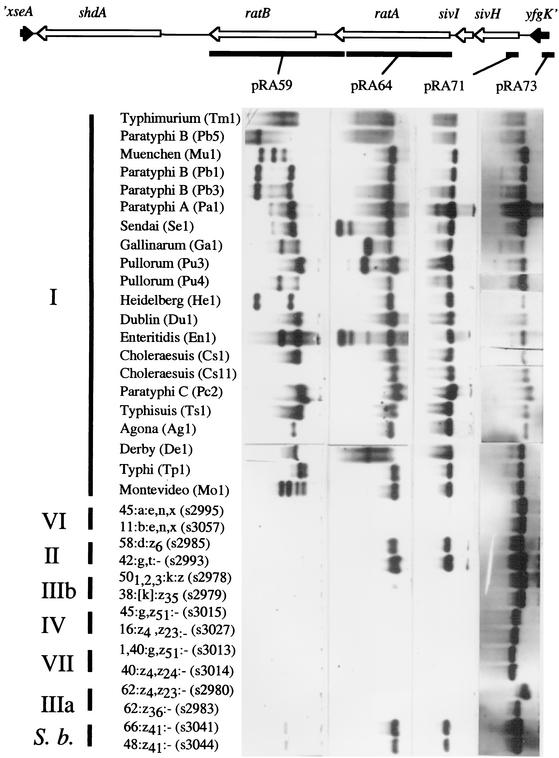
Hybridization analysis with Southern blots was employed to determine the distribution of sequences in the CS54 island of Salmonella serotype Typhimurium by using a collection of serotypes representing the full range of genetic variation within the genus Salmonella. This collection comprised 21 isolates from S. enterica subsp. I (from the Salmonella reference collection B), representing 17 different serotypes, and 14 isolates from S. enterica subsp. II, IIIa, IIIb, IV, VI, and VII and S. bongori (from the Salmonella reference collection C). The distributions of the xseA and shdA genes within this collection have been previously described (29). We extended the hybridization analysis upstream from the shdA gene by using nucleic acid probes generated from the pRK824 restriction fragments shown in Fig. 3. A probe overlapping the ratB gene (pRA59) hybridized with genomic DNA of all serotypes of S. enterica subsp. I (except one isolate [Cs11] of S. enterica subsp. I serotype Choleraesuis) but not with serotypes from S. enterica subsp. II to VII or S. bongori. In contrast, probes specific for ratA (pRA64) and sivH (pRA71) hybridized with genomic DNA from serotypes of S. enterica subsp. II, S. bongori, and all serotypes of S. enterica subsp. I. No signal was detected with genomic DNA prepared from serotypes of S. enterica subsp. IIIa, IIIb, IV, VI, or VII. The DNA probe specific for yfgK (pRA73) hybridized with serotypes from all lineages of the genus Salmonella, indicating the position of the right border of the CS54 island.
FIG. 3.
Phylogenetic distribution of the ratB, ratA, sivI, and sivH genes within the genus Salmonella. Southern blot analysis with representative serotypes of S. enterica (subspecies are indicated in roman numerals) and S. bongori (S.b.) is shown. Genomic DNA prepared from serotypes indicated on the left (strain designations are indicated in parentheses) was hybridized with DNA probes pRA59, pRA64, pRA71, and pRA73. The locations of these DNA probes (closed bars) relative to the ORFs of the CS54 island (arrows) are indicated on the map shown at the top.
In summary, these data indicated that shdA, ratA, ratB, sivI, and sivH are carried by all serotypes of S. enterica subsp. I tested, with the notable exception of the absence of ratB from the serotype Choleraesuis isolate Cs11. While shdA and ratB were absent from non-S. enterica subsp. I serotypes, sivH-, sivI-, and ratA-hybridizing sequences were detected in serotypes of S. enterica subsp. II and S. bongori. The presence of sivH, sivI, and ratA in S. enterica subsp. II and S. bongori serotypes in addition to S. enterica subsp. I suggested a complex evolutionary history of the CS54 island, involving multiple horizontal transfer or deletion events.
Contribution of the genes carried on the CS54 island to organ colonization of the BALB/c mouse.
Inbred mouse strains that are genetically susceptible to Salmonella serotype Typhimurium infection are frequently used to model typhoid fever caused by S. enterica serotype Typhi. Strains of Salmonella serotype Typhimurium were constructed in which shdA (strain AH9), ratA (strain AH8), ratB (AH12), sivI (strain AH10), or sivH (strain RAK19) were deleted or interrupted by insertion of a kanamycin resistance cassette (Fig. 1). The phenotypes of these strains were characterized by using competitive infection experiments with the inbred mouse strain BALB/c.
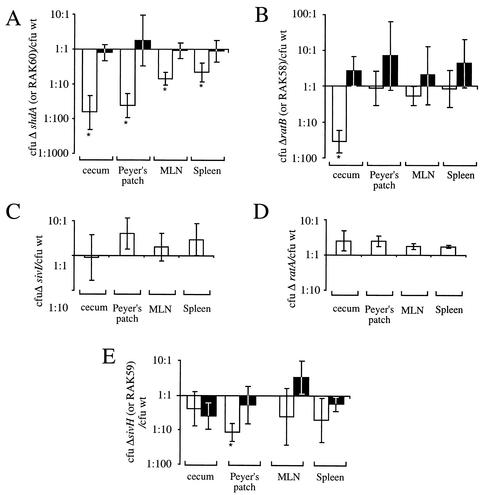
We previously reported the phenotype of a Salmonella serotype Typhimurium strain RAK1, in which the shdA gene was interrupted by the chloramphenicol acetyltransferase (cat) gene following oral inoculation of BALB/c mice. To further characterize the shdA colonization defect, we constructed a strain with a precise deletion of the shdA ORF (AH9) by allelic exchange (Fig. 1). Five BALB/c mice were inoculated with an equal mixture of AH9 (ΔshdA::Kmr) and its isogenic wild-type strain, IR715, and 5 days later each strain was enumerated in the cecum, Peyer's patches, mesenteric lymph nodes, and spleen (Fig. 4A). Significantly more CFU of the wild type (IR715) than of the shdA deletion mutant (AH9) were recovered from the cecum (P < 0.01), Peyer's patches (P < 0.005), mesenteric lymph nodes (P < 0.005), and spleen (P < 0.05). The colonization defect was greater in the cecum and Peyer's patches of the ileum (50- to 100-fold more CFU of IR715 than of AH9) than in the mesenteric lymph nodes and spleen (5- to 10-fold more CFU of IR715 than of AH9).
FIG. 4.
Recovery of bacteria from the cecum, Peyer's patch, mesenteric lymph node (MLN), and spleen of BALB/c mice 5 days post-oral inoculation with an equal mixture of AH9 (ΔshdA::Kmr, open bars) or RAK60 (AH9 complemented, filled bars) and IR715 (wild type [wt]) (A), AH12 (ΔratB, open bars) or RAK58 (AH12 complemented, filled bars) and AJB715 (wt) (B), AH8 (ΔratA::Kmr) and IR715 (wt) (C), AH10 (ΔsivI::Kmr) and IR715 (wt) (D), or RAK19 (sivH::Kmr, open bars) or RAK59 (RAK19 complemented, filled bars) and IR715 (wt) (E). The ratio of the two strains present is given as the mean ± standard error. An asterisk indicates that the output ratio was significantly different (P < 0.05) from that present in the inoculum.
In order to determine whether other genes carried on the CS54 island have functions related to that of shdA, we characterized strains AH12 (ΔratB), AH8 (ΔratA::Kmr), AH10 (ΔsivI::Kmr), and RAK19 (sivH::Kmr) in competitive infection experiments with the wild type (IR715). Four mice were each inoculated orally with an equal mixture of IR715 and either AH10 (ΔsivI::Kmr), AH8 (ΔratA::Kmr), AH12 (ΔratB), or RAK19 (sivH::Kmr). Five days postinoculation, the number of CFU of the wild type and mutant in homogenates of the cecum, Peyer's patches, mesenteric lymph nodes, and spleen were determined (Fig. 4B to E). Strains AH8 (ΔratA::Kmr) and AH10 (ΔsivI::Kmr) did not exhibit a colonization defect in the cecum, Peyer's patches, mesenteric lymph nodes, or spleen (Fig. 4C and D). RAK19 (ΔsivH::Kmr) was not recovered from the Peyer's patches of the terminal ileum (the limit of detection was 5 CFU), indicating that IR715 was present in this tissue at >10-fold-greater numbers than it was in RAK19 (Fig. 4E). IR715 (wild type) was recovered in significantly (P < 0.005) greater numbers (40-fold) from the cecum than was strain AH12 (ΔratB) (Fig. 4B). These data suggested that RatB is required for the colonization of the cecum while SivH contributes to the colonization of the Peyer's patches.
The role of shdA, sivH, and ratB in long-term shedding from CBA/J mice.
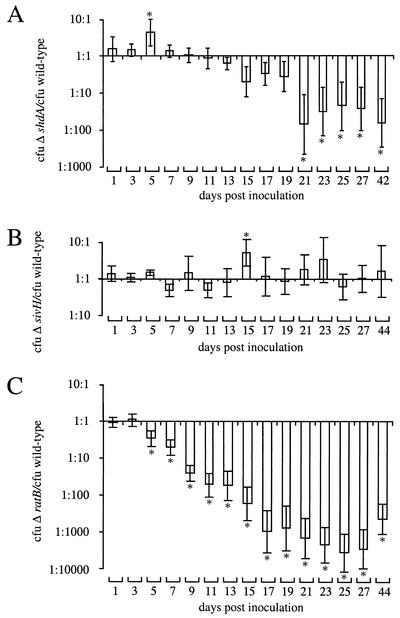
A Salmonella serotype Typhimurium shdA aroA mutant was used previously to investigate the role of the shdA gene in fecal shedding of bacteria from BALB/c mice at time points beyond day 5 postinoculation (29). The introduction of an aroA mutation was necessary in these experiments, since BALB/c mice show signs of lethal morbidity when infected with virulent Salmonella serotype Typhimurium within 4 to 6 days while the shdA phenotype becomes most pronounced at later times postinoculation. To study the role of shdA under more natural conditions (i.e., in a fully virulent Salmonella serotype Typhimurium strain background) we used a resistant mouse lineage, CBA/J. This strain of mouse does not succumb to infection by Salmonella serotype Typhimurium, but the intestine becomes colonized at a high level (104 to 105 CFU/mg of feces) for several weeks (34). To assess the effect of a mutation in shdA on bacterial shedding, 8 mice were inoculated orally with an equal mixture of AH9 (ΔshdA::Kmr) and IR715 (wild type). The presence of each strain in fecal pellets was enumerated over a 42-day period (Fig. 5A). The mean ratio of AH9 to IR715 was not significantly different from the inoculum ratio (1:1) in the first 19 days postinoculation, with the exception of one occasion (day 5 postinoculation) in which AH9 (ΔshdA::Kmr) was recovered in significantly (P < 0.05) greater numbers (threefold) than IR715 (wild type). On day 21 and on subsequent days postinoculation, IR715 (wild type) was recovered in significantly (P < 0.05) greater numbers (10- to 500-fold) than AH9 (ΔshdA::Kmr). These results were consistent with data reported previously that a Salmonella serotype Typhimurium shdA aroA mutant is recovered at lower numbers from fecal pellets of BALB/c mice at late time points postinoculation than an isogenic Salmonella serotype Typhimurium aroA mutant (29).
FIG. 5.
Recovery of bacteria from fecal pellets collected after inoculation of CBA/J mice with an equal mixture of AH9 (ΔshdA::Kmr) (A), RAK19 (sivH::Kmr) (B), or AH12 (ΔratB) and IR715 (wild type) (C) Salmonella serotype Typhimurium strains. The ratio of the two strains present in the fecal pellets is given as the mean ± standard error. An asterisk indicates that the output ratio was significantly different (P < 0.05) from that present in the inoculum.
We next characterized the shedding phenotype of Salmonella serotype Typhimurium strains containing deletions of the sivH gene or the ratB gene in groups of 6 or 9 CBA/J mice, respectively, during competitive infection experiments with the Salmonella serotype Typhimurium wild type (IR715). The mean ratio of RAK19 to IR715 was not significantly different from the inoculum ratio (1:1) on any day, with the exception of day 5 postinoculation, at which point RAK19 (sivH::Kmr) was recovered in significantly (P < 0.05) greater numbers (twofold) than IR715 (wild type) (Fig. 5B). In contrast, the ratio of AH12 to IR715 decreased over time. On day 5 postinoculation, and on all subsequent days investigated, significantly (P < 0.05) greater numbers of AJB715 CFU were shed with the feces than CFU of the ratB mutant (AH12) (Fig. 5C). On day 17 postinoculation and on subsequent days, AJB715 was recovered in >1,000-fold-higher numbers than AH12 (ratB). These results showed that, in contrast to a mutation in sivH, a deletion of the ratB gene reduced the magnitude of fecal shedding of Salmonella serotype Typhimurium from mice.
Correlation of cecal colonization and fecal shedding.
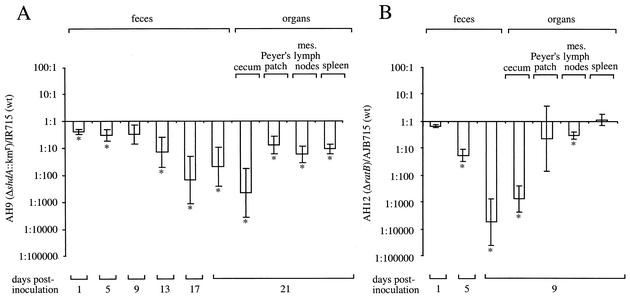
The characterization of the role of CS54 island-borne genes in the colonization of BALB/c mice and shedding with the feces of CBA/J mice revealed a correlation between the role of cecal colonization in BALB/c mice and shedding with the feces following oral inoculation of CBA/J mice. However, these observations were made in different inbred mouse strains. To further study this correlation, we investigated the role of the shdA and ratB genes in fecal shedding and organ colonization in groups of 5 CBA/J mice. Mice were inoculated orally with a mixture of Salmonella serotype Typhimurium strains AH9 (ΔshdA::Kmr) and AH12 (ΔratB) and the wild-type strains IR715 and AJB715, respectively. The deletion strains exhibited a similar colonization phenotype upon infection of CBA/J mice to that observed with BALB/c mice. That is, the shdA mutant was recovered in significantly lower numbers than the wild type from the Peyer's patches, cecum, mesenteric lymph nodes, spleen, and fecal pellets (Fig. 6A). The greatest colonization defect was observed in the cecum and fecal pellets 21 days postinfection, at which stage the wild-type strain (IR715) was recovered in approximately 100-fold-higher numbers than the ΔshdA mutant (AH9). With the exception of a small defect in colonization of the mesenteric lymph nodes, the ratB mutant (AH12) exhibited a colonization defect only in the cecum and fecal pellets (Fig. 6B). In this case, the wild-type strain (AJB715) was recovered in approximately 1,000-fold-higher numbers than the ΔratB mutant from these sites.
FIG. 6.
Recovery of bacteria from fecal pellets, the cecum, and the Peyer's patches of BALB/c mice following oral inoculation with an equal mixture of AH9 (ΔshdA::Kmr) and IR715 (wild type [wt]) (A), AH12 (ΔratB) and AJB715 (wt) (B), or RAK19 (sivH::Kmr) and IR715 (wt) (C). The ratio of the two strains present is given as the mean ± standard error. An asterisk indicates that the output ratio was significantly different (P < 0.05) from that present in the inoculum.
Next, we investigated whether the role of the shdA, ratB, and sivH genes in the colonization of the cecum correlates with a role in shedding with the feces during infection of BALB/c mice following oral inoculation. Groups of 5 mice were inoculated with an equal mixture of the wild-type strain (IR715 or AJB715) and either the ΔshdA mutant (AH9), the ΔratB mutant (AH12), or the sivH mutant (RAK19). The CFU of Salmonella serotype Typhimurium strains in approximately 100 mg of feces were enumerated on days 1, 3, and 5 postinoculation and in the cecum and Peyer's patches on day 5 postinoculation. On day 5 postinoculation, significantly greater numbers of wild-type CFU were recovered in the feces than were CFU of the shdA and ratB mutants. In contrast, the sivH mutant was recovered in numbers similar to those of the wild type (Fig. 7). The shedding defect observed for the ΔshdA mutant (AH9) and the ΔratB mutant (AH12) during competitive infection with the wild type correlated with the colonization defect exhibited in the cecum. Significantly greater numbers of wild-type CFU were recovered in the cecum than were CFU of the ΔshdA and ΔratB mutants. In contrast, the sivH mutant was recovered in numbers similar to those of the wild type. Both the sivH mutant and the ΔshdA mutant were recovered in lower numbers from the Peyer's patches than was the wild type (Fig. 7A and C). These data further supported the idea that there is a direct correlation between the ability of Salmonella serotype Typhimurium to colonize the cecum and its ability to be shed with the feces.
FIG. 7.
Recovery of bacteria from fecal pellets, the cecum, and the Peyer's patches of BALB/c mice following oral inoculation with an equal mixture of AH9 (ΔshdA::Kmr) and IR715 (wild type [wt]) (A), AH12 (ΔratB) and AJB715 (wt) (B), or RAK19 (sivH::Kmr) and IR715 (wt) (C). The ratio of the two strains present is given as the mean ± standard error. An asterisk indicates that the output ratio was significantly different (P < 0.05) from that present in the inoculum.
DISCUSSION
A common feature of many pathogenicity islands of enteric pathogens is their insertion at tRNA loci (23). For example, the selC tRNA locus is the site of SPI-3 integration in Salmonella serotype Typhimurium (5) and the site of the LEE pathogenicity island integration in enteropathogenic E. coli strains (37). This was not the case for the CS54 islands of Salmonella serotype Typhimurium. Instead, the island was found to be carried between the xseA gene, which encodes the exonuclease VII subunit, and the yfgK gene, whose function is currently unknown. Atypical GC content (often low GC content) is frequently an indication of horizontal acquisition (23). The shdA, ratB, and ratA genes have a G+C content of between 58.0 and 59.9%, which is higher than the average G+C content of 53% determined for the Salmonella serotype Typhimurium genome (31). The sivI and sivH ORFs, on the other hand, have a G+C content which is similar to the genomic average. The presence of remnants of an insertion element in the shdA-ratB intergenic region suggests that mobile genetic elements may be responsible for the current distribution of the CS54 island within the genus Salmonella.
We describe the complete coding sequence of the CS54 island of Salmonella serotype Typhimurium strain ATCC 14028. Including the previously described shdA locus, a total of 5 ORFs and the remnants of an insertion element were identified in the region. The sequence of strain 14028 differed from that determined for Salmonella serotype Typhimurium strain LT2 in that the ratB ORF was truncated by a point mutation. As a result, RatB encoded by strain LT2 is predicted to be composed of eight imperfect direct repeats of approximately 200 amino acids while RatB of strain 14028 contains just the first six of these repeats (Fig. 2).
The absence of the CS54 island from the E. coli K-12 genome suggested that this region was acquired by horizontal transfer or lost from the E. coli lineage by deletion. The shdA gene was previously reported to be present in serotypes of S. enterica subsp. I and absent from S. enterica subsp. II to VII and S. bongori (29). Hybridization of genomic DNA from one S. enterica subsp. IIIa isolate and one S. bongori isolate with an DNA array of Salmonella serotype Typhimurium strain LT2 indicates that the ratB gene is absent from both strains while sivI is present in S. bongori but absent from S. enterica subsp. IIIa (31). The distribution of ratA and sivH was not previously investigated since the LT2 DNA array of McClelland and coworkers does not contain PCR products specific for these ORFs (31). Here, we show that the ratB ORF exhibited an identical distribution within the genus to shdA. However, Southern blot analysis suggested a wider distribution of sivH and ratA, as homologous sequences were detected in S. enterica subsp. II and S. bongori serotypes in addition to serotypes of S. enterica subsp. I (Fig. 3). Together, these data suggest a complex evolutionary history of the CS54 island involving multiple horizontal transfer and/or deletion events.
The phylogenetic distribution of shdA and ratB is of significance because serotypes of S. bongori or S. enterica subsp. II, IIIa, IIIb, IV, VI, and VII are mainly reptile associated while serotypes of S. enterica subsp. I are frequently isolated from livestock and domestic fowl (38). The analysis of genes present in serotypes of S. enterica subsp. I but absent from serotypes of S. bongori and S. enterica subsp. II, IIIa, IIIb, IV, VI, and VII may provide an opportunity to gain new insights into mechanisms required for persistence of food-borne pathogens in populations of livestock and domestic fowl. Analysis of fecal samples shows that between 1 and 6% of apparently healthy food animals in the United States shed S. enterica subsp. I serotypes with their feces (11, 15, 17-19, 44). Before slaughter, the prevalence of S. enterica subsp. I serotypes in fecal samples or the intestine increases further, with 10 to 15% of animals testing positive (13, 44). A body of evidence shows that the main factor responsible for spreading the infection among animals on the farm or among animals during transport to slaughter is fecal contamination of the environment (14, 16, 25, 32, 45, 46). These investigations suggest that fecal shedding of S. enterica subsp. I serotypes is of prime importance for persistence of these pathogens within populations of livestock and domestic fowl. It is therefore significant that shdA and ratB, two genes whose presence is restricted to serotypes of S. enterica subsp. I, both contributed to the ability of Salmonella serotype Typhimurium to be persistently shed with the feces of mice (Fig. 5). It is interesting in this context that a Salmonella serotype Typhi vaccine strain colonizes and persists in the feces of experimentally infected human subjects at a lower level than a Salmonella serotype Typhimurium vaccine strain, although both vaccine strains were attenuated by the same mutation (24). It remains to be seen whether reduced persistence of the Salmonella serotype Typhi vaccine strain is related to the fact that shdA and ratB are pseudogenes in this serotype (36).
Isolation of a Salmonella serotype from the feces is commonly accompanied by intestinal colonization; however, the exact origin of bacteria exiting from the host to ensure transmission by the fecal-oral route is not well defined. Studies with S. enterica subsp. I serotype Enteritidis in the mouse show that the bulk of luminal bacteria is localized in the cecum, suggesting that this organ is an important reservoir for fecal shedding in this animal model (12). Mutations in shdA, ratB, and sivH resulted in a reduced ability of Salmonella serotype Typhimurium to colonize intestinal tissues in BALB/c mice (Fig. 4). Inactivation of sivH resulted in a reduced ability to colonize Peyer's patches (Fig. 4) but did not alter bacterial numbers shed with the feces of CBA/J mice over a period of 44 days (Fig. 5). Similarly, a mutation in invA results in a reduced ability of Salmonella serotype Typhimurium to colonize Peyer's patches (3, 21), but this mutation does not alter the ability of the pathogen to be shed with the feces of mice (29). In contrast, mutations in shdA and ratB both reduced the ability of Salmonella serotype Typhimurium to colonize the cecum (Fig. 4 and 6), and both mutations resulted in a significant reduction of bacterial numbers shed with the feces of mice over a period of 42 days (Fig. 5). These data further support the notion that the cecum is the main reservoir for fecal shedding of Salmonella serotypes from mice.
Acknowledgments
Work in A.J.B.'s laboratory is supported by USDA NRICGP grant no. 2002-35204-12247 and Public Health Service grants no. AI40124 and no. AI44170.
We thank Heather Briggs for constructing strain AJB715.
Editor: A. D. O'Brien
REFERENCES
- 1.Aleksic, S., F. Heinzerling, and J. Bockemühl. 1996. Human infection caused by salmonellae of subspecies II to VI in Germany, 1977-1992. Zentbl. Bakteriol. 283:391-398. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Bäumler, A. J., R. M. Tsolis, P. J. Valentine, T. A. Ficht, and F. Heffron. 1997. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect. Immun. 65:2254-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäumler, A. J., R. M. Tsolis, A. W. M. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 193:207-213. [DOI] [PubMed] [Google Scholar]
- 5.Blanc-Potard, A.-B., and E. A. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Colladovides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Bochner, B. R. 1984. Curing bacterial cells of lysogenic viruses by using UCB indicator plates. BioTechniques 2:234-240. [Google Scholar]
- 8.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 9.Boyd, E. F., F.-S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 10.Boyd, E. F., F.-S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationship of the Salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrd, J. A., J. R. DeLoach, D. E. Corrier, D. J. Nisbet, and L. H. Stanker. 1999. Evaluation of Salmonella serotype distributions from commercial broiler hatcheries and grower houses. Avian Dis. 43:39-47. [PubMed] [Google Scholar]
- 12.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corrier, D. E., J. A. Byrd, B. M. Hargis, M. E. Hume, R. H. Bailey, and L. H. Stanker. 1999. Presence of Salmonella in the crop and ceca of broiler chickens before and after preslaughter feed withdrawal. Poult. Sci. 78:45-49. [DOI] [PubMed] [Google Scholar]
- 14.Dahl, J., A. Wingstrand, B. Nielsen, and D. L. Baggesen. 1997. Elimination of Salmonella typhimurium infection by the strategic movement of pigs. Vet. Rec. 140:679-681. [DOI] [PubMed] [Google Scholar]
- 15.Dargatz, D. A., P. J. Fedorka-Cray, S. R. Ladely, and K. E. Ferris. 2000. Survey of Salmonella serotypes shed in feces of beef cows and their antimicrobial susceptibility patterns. J. Food Prot. 63:1648-1653. [DOI] [PubMed] [Google Scholar]
- 16.Davies, P. R., W. E. Morrow, F. T. Jones, J. Deen, P. J. Fedorka-Cray, and J. T. Gray. 1997. Risk of shedding Salmonella organisms by market-age hogs in a barn with open-flush gutters. J. Am. Vet. Med. Assoc. 210:386-389. [PubMed] [Google Scholar]
- 17.Ebel, E. D., M. J. David, and J. Mason. 1992. Occurrence of Salmonella enteritidis in the U.S. commercial egg industry: report on a national spent hen survey. Avian Dis. 36(3):646-654. [PubMed] [Google Scholar]
- 18.Fedorka-Cray, P. J., D. A. Dargatz, L. A. Thomas, and J. T. Gray. 1998. Survey of Salmonella serotypes in feedlot cattle. J. Food Prot. 61:525-530. [DOI] [PubMed] [Google Scholar]
- 19.Fedorka-Cray, P. J., J. T. Gray, and C. Wray. 2000. Salmonella infections in pigs, p. 191-207. In C. Wray and A. Wray (ed.), Salmonella in domestic animals. CABI Publishing, New York, N.Y.
- 20.Frankel, G., D. C. Candy, P. Everest, and G. Dougan. 1994. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect. Immun. 62:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 24.Hindle, Z., S. N. Chatfield, J. Phillimore, M. Bentley, J. Johnson, C. A. Cosgrove, M. Ghaem-Maghami, A. Sexton, M. Khan, F. R. Brennan, P. Everest, T. Wu, D. Pickard, D. W. Holden, G. Dougan, G. E. Griffin, D. House, J. D. Santangelo, S. A. Khan, J. E. Shea, R. G. Feldman, and D. J. Lewis. 2002. Characterization of Salmonella enterica derivatives harboring defined aroC and Salmonella pathogenicity island 2 type III secretion system (ssaV) mutations by immunization of healthy volunteers. Infect. Immun. 70:3457-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurd, H. S., J. K. Gailey, J. D. McKean, and M. H. Rostagno. 2001. Experimental rapid infection in market swine following exposure to a Salmonella contaminated environment. Berl. Munch. Tierarztl. Wochenschr. 114:382-384. [PubMed] [Google Scholar]
- 26.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R-M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 27.Kingsley, R. A., R. Reissbrodt, W. Rabsch, J. M. Ketley, R. M. Tsolis, P. Everest, G. Dougan, A. J. Bäumler, M. Roberts, and P. H. Williams. 1999. Ferrioxamine-mediated iron(III) utilization by Salmonella enterica. Appl. Environ. Microbiol. 65:1610-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingsley, R. A., R. L. Santos, A. M. Keestra, L. G. Adams, and A. J. Bäumler. 2002. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol. Microbiol. 43:895-905. [DOI] [PubMed] [Google Scholar]
- 29.Kingsley, R. A., K. van Amsterdam, N. Kramer, and A. J. Bäumler. 2000. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect. Immun. 68:2720-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong, J. M., R. S. Fournier, and R. R. Isberg. 1990. Identification of the integrin binding domain of the Yersinia pseudotuberculosis invasin protein. EMBO J. 9:1979-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 32.McLaren, I. M., and C. Wray. 1991. Epidemiology of Salmonella typhimurium infection in calves: persistence of salmonellae on calf units. Vet. Rec. 129:461-462. [DOI] [PubMed] [Google Scholar]
- 33.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson, T. L., and A. J. Baumler. 2001. Salmonella enterica serotype Typhimurium elicits cross-immunity against a Salmonella enterica serotype enteritidis strain expressing LP fimbriae from the lac promoter. Infect. Immun. 69:204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyman, K., K. Nakamura, H. Ohtsubo, and E. Ohtsubo. 1981. Distribution of the insertion sequence IS1 in gram-negative bacteria. Nature 289:609-612. [DOI] [PubMed] [Google Scholar]
- 36.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 37.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popoff, M. Y., and L. Le Minor. 1992. Antigenic formulas of the Salmonella serovars, 5th ed. W. H. O. Collaborating Center for Reference and Research on Salmonella, Institute Pasteur, Paris, France.
- 39.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds, B. L., and D. Rowley. 1969. Sensitization of complement resistant bacterial strains. Nature 221:1259-1261. [DOI] [PubMed] [Google Scholar]
- 41.Schlosser, W., A. Hogue, E. Ebel, B. Rose, R. Umholtz, K. Ferris, and W. James. 2000. Analysis of Salmonella serotypes from selected carcasses and raw ground products sampled prior to implementation of the Pathogen Reduction; Hazard Analysis and Critical Control Point Final Rule in the US. Int J. Food Microbiol. 58:107-111. [DOI] [PubMed] [Google Scholar]
- 42.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 43.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wells, S. J., P. J. Fedorka-Cray, D. A. Dargatz, K. Ferris, and A. Green. 2001. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J. Food Prot. 64:3-11. [DOI] [PubMed] [Google Scholar]
- 45.Wray, C., J. N. Todd, and M. Hinton. 1987. Epidemiology of Salmonella typhimurium infection in calves: excretion of S. typhimurium in the faeces of calves in different management systems. Vet. Rec. 121:293-296. [DOI] [PubMed] [Google Scholar]
- 46.Wray, C., N. Todd, I. M. McLaren, and Y. E. Beedell. 1991. The epidemiology of Salmonella in calves: the role of markets and vehicles. Epidemiol Infect. 107:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]