Abstract
The gene encoding Clostridium sordellii phospholipase C (Csp) was cloned and expressed as a histidine-tagged (His-tag) protein, and the protein was purified to compare its enzymatic and biological activities with those of Clostridium perfringens phospholipase C (Cpa) and Clostridium bifermentans phospholipase C (Cbp). Csp was found to consist of 371 amino acid residues in the mature form and to be more homologous to Cbp than to Cpa. The egg yolk phospholipid hydrolysis activity of the His-tag Csp was about one-third of that of His-tag Cpa, but the hemolytic activity was less than 1% of that of His-tag Cpa. His-tag Csp was nontoxic to mice. Immunization of mice with His-tag Cbp or His-tag Csp did not provide effective protection against the lethal activity of His-tag Cpa. These results indicate that Csp possesses similar molecular properties to Cbp and suggest that comparative analysis of toxic and nontoxic clostridial phospholipases is helpful for characterization of the toxic properties of clostridial phospholipases.
Clostridium perfringens elaborates lecithinase, known as alpha-toxin (Cpa), which is the best characterized of all clostridial lecithinases (10, 29, 30). Cpa is a phospholipase C enzyme (30). It is toxic to mammals and is considered to be one of the major virulence factors produced by C. perfringens (25, 29, 30). However, there are still many lecithinases produced by other clostridia that are poorly characterized, and their roles in the pathogenesis of disease have not yet been determined (29, 30). Clostridial lecithinases whose primary structures have been determined are limited to only Cpa (13, 22, 23, 26, 32), Clostridium bifermentans phospholipase C (Cbp) (32), and Clostridium novyi type A phospholipase C (Cnp) (33). Additionally, clostridial lecithinases that have been purified and characterized are limited to Cpa and Cbp.
C. bifermentans and C. sordellii resemble each other in their cultural and biological properties, but they have been determined to be genetically different species (19). C. sordellii lecithinase is one of the clostridial lecithinases whose molecular properties are not yet understood (28). Here we report the cloning of the C. sordellii lecithinase (Csp) gene, expression of its product using purified histidine-tagged (His-tag) proteins, and comparison of the enzymatic and biological activities of Csp with those of Cpa and Cbp.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture.
C. sordellii NCIB10717 (ATCC 9714), C. perfringens KZ 221 (33), and C. bifermentans KZ 1012 (SJ2) were used to isolate the lecithinase genes. To investigate the occurrence of the csp gene, 23 C. sordellii strains kept at our laboratory were used. Esherichia coli TOP10F' (Invitrogen) was used for transformation. PCRII-TOPO (Invitrogen) and pKF3 (Takara Shuzo) plasmid vectors were used. Clostridia were grown by using home-made liver broth, brain heart infusion (BHI; Becton Dickinson Microbiology Systems), BHI agar plate, or 10% (vol/vol) egg yolk BHI agar plate under anaerobic conditions at 37°C. E. coli was grown by using 2× YT broth (24), 2× YT agar plate, or 10% egg yolk-2× YT agar plate at 37°C.
Extraction of total DNA.
Total DNA was extracted as previously described (37). Bacterial cultures were centrifuged at 5,000 × g for 15 min to collect cells. The cells were resuspended with 400 μl of TE buffer (10 mM Tris [pH 7.4], 1 mM EDTA), incubated at 37°C for 15 min with 25 U of mutanolysin (Nacalai Tesque, Kyoto, Japan), and digested with 25 μl of proteinase K (20 mg/ml) for 15 min. The cells were then incubated with 1% sodium dodecyl sulfate and 1 μl of RNase (10 mg/ml) at 37°C for 15 min. The cell lysate was treated with an equal volume of phenol and then with an equal volume of chloroform-isoamyl alcohol (24:1 [vol/vol]). The DNA was precipitated with isopropanol, rinsed with 70% ethanol, and finally resuspended with 200 μl of TE buffer. For the PCR assay to survey the csp gene, a simple DNA extraction method was employed. Bacterial cells collected from 2 ml of culture were suspended with 0.3 ml of TE buffer and boiled for 5 min. The supernatant was extracted with 0.3 ml of phenol-chloroform-isoamyl alcohol (25:24:1) and then precipitated with ethanol. The DNA was finally resuspended with 100 μl of TE buffer.
PCR.
The KAG209 (5′ TGGGATGGAAAAGATTGATGGAACAGG) and KAG210 (5′ TTTCTCTTTTCTTATCCACATATTCTTGTATATC) primers were designed based on the highly conserved regions among Cbp, Cnp, and Cpa (see Results). mF2 (GAGCTCGTAAAGTGGCCAAATCTAACGT) corresponds to nucleotides 35 to 62 of pKF3 (DDBJ/EMBL/GenBank accession no. D14641). KAG211 (5′CTGCAGTAGATAGTCCAGGTCATGT), KAG212 (5′CCTGTATCTGGGTCAAAGAAATGGTC), and KAG213 (5′CTGCAGACAATGAATATGCAGGAAC) correspond to nucleotides 768 to 792, 545 to 570, and 1134 to 1158 of the csp gene (DDBJ/EMBL/GenBank accession no. AB061868), respectively. PCR amplifications were performed by using Takara Ex Taq (Takara Shuzo) on a GeneAmp 9700 apparatus (Applied Biosystems) or a Touch Down thermal cycler (Hybaid) under a PCR profile of preheating at 94°C for 1 min followed by 30 cycles at 94°C for 20 s, 60°C for 15 s, and 72°C for 30 s, unless otherwise noted. The PCR assay was optimized to survey the csp gene by examining several primers, PCR profiles, and several clostridial species (data not shown). The PCR assay employed involved using 2 μl of DNA samples; 20-s incubations at 94, 50, and 72°C for 25 cycles; and KAG211 and KAG210 primers, which produced a 569 bp-fragment of csp.
Determination of the nucleotide sequences.
The nucleotide sequences were determined on an ABI PRISM 310 genetic analyser (Applied Biosystems) by using a BigDye Terminator cycle-sequencing ready reaction kit (Applied Biosystems).
His tag phospholipases and protein purification.
A His tag Csp plasmid was constructed for protein expression and purification. A PCR amplicon with KAG243 (5′ GAGAGGGCCCGCACAAAGGGCTTTTAAGGTATT [the underlined sequence corresponds to nucleotides 220 to 242; the putative ribosome binding site is at positions 248 to 254]) and KAG244 (5′GAGAGGTACCTAATTAGTGATGGTGATGGTGATGTTTGTTTATATCATAAACTTGATTACCTGAAAATAC [the underlined sequence corresponds to nucleotides 1424 to 1459]) was digested with ApaI and KpnI and ligated in the ApaI-KpnI site of pCRII-TOPO. The E. coli transformant cells grown in 2× YT broth were centrifuged, suspended with B-PER (Pierce) containing 20 mM imidazole and a protein inhibitor cocktail (Complete EDTA-free; Roche Diagnostic), and treated as described in the instruction manual for B-PER. Protein purification was performed by two-step column chromatography, in which the first and second steps were operated with a syringe and with a SMART System (Amersham Pharmacia Biotech), respectively. The supernatant was loaded on a HisTrap column (Amersham Pharmacia Biotech). The column was washed with 50 mM NaH2 PO4-300 mM NaCl-20 mM imidazole (pH 8), and the protein was eluted with 50 mM of NaH2 PO4-300 mM NaCl-250 mM imidazole (pH 8). The eluate was concentrated, buffer exchanged to 50 mM of sodium phosphate buffer (pH 7.5) with Centriprep YM-10 (Millipore), and loaded on a Mono Q column (Amersham Pharmacia Biotech). The column was washed with 50 mM of sodium phosphate buffer (pH 7.5), and proteins were eluted with a linear gradient of 0 to 1 M NaCl in the same buffer. His tag Cpa and His tag Cbp vectors were constructed by ligating PCR amplicons with KAG245 (5′GAGAGGGCCCAAATTAACGGGGGATATAAAAATGAAAAGAAAGA [the underlined sequence corresponds to nucleotides 147 to 180, under DDBJ/EMBL/GenBank accession no. D32124]) and KAG246 (5′GAGAGGTACCTAATTAGTGATGGTGATGGTGATGTTTTATATTATAAGTTGAATTTCCTGAAATCCA [the underlined sequence corresponds to nucleotides 1329 to 1361]) derived from C. perfringens KZ 221 (33) and with KAG241 (5′ GAGAGGGCCCGCAATGCAAGATTAGAGGATATTAG [the underlined sequence corresponds to nucleotides 1 to 25, under DDBJ/EMBL/GenBank accession no. AB061869]) and KAG242 (5′GAGAGGTACCTAATTAGTGATGGTGATGGTGATGTTTATTTATGTAATAAGTTTCGTTACCTGT [the underlined sequence corresponds to nucleotides 1202 to 1231]) derived from C. bifermentans KZ 1012 to pCRII-TOPO, respectively. The protein concentration was determined by the Bradford method, using albumin as a standard. The His tag Cpa, His tag Csp, and His tag Cbp plasmid constructs do not contain the natural promoter region of the plc gene (12, 30) or the predictive promoter regions of the csp and the cbp genes.
Enzymatic and biological activities.
Egg yolk solution (10%, vol/vol) was prepared by diluting freshly obtained egg yolk with Dulbecco's phosphate-buffered saline without calcium and magnesium (PBS) and by filtering with a Millex HA (pore size, 0.45 μm; [Millipore]). Samples were twofold serially diluted (0.8 to 100 ng) on 96-well microtiter plates in 100-μl volumes of PBS, and after the addition of 100 μl of 10% (vol/vol) egg yolk solution to each well, the plate was incubated at 37°C for 3 h. The optical density at 620 nm was monitored. One unit of the egg yolk-phospholipid hydrolysis activity was defined as an optical density increase of 1.0 unit. For the hemolysis assay, samples were diluted with PBS containing 1 mM CaCl2 in 100-μl volumes, mixed with 100 μl of 1% (vol/vol) mouse whole blood suspended in PBS (final concentration, 0.5% mouse blood), and incubated at 37°C for 1 h. The mixture was centrifuged, and the absorbance at 540 nm of the supernatant was measured. Several concentrations of each lecithinase were tested, and the concentration which showed 50% hemolytic activity against 0.5% mouse blood was calculated. The p-nitrophenylphosphorylcholine (NPPC) hydrolysis assay was performed as described previously (12). Samples (0.4 to 1.7 μg) were incubated at 35°C for 2 h in a 1-ml reaction mixture containing 20 mM NPPC, 0.25 M Tris-HCl (pH 7.2), and 60% glycerol, and the absorbance at 410 nm was monitored. To calculate the amount of p-nitrophenol produced, a molar extinction coefficient of 1.51 × 104 (12) was used.
ELISA.
The antibody response of immunized mice was determined by enzyme-linked immunosorbent assay (ELISA). A 100-μl volume of purified His tag phospholipases at 1 ng/ml in coating buffer (30 mM Na2CO3, 70 mM NaHCO3, 0.02% NaN3 [pH 9.6]) was dispensed into each well of a 96-well plate (Immuno-Plate C I Maxisorp; Nunc) and incubated overnight at 4°C. After the wells were washed with PBS containing 0.15% Tween 20 (PBST), 200 μl of blocking buffer (PBS containing 3% skim milk) was dispensed into the wells, which were incubated for 1 h at room temperature (RT). After the wells were washed with PBST, 100 μl of serum samples twofold serially diluted in blocking buffer were dispensed and incubated for 2 h at RT. After the wells were washed with PBST, 100 μl of anti-mouse goat immunoglobulin G labelled with horseradish peroxidase (Dako) diluted 1/2,000 in blocking buffer was added per well and incubate for 1 h at RT. After the wells were washed with PBST, 100-μl portions of substrates (1-Step Turbo TMB-ELISA; Pierce) were dispensed and incubated for 30 min at RT, and the reaction was stopped by adding 100 μl of 2 M H2SO4. The serum dilution for which an absorbance reading of twice (450/620 nm) the background was recorded was considered the titer of this serum.
Immunization of mice.
Male ddY mice (4 weeks) were injected subcutaneouslly three times (on days 0, 14, and 28) with either 2 μg of purified His tag Csp in 100 μl of PBS mixed with 20 μl of ImmunoEasy mouse adjuvant (Qiagen), 2 μg of purified His tag Cbp in 100 μl of PBS mixed with 20 μl of the adjuvant, or 0.5 μg of purified His tag Cpa in 100 μl of PBS mixed with 20 μl of the adjuvant. Serum samples were taken on days 7, 14, 21, 28, and 31, and the antibody titers were determined by ELISA. Serum samples were taken before boosting was performed on days 14 and 28. On day 35, immunized mice were challenged with His tag Cpa.
Mouse toxicity assay.
Mice were injected intraperitoneally with purified His tag phospholipases in 100 μl of PBS and monitored for 24 h after injection. The survival data were compared using Fisher's exact test (extended).
Nucleotide sequence accession numbers.
The nucleotide sequences of Csp and Cbp in this report have been deposited in the DDBJ/EMBL/GenBank databases under the accession numbers AB061868 and AB061869, respectively.
RESULTS
Cloning of the gene encoding C. sordellii phospholipase.
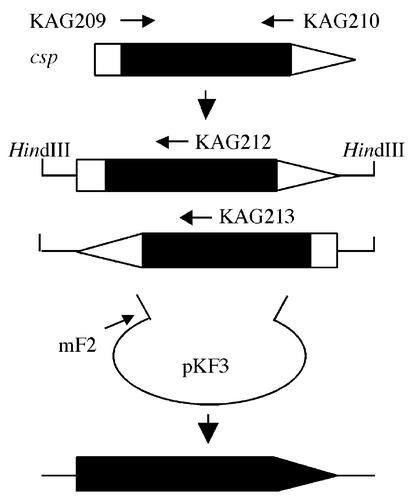
We compared the amino acid sequences of Cbp, Cnp, and Cpa, and designed PCR primers KAG209 and KAG210 in highly conserved regions among the three phospholipases (Fig. 1 and 2). A 1-kb DNA fragment of the expected size was amplified with KAG209 and KAG210 (45°C annealing) using total DNA of C. sordellii NCIB 10717. Since the nucleotide sequence of the 1-kb DNA fragment was similar to the sequence of the part of the genes thought to encode clostridial phospholipases, the single specific primer PCR (24) was performed to obtain a full open reading frame (Fig. 1). Digested fragments of C. sordellii NCIB10717 total DNA (1- to 7-kbp HindIII digestion products) were ligated with pKF3. PCR was conducted with mF2 located on pKF3 and with KAG212 or KAG213, located on the known 1-kb fragment (Fig. 1 and 2), and resulted in successful amplification. Nucleotide sequences of the upstream, the known 1-kb, and the downstream DNA fragments were consistent with the sequence of the Csp open reading frame.
FIG. 1.
Schematic presentation of the single specific primer PCR used to clone the csp gene. A 1-kb partial fragment of the csp gene was amplified from C. sordellii NCIB10717 total DNA with KAG209 and KAG210 PCR primers, designed in highly conserved regions among Cbp, Cnp, and Cpa. Using a HindIII DNA library of C. sordellii NCIB10717 as a template, PCR was conducted with mF2, located on pKF3, and with KAG212 or KAG213, located on the known 1-kb fragment. Nucleotide sequences of the upstream, the known 1-kb, and the downstream DNA fragments showed an open reading frame of Csp. The thick arrowed line depicts the csp gene. The filled and open areas indicate nucleotide sequence-determined (known) and -nondetermined (unknown) regions, respectively.
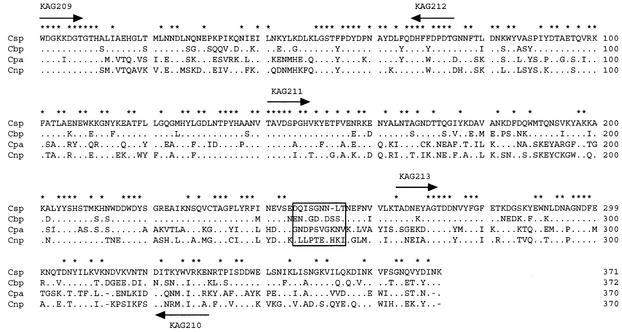
FIG. 2.
Amino acid alignment of clostridial phospholipases from C. sordellii (Csp), C. bifermentans (Cbp), C. perfringens (Cpa), and C. novyi (Cnp). Identical amino acid residues and gapped residues are depicted by dots and dashes, respectively. The residues conserved in all of the phospholipases are marked with asterisks. The peptide linker between the N-terminal and the C-terminal domains is boxed. The location of PCR primers is depicted with arrows. Data for Csp and Cbp are from this study, and data for Cpa and Cnp are from reference 35.
Comparison of the primary structure.
The csp gene consisted of 1,197 nucleotides encoding 399 amino acid residues (Fig. 2). In comparison with the other clostridial phospholipases (9, 32), the signal peptide and the mature protein were predicted to consist of 28 and 371 amino acid residues, respectively. The alignment and amino acid identity of the primary structure of clostridial phospholipases are shown in Fig. 2 and Table 1. In the overall sequence, Csp was more homologous to Cbp (77.4% identity) than to Cpa (53.4%) or Cnp (56.7%). Between Csp and Cbp, the amino acid identities of the N-terminal and C-terminal domains (see below) were 81.7 and 71.8%, respectively. Among the four clostridial phospholipases, the N-terminal domains were more homologous than the C-terminal domains.
TABLE 1.
Comparison of amino acid sequences of Csp, Cbp, Cpa, and Cnpa
| Phospholipase | % Identity (N-terminal domain/C-terminal domain) to:
|
|||
|---|---|---|---|---|
| Csp | Cbp | Cpa | Cnp | |
| Csp | 77.4 (81.7/71.8) | 53.4 (58.1/45.3) | 56.7 (59.8/53.9) | |
| Cbp | 77.4 (81.7/71.8) | 53.0 (57.7/46.2) | 54.6 (59.4/48.7) | |
| Cpa | 53.4 (58.1/45.3) | 53.0 (57.7/46.2) | 61.9 (68.7/51.3) | |
| Cnp | 56.7 (59.8/53.9) | 54.6 (59.4/48.7) | 61.9 (68.7/51.3) | |
Data for Csp and Cbp are from this study, and data for Cpa and Cnp are from reference 35.
Occurrence of the csp gene.
Of 22 test strains, 21 (95.9%) exhibited the lecithinase reaction on egg yolk-agar plates, and all of the reaction-positive strains carried the csp gene. Meanwhile, one strain which did not exhibit the lecithinase reaction was PCR negative.
Enzymatic and hemolytic activities.
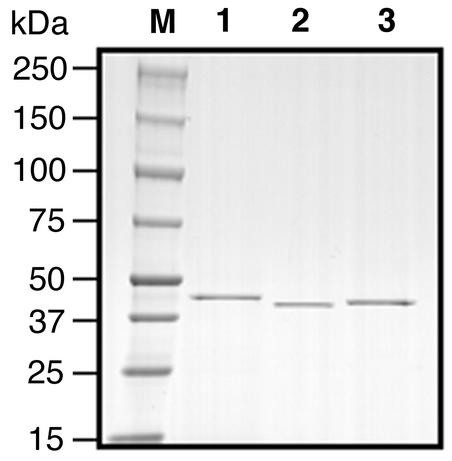
Figure 3 shows the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (5 to 20% gradient) of purified His tag proteins under a reduced condition. Purified proteins of Cbp, Cpa, and Csp were used in the egg yolk-phospholipid hydrolysis assay, the hemolysis assay, and the NPPC hydrolysis assay; the results are shown in Table 2. His tag Cpa showed greater activity than did His tag Csp or His tag Cbp in all of the assays. The egg yolk-phospholipid hydrolysis activities of His tag Csp and His tag Cbp were 38.2 and 6.1% that of the His tag Cpa, respectively. Likewise, the NPPC hydrolysis activities of His tag Csp and His tag Cbp were 7.6 and 2.5% that of His tag Cpa, respectively. His tag Cpa showed a 50% hemolysis when 29.8 ng of His tag Cpa was added to the reaction mixture, while 3 μg of His tag Csp and 3 μg of His tag Cbp caused less than 10% hemolysis only, indicating that the hemolytic activity of His tag Cpa was more than 100-fold stronger than those of His tag Csp and His tag Cbp. When 1, 2, and 3 μg of His tag Cpa was injected into six male ddY mice (4 weeks old) intraperitoneally, the numbers of survivers were 6, 0, and 0, respectively, indicating that the 100% lethal dose (LD100) of His tag Cpa was 2 μg. However, even when 20 μg of His tag Csp or His tag Cbp was administered, the mice (n = 3 each) did not die.
FIG. 3.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of His tag Cbp (lane 1), His tag Cpa (lane 2), and His tag Csp (lane 3). Each purified protein (1 μg) was electrophoresed and stained with Coomassie brilliant blue R250. M, protein standard marker.
TABLE 2.
Enzymatic activities of Cbp, Csp, and Cpa
| Phospholipase | egg yolk-phospholipid hydrolysis (U/mg/min)(% activity relative to Cpa) | NPPC hydrolysisa (nmol/mg/min) (% activity relative to Cpa) | Dose giving 50% hemolysisb (ng/0.2 ml) (% activity relative to Cpa) |
|---|---|---|---|
| Cbp | 7.41 ± 0.13 (6.1) | 2.37 ± 1.94 (2.5) | >3,000 (<1) |
| Cpa | 122 ± 0.39 (100) | 94.7 ± 2.27 (100) | 29.8 (100) |
| Csp | 46.6 ± 13.6 (38.2) | 7.16 ± 2.88 (7.6) | >3,000 (<1) |
Experiments were carried out at least three times, and the numbers are mean ± standard deviation.
Experiments were carried out at least three times, and the doses which shows 50% hemolysis in the assay were calculated.
Immunization and His tag Cpa challenge.
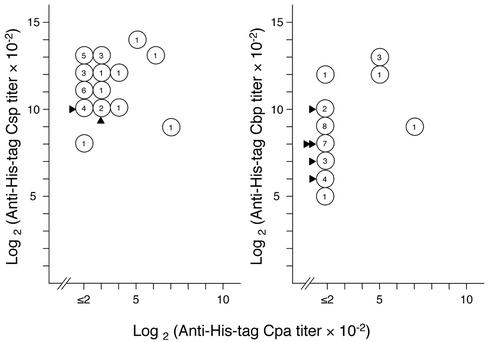
We determined the antibody titers of sera obtained from His tag phospholipase-immunized mice (Fig. 4). Almost all of the immunized mice showed plateau titers on day 28 (data not shown). The log2 values (10−2 anti-His tag Csp titer) (on day 31) for His tag Csp-immunized mice (n = 31) ranged between 8 and 14 for His tag Csp and between ≤2 and 7 for His tag Cpa. The log2 values (10−2 anti His tag Cbp titer) for His tag Cbp-immunized mice (n = 31) ranged between 5 and 13 for His tag Cbp and between ≤2 and 7 for His tag Cpa. The log2 values (10−2 anti-His tag Cpa titer) for His tag Cpa-immunized mice (n = 6) ranged between 6 and 12 for His tag Cpa. In general, mice with high titers to His tag Cpa also had high titers to the immunized proteins, but the strong association between the titer to His tag Cpa and those to the immunized proteins was not observed. Finally, immunized mice received a His tag Cpa challenge (5 μg, intraperitoneally) (Fig. 4). All His tag Cpa-immunized mice survived, and all nonimmunized control mice (n = 10) died. Five His tag Cbp-immunized mice and two His tag Csp-immunized mice survived, but immunization with Cbp and that with Csp did not confer a significant increase in the number of survivors. The surviving immunized mice had log2 values (10−2 anti-His tag Cpa titer) of ≤2 and 8.
FIG. 4.
Antibody responses of His tag Csp-immunized (left) and His tag Cbp-immunized (right) mice and the result of the His tag Cpa challenge. Ttiters for day 31 are presented. Numbers in open circles indicate the number of mice that had the indicated titer. Log2 values (10−2 anti-His tag Cpa titer) of His tag Cpa-immunized mice were 6 (n = 2), 9 (n = 2), 10 (n = 1), and 12 (n = 1). In His tag Cpa challenge, 5 μg of Cpa was injected intraperitoneally. All nonimunized mice (n = 10) died, and all His tag Cpa-immunized mice survived. The number of solid arrowheads indicates the number of survivors.
DISCUSSION
In the present study, structural and functional analyses revealed that Csp was more similar to Cbp than to Cpa and Cnp. The overall charasteristics of clostridial phospholipases, including those of Csp presented here, suggest that phospholipase-producing clostridia possess phospholipases which are similar to each other but unique to the species with respect to structure and function.
Nagahama et al. (15-18) and Guillouard et al. (7) investigated the roles of amino acid residues of Cpa by site-directed mutagenesis and reported that W1, H68, H126, H136, H148, and E152 were essential for zinc binding, D56 and T74 were required for catalitic activity, and D130 and T272 were required for maintainance of structure. All these residues are also conserved in Cbp, Cnp, and Csp (Fig. 2), suggesting that they are critical for maintaining the fundamental structure and function of clostridial phospholipases.
In the present study, we used His tag phospholipases to investigate enzymatic and biological activities. Two research groups have reported comparisons of enzymatic and biological activities between Cpa and Cbp by using purified “unfused” proteins (9, 34). The present results for enzymatic activities were compatible with the results of Tso and Siebel (34) (egg yolk-phospholipid hydrolysis and hemolysis) and with the results of Jepson et al. (9) (NPPC hydrolysis and hemolysis) with respect to the relative activities between Cpa and Cbp. Furthermore, the mouse LD100 of Cpa in intraperitoneal injections appeared to be essentially the same in the present study (2 μg) and in the study by Jepson et al. (1 μg). These comparisons suggest that the His tag does not inhibit the enzymatic and biological activities of clostridial phospholipases, but the possibility of some interference cannot be formally excluded.
Numerous studies have identified Cpa as one of the principal toxins involved in the pathophysiology of gas gangrene (25), and recent genetic studies have provided additional evidence that Cpa is an essential virulence factor in gas gangrene (3, 21, 36). Moreover, the pathophysiology in the progression of tissue destruction and in the cardiovascular system from gas gangrene has been elucidated. Asmuth et al. (2) suggested that Cpa-induced myocardial dysfunction, which may be mediated by the direct toxic effect, contributes to shock in C. perfringens infection. More recently, Bryant et al. (4, 5) concluded that tissue destruction in gas gangrene is related to a profound attenuation of blood flow caused by Cpa-induced, gpIIbIIIa-mediated formation of heterotypic platelet-polymorphonuclear leukocyte aggregates. Bunting et al. (6) demonstrated that human endothelial cells synthesize two vasoactive lipids, platelet-activating factor and prostacyclin, in response to Cpa treatment and suggest that this process may contribute to localized and systemic manifestations of gas gangrene including enhanced vascular permeability, localized neutrophil accumulation, and myocardial dysfunction. These results indicate that Cpa exhibits a number of biological activities, some of which might be produced as a result of a cascade of products such as platelet-activating factor, leukotrienes, and prostaglandins. Whether the direct activity of the phospholipase can account for these activities remains to be determined.
Clues to clarify the molecular mechanism by which Cpa exerts its strong toxic activity (e.g., hemolytic activity and lethality) may lie in the comparative analysis of the structure-function relationship between toxic and nontoxic clostridial phospholipases. The crystal structure of Cpa reveals a two-domain protein with the domains linked by a flexible peptide (19). The N-terminal domain has phospholipase C activity, while the C-terminal domain is a calcium-dependent putative phospholipid binding domain (8, 19, 20). The C-terminal domain is essential for sphingomyelinase, hemolytic, and lethal activities (27, 29, 31). Jepson et al. (9) made a hybrid toxin comprising the N-terminal domain of Cbp and the C-terminal domain of Cpa, which resulted in increased hemolytic and toxic activities compared with Cbp but did not restore the same level of hemolytic and toxic activities as did Cpa. They suggested that structural motifs in the N-terminal domain, as well as in the C-terminal domain, are thought to play a role in the recognition of membrane phospholipids, i.e., expression of biological activities. D293, D305, and K330 of Cpa are suggested to play a role in the recognition of membrane phospholipids and in the expression of hemolytic, cytotoxic, and myotoxic activities (34). However, the present study showed that residues which correspond to D293, D305, and K330 are conserved in Csp and/or Cbp, suggesting that other residues may be required for the strong biological activities of Cpa. Alape-Giron et al. (1) reported that D269, Y275, Y307, Y331, and D336 of Cpa were critical for toxicity, and Jepson et al. (11) reported that Y331 and F334 of Cpa were essential for it. Among the six residues, those that correspond to Y307 and D336 are conserved in both Csp and Cbp but those that correspond to D269, Y275, Y331, and F334 are substituted for Y, N, R, and I, respectively, in Csp and Y, N, Y, and I, respectively, in Cbp. As discussed in the literature on the subject (1, 11), amino acid residues which are specific to Cpa and not to other nontoxic phospholipases should be responsible for the toxicity. The C-terminal domain of Cpa possesses three calcium-binding sites, termed Ca1 (E32, D269, E271, D336, and A337), Ca2 (D293, N294, G296, and D298), and Ca3 (T272, D273, N297, and D298) (21). The present work has shown that the amino acid residues involved in the three calcium-binding sites and in the neighboring regions are basically conserved in Csp, with the exception of Ca1 (K32, Y268, and D337), as well as in Cbp Ca1 (Q32, Y269, and D338) and Cnp Ca1 (A269 and D337), suggesting that the differences in toxic activities of these proteins are not due to differences in their calcium-mediated recognition of phospholipid (9). However, further precise studies are required to clarify the relationship between the toxic activities and the calcium-mediated recognition of phospholipid.
Williamson and Titball (38) demonstrated that immunization with a truncated protein of Cpa247-370 (the C-terminal region) provided protection in mouse models against doses of at least 10 LD100 of purified Cpa and viable cells of C. perfringens type A. We could not demonstrate that immunization with His tag Cbp or with His tag Csp afforded effective protection of mice against the lethal activity of His tag Cpa, suggesting that structural differences between the C-terminal regions of Csp, Cbp, and Cpa are critical for achieving protection against the lethal effects of Cpa. Thus, comparative studies of phospholipases from various clostridia should be important to clarify the sites active for Cpa lethality and to develop a highly effective component vaccine for gas gangrene caused by C. perfringens. Moreover, studies of a variety of clostridial phospholipases will contribute to the design of therapeutic compounds effective against gas gangrene caused not only by C. perfringens but by other clostridia such as C. sordellii, C. bifermentans, and C. novyi.
Acknowledgments
T. Karasawa and X. Wang contributed equally to this study.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and from the Ministry of Health, Labour and Welfare, Japan.
We are grateful to Shigeru Sakurai, Hiroshi Yamamoto, Yasuhiko Yamamoto, and Hideto Yonekura (Kanazawa University, Kanazawa, Japan) for technical assistance; to Joseph J. Ferretti (University of Oklahoma Health Sciences Center, Oklahoma City, Okla.) for reviewing the manuscript; and to Richard W. Titball (Defence Science and Technology Laboratory, Salisbury, United Kingdom) for critical manuscript reading.
Editor: D. L. Burns
REFERENCES
- 1.Alape-Giron, A., M. Flores-Diaz, I. Guillouard, C. E. Naylor, R. W. Titball, A. Rucavado, B. Lomonte, A. K. Basak, J. M. Gutierrez, S. T. Cole, and M. Thelestam. 2000. Identification of residues critical for toxicity in Clostridium perfringens phospholipase C, the key toxin in gas gangrene. Eur. J. Biochem. 267:5191-5197. [DOI] [PubMed] [Google Scholar]
- 2.Asmuth, D. M., R. D. Olson, S. P. Hackett, A. E. Bryant, R. K. Tweten, J. Y. Tso, T. Zollman, and D. L. Stevens. 1995. Effects of Clostridium perfringens recombinant and crude phospholipase C and theta-toxin on rabbit hemodynamic parameters. J. Infect. Dis. 172:1317-1323. [DOI] [PubMed] [Google Scholar]
- 3.Awad, M. M., A. E. Bryant, D. L. Stevens, and J. I. Rood. 1995. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15:191-202. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, A. E., R. Y. Chen, Y. Nagata, Y. Wang, C. H. Lee, S. Finegold, P. H. Guth, and D. L. Stevens. 2000. Clostridial gas gangrene. I. Cellular and molecular mechanisms of microvascular dysfunction induced by exotoxins of Clostridium perfringens. J. Infect. Dis. 182:799-807. [DOI] [PubMed] [Google Scholar]
- 5.Bryant, A. E., R. Y. Chen, Y. Nagata, Y. Wang, C. H. Lee, S. Finegold, P. H. Guth, and D. L. Stevens. 2000. Clostridial gas gangrene. II. Phospholipase C-induced activation of platelet gpIIbIIIa mediates vascular occlusion and myonecrosis in Clostridium perfringens gas gangrene. J. Infect. Dis. 182:808-815. [DOI] [PubMed] [Google Scholar]
- 6.Bunting, M., D. E. Lorant, A. E. Bryant, G. A. Zimmerman, T. M. McIntyre, D. L. Stevens, and S. M. Prescott. 1997. Alpha toxin from Clostridium perfringens induces proinflammatory changes in endothelial cells. J. Clin. Investig. 100:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guillouard, I., T. Garnier, and S. T. Cole. 1996. Use of site-directed mutagenesis to probe structure-function relationships of alpha-toxin from Clostridium perfringens. Infect. Immun. 64:2440-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guillouard, I., P. M. Alzari, B. Saliou, and S. T. Cole. 1997. The carboxy-terminal C2-like domain of the alpha-toxin from Clostridium perfringens mediates calcium-dependent membrane recognition. Mol. Microbiol. 26:867-876. [DOI] [PubMed] [Google Scholar]
- 9.Jepson, M., A. Howells, H. L. Bullifent, B. Bolgiano, D. Crane, J. Miller, J. Holley, P. Jayasekeera, and R. W. Titball. 1999. Differences in the carboxy-terminal (putative phospholipid binding) domains of Clostridium perfringens and Clostridium bifermentans phospholipases C influence the hemolytic and lethal properties of these enzymes. Infect. Immun. 67:3297-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jepson, M., and R. W. Titball. 2000. Structure and function of clostridial phospholipase C. Microb. Infect. 2:1277-1284. [DOI] [PubMed] [Google Scholar]
- 11.Jepson, M., H. L. Bullifent, D. Crane, M. Flores-Diaz, A. Alape-Giron, P. Jayasekeera, B. Lingard, D. Moss, and R. W. Titball. 2001. Tyrosine 331 and phenylalanine 334 in Clostridium perfringens α-toxin are essential for cytotoxic activity. FEBS Lett. 495:172-177. [DOI] [PubMed] [Google Scholar]
- 12.Katayama, S., O. Matsushita, C. M. Jung, J. Minami, and A. Okabe. 1999. Promoter upstream bent DNA activates the transcription of the Clostridium perfringens phospholipase C gene in a low temperature-dependent manner. EMBO J. 18:3442-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurioka, S., and M. Matsuda. 1976. Phospholipase C assay using p-nitrophenylphosphoryl-choline together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal. Biochem. 75:281-289. [DOI] [PubMed] [Google Scholar]
- 14.Leslie, D., N. Fairweather, D. Pickard, G. Dougan, and M. Kehoe. 1989. Phospholipase C and haemolytic activities of Clostridium perfringens alpha-toxin cloned in Escherichia coli: sequence and homology with a Bacillus cereus phospholipase C. Mol. Microbiol. 3:383-392. [DOI] [PubMed] [Google Scholar]
- 15.Nagahama, M., Y. Okagawa, T. Nakayama, E. Nishioka, and J. Sakurai. 1995. Site-directed mutagenesis of histidine residues in Clostridium perfringens alpha-toxin. J. Bacteriol. 177:1179-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagahama, M., and J. Sakurai. 1996. Threonine-74 is a key site for the activity of Clostridium perfringens alpha-toxin. Microbiol. Immunol. 40:189-193. [DOI] [PubMed] [Google Scholar]
- 17.Nagahama, M., T. Nakayama, K. Michiue, and J. Sakurai. 1997. Site-specific mutagenesis of Clostridium perfringens alpha-toxin: replacement of Asp-56, Asp-130, or Glu-152 causes loss of enzymatic and hemolytic activities. Infect. Immun. 65:3489-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagahama, M., M. Mukai, S. Ochi, and J. Sakurai. 2000. Role of tryptophan-1 in hemolytic and phospholipase C activities of Clostridium perfringens alpha-toxin. Microbiol. Immunol. 44:585-589. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura, S., T. Shimamura, H. Hayashi, and S. Nishida. 1975. Reinvestigation of the taxonomy of Clostridium bifermentans and Clostridium sordellii. J. Med. Microbiol. 8:299-309. [DOI] [PubMed] [Google Scholar]
- 20.Naylor, C. E., J. T. Eaton, A. Howells, N. Justin, D. S. Moss, R. W. Titball, and A. K. Basak. 1998. Structure of the key toxin in gas gangrene. Nat. Struct. Biol. 5:738-746. [DOI] [PubMed] [Google Scholar]
- 21.Naylor, C. E., M. Jepson, D. T. Crane, R. W. Titball, J. Miller, A. K. Basak, and B. Bolgiano. 1999. Characterisation of the calcium-binding C-terminal domain of Clostridium perfringens alpha-toxin. J. Mol. Biol. 294:757-770. [DOI] [PubMed] [Google Scholar]
- 22.Ninomiya, M., O. Matsushita, J. Minami, H. Sakamoto, M. Nakano, and A. Okabe. 1994. Role of alpha-toxin in Clostridium perfringens infection determined by using recombinants of C. perfringens and Bacillus subtilis. Infect. Immun. 62:5032-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okabe, A., T. Shimizu, and H. Hayashi. 1989. Cloning and sequencing of a phospholipase C gene of Clostridium perfringens. Biochem. Biophys. Res. Commun. 160:33-39. [DOI] [PubMed] [Google Scholar]
- 24.Saint-Joanis, B., T. Garnier, and S. T. Cole. 1989. Gene cloning shows the alpha-toxin of Clostridium perfringens to contain both sphingomyelinase and lecithinase activities. Mol. Gen. Genet. 219:453-460. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Shamala, V., and G. F.-L. Ames. 1993. Single specific primer-polymerase chain reaction (SSP-PCR) and genome walking. Methods Mol. Biol. 15:339-348. [DOI] [PubMed] [Google Scholar]
- 27.Stevens, D. L., and A. E. Bryant. 1999. The pathogenesis of shock and tissue injury in clostridial gas gangrene, p. 623-636. In J. E. Alouf and J. H. Freer (ed.), The comprehensive sourcebook of bacterial protein toxins, 2nd ed. Academic Press, Ltd., London, United Kingdom.
- 28.Titball, R. W., S. E. Hunter, K. L. Martin, B. C. Morris, A. D. Shuttleworth, T. Rubidge, D. W. Anderson, and D. C. Kelly. 1989. Molecular cloning and nucleotide sequence of the alpha-toxin (phospholipase C) of Clostridium perfringens. Infect. Immun. 57:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Titball, R. W., D. L. Leslie, S. Harvey, and D. Kelly. 1991. Hemolytic and sphingomyelinase activities of Clostridium perfringens alpha-toxin are dependent on a domain homologous to that of an enzyme from the human arachidonic acid pathway. Infect. Immun. 59:1872-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Titball, R. W., H. Yeoman, and S. E. C. Hunter. 1993. Gene cloning and organization of the alpha-toxin of of Clostridium perfringens, p. 211-226. In M. Sebald (ed.), Genetics and molecular biology of anaerobic bacteria. Springer-Verlag, New York, N.Y.
- 31.Titball, R. W., C. E. Naylor, and A. K. Basak. 1999. The Clostridium perfringens α-toxin. Anaerobe 5:51-64. [DOI] [PubMed] [Google Scholar]
- 32.Titball, R. W. 1999. Membrane-damaging and cytotoxic phospholipases, p. 310-329. In J. E. Alouf and J. H. Freer (ed.), The comprehensive sourcebook of bacterial protein toxins, 2nd ed. Academic Press, Ltd., London, United Kingdom.
- 33.Titball, R. W., C. E. Naylor, J. Miller, D. S. Moss, and A. K. Basak. 2000. Opening of the active site of Clostridium perfringens α-toxin may be triggered by membrane binding. Int. J. Med. Microbiol. 290:357-361. [DOI] [PubMed] [Google Scholar]
- 34.Tso, J. Y., and C. Siebel. 1989. Cloning and expression of the phospholipase C gene from Clostridium perfringens and Clostridium bifermentans. Infect. Immun. 57:468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsutsui, K., J. Minami, O. Matsushita, S. Katayama, Y. Taniguchi, S. Nakamura, M. Nishioka, and A. Okabe. 1995. Phylogenetic analysis of phospholipase C genes from Clostridium perfringens types A to E and Clostridium novyi. J. Bacteriol. 177:7164-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker, N., J. Holley, C. E. Naylor, M. Flores-Diaz, A. Alape-Giron, G. Carter, F. J. Carr, M. Thelestam, M. Keyte, D. S. Moss, A. K. Basak, J. Miller, and R. W. Titball. 2000. Identification of residues in the carboxy-terminal domain of Clostridium perfringens alpha-toxin (phospholipase C) which are required for its biological activities. Arch. Biochem. Biophys. 384:24-30. [DOI] [PubMed] [Google Scholar]
- 37.Wang, X., T. Maegawa, T. Karasawa, S. Kozaki, K. Tsukamoto, Y. Gyobu, K. Yamakawa, K. Oguma, Y. Sakaguchi and S. Nakamura. 2000. Genetic analysis of type E botulinum toxin-proucing Clostridium butyricum strains. Appl. Environ. Microbiol. 66:4992-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williamson, E. D., and R. W. Titball. 1993. A genetically engineered vaccine against the alpha-toxin of Clostridium perfringens protects mice against experimental gas gangrene. Vaccine 11:1253-1258. [DOI] [PubMed] [Google Scholar]