Abstract
Pathogenic mycobacteria survive and replicate within host macrophages, but the molecular mechanisms involved in this necessary step in the pathogenesis of infection are not completely understood. Mycobacterium marinum has recently been used as a model for aspects of the pathogenesis of tuberculosis because of its close genetic relationship to Mycobacterium tuberculosis and because of similarities in the pathology and course of infection caused by this organism in its natural hosts, fish and frogs, with tuberculosis in humans. In order to advance the utility of the M. marinum model, we have developed efficient transposon mutagenesis of the organism by using a Drosophila melanogaster mariner-based transposon. To determine the efficiency of transposition, we have analyzed pigmentation mutants from the transposon mutant library. In addition to insertions in four known genes in the pathway of pigment biosynthesis, two insertions in novel genes were identified in our mutant library. One of these is in a putative inhibitor of the carotenoid biosynthesis pathway. The second unexpected insertion is in an intergenic region between two genes homologous to Rv2603c and Rv2604c of M. tuberculosis. In addition to a pigmentation defect, this mutant showed increased susceptibility to singlet oxygen and grew poorly in murine macrophages. Complementation with M. tuberculosis genomic DNA encompassing Rv2603c to Rv2606c corrected the pigmentation and growth defects of the mutant. These data demonstrate the utility of mariner-based transposon mutagenesis of M. marinum and that M. marinum can be used to study the function of M. tuberculosis genes involved in intracellular survival and replication.
The global burden of Mycobacterium tuberculosis infection is overwhelming, with approximately one-third of the world's population infected and 2 million deaths each year (9). As an intracellular obligate human pathogen, M. tuberculosis has evolved a complex parasitic lifestyle and sophisticated mechanisms to combat host defense machinery. It modulates a number of host cell processes, including inhibition of acidification of the bacterium-containing phagosome and its maturation to a phagolysosome (7, 34), inhibition of host cell apoptosis to benefit intracellular bacterial growth and to evade detection and destruction by the host immune system (12, 17), and inhibition of the macrophage response to gamma interferon, thus altering antigen presentation (20, 24, 42). All of these mechanisms are thought to lead ultimately to enhanced survival of M. tuberculosis within macrophages. Despite this understanding of the exploitation of host cell processes by mycobacteria, the bacterial components responsible for mediating these processes are largely unknown. In addition to these basic issues in host-microbe interaction, efforts to understand the molecular bases for mycobacterial subversion of host defense has value for the development of new antibiotics that target these bacterial mediators. One direct and effective approach to this goal is to generate, isolate, and characterize defined mutations that affect steps in the pathogenesis of infection.
Transposon mutagenesis is a powerful technique for the identification of bacterial virulence factors which uses a traceable mobile element to randomly disrupt genes in the chromosome. Several transposable elements have been identified and isolated from Mycobacterium species, such as IS1096 (from Mycobacterium smegmatis) and IS6110 (from M. tuberculosis), and used to generate transposon mutants in mycobacteria (15, 21). However, these transposons do not function in all mycobacteria (22, 41) and seem to require recognition of relatively large sequences for insertion (21). Therefore, the development of a transposon that functions in a broad host range, recognizes a short sequence for insertion, and segregates from transposase upon chromosomal insertion should provide an extremely useful and potentially more-general tool for genetic studies of mycobacteria as well as other microorganisms. Such potentially valuable transposon systems have been identified, initially in insects, and include members of the mariner/Tc1 transposon superfamily (16, 27). Mos1 (from Drosophila melanogaster [32]) and Himar1 (from horn fly Haematobia irritans [19]) are among the best-studied mariner transposons that target insertion at a TA dinucleotide recognition site (27). mariner transposons are able to insert into diverse genomes of distantly related organisms, including both eukaryotes and prokaryotes (3, 11, 14, 19, 33, 36).
Mycobacterium marinum, a fish, amphibian, and opportunistic human pathogen, has recently been developed as a model system for studying M. tuberculosis pathogenesis (6, 28, 29, 31, 40). M. marinum is phylogenetically closest to members of the M. tuberculosis complex (43), and the infection it causes in its natural hosts manifests pathological hallmarks of tuberculosis, including granulomas (6, 31, 40). These observations suggest that many of the basic mechanisms of disease initiation are conserved between M. marinum and M. tuberculosis. M. marinum grows significantly faster than M. tuberculosis and other slow-growing mycobacteria, with a generation time of 4 h compared to 20 h for M. tuberculosis (28). Therefore, M. marinum offers an attractive and potentially valuable model for studying the molecular biology of mycobacterial pathogenesis. In the present work, we demonstrate that a mariner-based transposon system can be used for facile generation of random mutations in M. marinum (M. marinum mariner-based mutagenesis [M4]) and that these mutants can be used to identify and study genes involved in intracellular survival.
MATERIALS AND METHODS
Bacterial strains and media.
M. marinum strain M (the kind gift of Lalita Ramakrishnan, University of Washington, Seattle) was cultured either on Middlebrook 7H9 (Difco) supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% albumin-dextrose-catalase enrichment (7H9 broth) or on Middlebrook 7H10 agar (Difco), supplemented with 0.5% glycerol and 10% oleic acid-albumin-dextrose-catalase enrichment. The initial inoculum was cultured at 32°C in 7H9 broth to mid-log phase with slow shaking (100 rpm), and the culture was concentrated in 7H9 broth containing 30% glycerol, aliquoted, and frozen at −80°C (passage 1). Subsequent cultures were carried out by thawing frozen aliquots of passage 1 on ice and growing them in 50 ml of 7H9 broth for 4 days to reach an optical density at 600 nm (OD600) of ∼1.5 (passage 2). For infection or transformation experiments, passage 2 cultures were diluted 50 times and cultured for an additional 48 h to reach an OD600 of ∼1.8 to 1.9 (post-log phase) (passage 3). When required, the Middlebrook media were supplemented with the antibiotic kanamycin (KAN) or Zeocin (ZEO) (Invitrogen) at a concentration of 50 or 100 μg/ml, respectively. To test strains of M. marinum for sensitivity to light induction of methylene blue, passage 3 cultures were centrifuged at 1,000 × g for 5 min. The supernatants containing single bacilli were diluted, and small aliquots were spotted onto 7H10 plates containing 25 μM methylene blue and incubated in the presence of light. Escherichia coli DH5α (Invitrogen) was used for DNA cloning procedures. E. coli was grown in Luria-Bertani broth or on agar (Difco) at 37°C. When required, Luria-Bertani media were supplemented with the following antibiotics: carbenicillin at 50 μg/ml, KAN at 30 μg/ml, gentamicin at 15 μg/ml, or ZEO at 25 μg/ml. Standard molecular biology techniques involving cloning in E. coli were performed as previously described (35).
Construction of mariner-based transposon mutagenesis vectors.
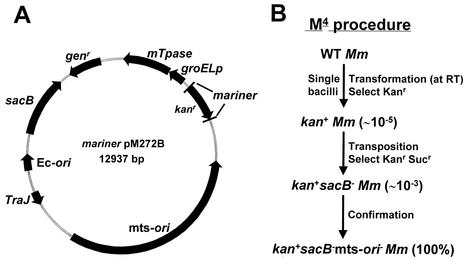
A series of mariner-based transposon mutagenesis vectors were constructed by inserting different mariner transposon cassettes into the plasmid backbone of pPR27 (26), a mycobacterial shuttle vector that contains the sacB gene from Bacillus subtilis (38), the Genr marker, an E. coli origin of replication, and a thermosensitive Mycobacterium ori (mts-ori). These mariner transposon cassettes contained the following elements constructed in different orientations: the 5′ and 3′ inverted repeats of Mos1 surrounding the Kanr cassette of Tn903 (from MosKΔCD [44]) and the Mos1 transposase (mTpase from pET3a-Tpase [44]), which was under the control of the mycobacterial groEL promoter (groELp from pSMT3 [13]). These mariner-based transposon vectors had four different orientations of the mariner Kanr and the groEL promoter-mariner transposase cassettes, namely, pM271A, pM271B, pM272A, and pM272B. The map of one of these vectors, pM272B, is shown in Fig. 1A.
FIG. 1.
Map of the mariner transposon mutagenesis vector pM272B (A) and the M4 procedure (B). (B) Transposon insertion mutants were isolated by a two-step selection procedure, first for Kanr and second for Kanr and Sucr. The numbers in parentheses indicate the frequencies of the occurrence of resistant colonies. Mm, M. marinum; Ec-ori, E. coli origin of replication; WT, wild type; RT, room temperature; sacB−, sacB deficient; mts-ori−, mts-ori deficient.
Transformation of M. marinum with mariner-based transposon mutagenesis vectors and selection for transposition mutations.
A passage 3 culture of the wild-type M. marinum strain M was washed three times in a wash buffer containing 10% glycerol and 0.05% Tween 80 and resuspended in an appropriate volume of wash buffer to an OD600 of ∼1.5 (competent bacteria) prior to electroporation. In preliminary experiments, we examined growth stage, bacterial clumping, electroporation conditions, and temperature as variables that could affect the efficiency of transformation. In addition, we tested one-step selection against sequential positive and negative selections for transposition of the mini-transposon into the bacterial chromosome. Finally, we compared the four transposon mutagenesis vectors described above for transformation and transposition efficiencies. Our final M4 protocol for efficient transformation and transposition is illustrated in Fig. 1B. M. marinum bacteria grown to an OD600 of ∼1.9 were repeatedly passed through a 26-gauge needle and then centrifuged at 500 × g to remove clumps. All procedures were carried out at room temperature. Competent bacteria (200 μl) were mixed with ∼2 μg of plasmid pM272B, incubated for 10 min to facilitate DNA adsorption, transferred into a 0.2-cm-gap electroporation cuvette, and subjected to a pulse of 2.5 kV, 25 μF, and 1,000 Ω (Gene Pulser II; Bio-Rad). Electroporated bacteria were incubated in 10 ml of 7H9 broth at 30°C for 6 h or overnight before being plated onto 7H10 agar containing KAN (positive selection) at ∼108 bacteria/plate. Transformants were observed after 6 to 7 days of incubation at 32°C. As a negative control to estimate the level of spontaneous resistance to KAN, competent bacteria were electroporated with the transposon-free vector pPR27 and further handled in the same way. The transformation efficiency for pM272B was ∼1.2 × 104 CFU/μg of DNA/109 mycobacteria electroporated compared to an extremely low background of ∼10 CFU/μg of DNA/109 mycobacteria electroporated for pPR27. To enrich and select for transposition mutations, the transformants obtained after 7 to 10 days of incubation were pooled (∼100 colonies/pool) and cultured in 50 ml of 7H9 broth containing KAN to an OD600 of ∼1.5. Single bacteria were obtained as described above and plated onto multiple 7H10 plates containing KAN and 10% sucrose (Suc) (negative selection) at ∼105 bacteria/plate. Transposon insertion mutants were obtained after 7 to 10 days of incubation at 32°C. The transposition efficiency was approximately 3 × 102 Kanr and Sucr bacteria/105 Kanr bacteria. To ensure that each colony arose from a single mutant, bacteria were resuspended, passed through a 26-gauge needle, recentrifuged, and replated. Individual mutant colonies were cultured in 1 ml of 7H9 broth containing KAN to late-log phase, mixed with glycerol (30%), transferred to 96-well plates, and frozen at −80°C (passage 1).
Detection of mariner-based transposon insertions by PCR.
Transposon insertion mutants selected on 7H10 agar containing KAN and Suc were initially characterized by PCR designed to amplify elements that define transposon insertion into the chromosome, including Kanr, sacB, and mts-ori. The primers used for detecting Kanr were KN-15′ (5′ GAGGCAGTTCCATAGGATGG) and KN-23′ (5′ TCAGGTGCGACAATCTATCG). The primers used for detecting sacB were SACB-15′ (5′ ACCCATCACATATACCTGCC) and SACB-23′ (5′ ATCGTTAGACGAAATGCCGT). The primers used for detecting essential open reading frames (ORFs) 1 and 2 of mts-ori were MORI-15′ (5′ AGGACCGCAAAGATGTTCCG) and MORI-23′ (5′ GATCTATGCCGAGTGCCACG). Transposon insertion mutant bacteria were resuspended in H2O, and ∼105 bacteria were added to each PCR as a source of template DNA. The PCR products were examined by agarose gel electrophoresis.
Southern blot analysis.
Passage 3 cultures (OD600 of ∼1.0, 50 ml) of strains of M. marinum were used for isolation of genomic DNA by the cetyltrimethylammonium bromide method (21, 23) with minor modifications. After delipidation with 1 ml of chloroform-methanol (2:1) for 5 min, the bacteria were resuspended in 10 mM Tris-1 mM EDTA (pH 8.0), digested with lysozyme (10 mg/ml) at 37°C for 5 h, and further digested with proteinase K (200 μg/ml) in 1% sodium dodecyl sulfate at 55°C for an additional 3 h. Fully digested bacteria were extracted once with cetyltrimethylammonium bromide-NaCl and then twice with phenol-chloroform-isoamyl alcohol (25:24:1) before precipitation of genomic DNA with isopropanol. Approximately 1 μg of Tris-HCl-dissolved genomic DNA was digested with PstI or BamHI and subjected to Southern blotting by following standard procedures (32). The probe was labeled and hybridization was detected by using an enhanced chemiluminescence random prime labeling and detection system (Amersham Pharmacia, Piscataway, N.J.) according to the manufacturer's instructions.
Recovery of mariner transposon insertion sites from mutants and sequencing of the insertion junction.
Genomic DNA (∼200 ng) of transposon insertion mutants was digested with PstI or BamHI, and the restriction fragments were self-ligated. The genomic sequence surrounding the transposon was amplified by performing inverse PCR with the outward primers 821A (5′ CGCATCTTCCCGACAACGCAGACCGTTCC) and 822A (5′ TAATCGCGGCCTCGAGCAAGACGTTTCCCG). Inverse PCR products were confirmed for the appropriate sizes by agarose gel electrophoresis or verified by Southern blotting and then sequenced with either primer 821A or primer 822A.
Isolation of pigmentation mutants of M. marinum and localization of the transposon insertions.
A pool of ∼15,000 transposon insertion mutants was screened for the isolation of pigmentation mutants. After growth of the mutant colonies to a visible size in the dark, the agar plates were exposed to light on the bench top. Wild-type M. marinum turned gold upon exposure to light in approximately 48 h. Pigmentation variants were identified that either did not produce colored pigment or were red rather than gold. The chromosomal location of the transposon insertion for mutant MmR03 relative to those of other pigment mutants was determined by PCR with genomic DNA from an individual mutant as a template and a combination of primer R03F1 (5′ GTGCCACCGTTCCAGTGGTTTGGGTGCCC) or R03R1 (5′ GGGCACCCAAACCACTGGAACGGTGGCAC) with 821A or 822A. The R03F1 and R03R1 primers were designed based on the genomic sequences surrounding the transposon insertion in MmR03.
Construction of mycobacterial expression vector pLYG204.Zeo.
A mycobacterial expression vector containing a Zeor determinant was constructed based on pMV261.kan (39). The Zeor gene was PCR amplified from pEM7/Zeo (Invitrogen) and ligated into NheI- and SpeI-digested pMV261.kan to generate pLYG204.Zeo, replacing the Kanr gene of pMV261 with the Zeor gene.
Complementation of M. marinum pigmentation mutants.
To complement the crtB defect of mutant MmW01, the wild-type crtB gene coding sequence (30) was PCR amplified with M. marinum genomic DNA as the template and primers mcrtBF1 (5′ GCTACGTATTCGCAGTGAATTGGACGCCGCCG, with an engineered SnaBI site underlined) and mcrtBR1 (5′ GGAATTCGCTCATGCGGCACCGTACTCGGAG, with an engineered EcoRI site underlined). The PCR product was cloned into EcoRV-digested pBluescript II KS(+) to generate pBSmcrtB. The SnaBI-to-EcoRI fragment of pBSmcrtB was subsequently cloned into MscI- and EcoRI-digested pLYG204.Zeo to create pLYG601, in which the 5′ end of the crtB coding sequence was placed in frame with the plasmid-derived start codon following the groEL promoter. pLYG601 was electroporated into MmW01 and MmR02, and the transformants were selected on 7H10 plates containing ZEO. The empty vector pLYG204.Zeo was used as a negative control for complementation.
To restore the defects of mutant MmW04, the M. tuberculosis genomic fragment encompassing Rv2603c to Rv2606c (corresponding to bases 2930803 to 2934068 of the published H37Rv sequence [8]) was PCR amplified from genomic DNA of the Erdman strain and ultimately cloned into pLYG204.Zeo to create pLYG602. The electroporation of pLYG602 into MmW04 and the selection of transformants were performed as described above.
Infection of J774 macrophages by M. marinum.
The mouse macrophage-like cell line J774 (ATCC TIB67) was maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone). Immediately prior to infection, J774 monolayers were washed once with FBS-free DMEM. Single disaggregated bacilli of passage 3 cultures of wild-type, mutant, or complemented M. marinum (OD600, ∼1.9) were added to J774 monolayers at a multiplicity of infection of 3 and incubated for 2 h at 32°C and 5% CO2. At the end of the 2-h incubation period, infected monolayers were washed twice with DMEM and further incubated in DMEM containing 2% FBS and 200 μg of amikacin/ml for 2 h to kill extracellular bacteria. At the end of the antibiotic treatment, monolayers were washed twice with DMEM and incubated in DMEM containing 1% FBS at 32°C and 5% CO2. This was designated time zero. The incubation medium was changed at 48 h. At 96 h after infection, bacteria were enumerated by combining supernatants and hypotonically lysed cell monolayers for quantitation of CFU. M. marinum grows poorly in tissue culture medium containing small amounts of FBS (data not shown), and at 96 h following infection, the cells infected with wild-type M. marinum showed some cytotoxicity. Therefore, the bacteria in the supernatant (always <10% of the total) represent bacteria that have escaped from macrophages, probably after lysis.
Similarity searches.
Similarity searches were performed by using BLASTp and BLASTn (2), as appropriate, at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/). Similarity searches against the incomplete M. marinum genome sequences were performed at the Sanger Center website (http://www.sanger.ac.uk/Projects/M_marinum/). Domain similarities were found by using the conserved domain search at the National Center for Biotechnology Information.
RESULTS
Molecular characterization of transposon insertion into the chromosome of M. marinum.
M. marinum strain M was transformed with the mariner-based transposon mutagenesis vector pM272B (Fig. 1A), and the transposition mutants were selected by using the M4 protocol described in Materials and Methods and illustrated in Fig. 1B. Since the vector contains the mariner transposon with a Kanr gene for positive selection and a sacB gene for counterselection, the Kanr and Sucr bacteria potentially had a chromosomal insertion(s) by mariner, with loss of the delivery vector. To test this, we examined the presence of the Kanr gene, sacB, and mts-ori by PCR in 15 randomly picked Kanr and Sucr colonies (data not shown). All contained the Kanr gene but had lost mts-ori and sacB, suggesting the integration of the transposon to the chromosome. Southern blots of PstI-digested or BamHI-digested genomic DNA probed with the transposon sequences yielded a single hybridization fragment for each of the colonies, suggesting a single transposition event for each colony, and each hybridization fragment had a unique size, indicating that the transposon had inserted at different sites of the chromosome (data not shown). There was no hybridization with sequences from the plasmid backbone (data not shown), indicating loss of the delivery vector sequence. In subsequent Southern blot analyses of over 50 colonies, only 2 have shown evidence of more than one insertion, suggesting that the vast majority of colonies arose from single transposition events. Additional proof for transposon insertion into the chromosome was obtained by hybridization of undigested genomic DNA from the Kanr and Sucr bacteria with the Kanr gene probe: hybridization was detected only at the size range for genomic DNA (data not shown). Definitive proof for transposon insertion into the chromosome was obtained by sequencing across the transposon insertion junctions outwards from mariner. All sequences (now from 40 independent colonies) have a TA dinucleotide at the transposon-chromosome junction, followed by diverse GC-rich genomic sequences. Taken together, these results prove that M4 leads to integration of the mariner transposon into the chromosome of M. marinum at random sites, usually as a single copy per genome, and that the delivery vector is completely lost upon transposition.
Identification and characterization of transposon insertion within genes of the carotenoid biosynthesis pathway.
Using the M4 protocol, we obtained a M. marinum mutant library consisting of ∼15,000 individual transposon insertion mutants from a single electroporation. To evaluate the randomness of mariner transposition, we screened for pigmentation mutants. Assuming that M. marinum has about the same number of genes as M. tuberculosis (∼4,000), if transposition is random, the Poisson statistic predicts that the likelihood of failure to find in this library at least one insertion in any gene involved in pigmentation is approximately 2%. Normally, M. marinum is gold when exposed to light because of the production of carotenoid pigments (30, 45). We identified eight colonies that demonstrated pigmentation variations: four of them lost pigment, resulting in white colonies (MmW01 to MmW04); three others showed a red colony color upon exposure to light (MmR01, MmR02, and MmR04); and one appeared red in the dark and developed a gold color after exposure to light (MmR03) (Fig. 2). The sequences of the transposon insertion junctions for all eight mutants showed that seven of them were unique insertions. MmW01 and MmW02 showed the same sequence, which could be due either to duplication of siblings or to independent insertions at the same site.
FIG. 2.
Pigmentation of M. marinum mutants in the presence or absence of light. M. marinum colonies were grown in the dark (left) and then exposed to light for 48 h (right). Wild-type (WT) M. marinum is pigmented only after light exposure. R01, MmR01; R02, MmR02; R03, MmR03; R04, MmR04; W01, MmW01; W03, MmW03; W04, MmW04.
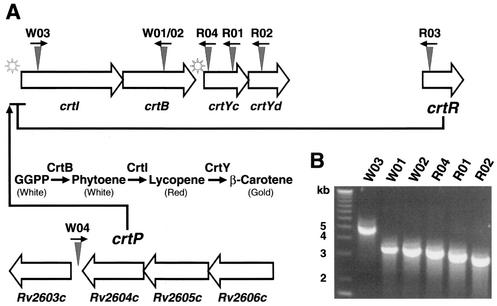
BLAST searches performed for these seven unique insertion junction sequences revealed that five of them matched segments of four previously identified contiguous ORFs of M. marinum in a carotenoid biosynthesis locus (30). As shown in Fig. 3A, MmW01 and MmW02 had an insertion in the crtB gene and MmW03 had an insertion in the crtI gene. MmR01, MmR02, and MmR04 had independent insertions in two previously identified (30) but uncharacterized ORFs (ORF-3 and ORF-4) of this M. marinum locus. We found that ORF-3 and ORF-4 have significant homology to crtYc and crtYd, respectively, of Mycobacterium aurum and Brevibacterium linens (18, 30, 45). Both the sequence and genomic organization of crtI, crtB, crtYc, and crtYd are highly conserved among M. marinum, M. aurum, and B. linens (18, 30, 45). As diagrammed in Fig. 3A, our transposon mutant library contained at least one mutation in each of these ORFs identified in the M. marinum carotenoid biosynthesis gene locus. Although only one locus was analyzed, this frequency is consistent with the generation of random mutations throughout the M. marinum genome by mariner.
FIG. 3.
M. marinum carotenoid biosynthesis and regulatory genes, their genomic organization, and location of transposon insertions. (A) The known carotenoid biosynthesis locus in M. marinum is shown as a gene cluster from crtI to ORF-4. Below these genes the biosynthetic pathway is depicted; GGPP represents the initial substrate, geranylgeranylpyrophosphate. The novel crtR and crtP genes encode a putative repressor and a positive regulator, respectively, for the carotenoid biosynthesis locus. crtP indicates the locus containing the Rv2606c to Rv2603c homologues. The star symbols represent putative light-inducible promoters. Triangles indicate sites of transposon insertion, and arrows represent the orientation of the Kanr gene in the chromosome. R01, MmR01; R02, MmR02; R03, MmR03; R04, MmR04; W01, MmW01; W02, MmW02; W03, MmW03; W04, MmW04. (B) The location of transposon insertion in mutant MmR03 is shown relative to those in other pigmentation mutants as detected by a PCR that amplifies the genomic sequence between insertions. The distance of the transposon insertion in MmW04 from others in the carotenoid biosynthesis locus is insufficiently detected by PCR, and the results were not shown.
CrtB (phytoene synthase) and CrtI (phytoene desaturase) are thought to catalyze two consecutive biochemical reactions leading to the synthesis of lycopene, which has a red pigment (Fig. 3A) (45). Mutations in either of these genes in MmW01/MmW02 and MmW03 produced no colored pigments (intermediates before lycopene in the pathway are not bright colored) (Fig. 2). MmR01, MmR02, and MmR04 had mutations in the crtY locus, which controls the conversion of lycopene (red) to beta carotene (gold) (18, 45). As predicted, these mutants accumulated lycopene and were therefore red (Fig. 2).
In addition to these five insertions within the locus known to be involved in carotene biosynthesis, we identified two mutants, MmR03 and MmW04, that had insertions within novel genes. The sequence at the transposon insertion junction of MmR03 was used in a BLAST search of the M. marinum database at the Sanger Center (http://www.sanger.ac.uk/Projects/M_marinum/). The insertion junction sequence was identical in reverse orientation to bases 77208 to 77843 of the assembled contig sequence mar502f07.plk. This sequence is within a hypothetical ORF that contains a domain homologous to the helix-turn-helix MarR family of transcription repressors (1). Thus, the gene interrupted by the transposon in MmR03 (crtR) is likely a repressor of the crt locus, consistent with production of red pigment in the mutant in the absence of light induction. We examined the chromosomal location of the MmR03 transposon insertion relative to the known carotenoid biosynthesis genes by a PCR protocol that amplified sequences between the transposon inserts. As shown in Fig. 3B, the MmR03 insertion is approximately 2.8 kb downstream from that of MmR02.
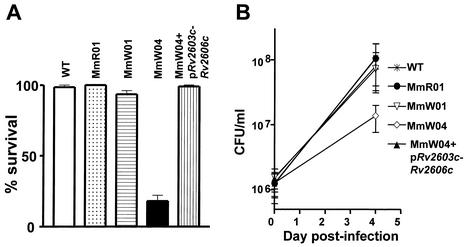
The second unanticipated transposon insertion (in MmW04) was in an intergenic sequence immediately downstream of a coding sequence homologous to Rv2604c of M. tuberculosis (8) (Fig. 3A). In the M. tuberculosis genome, Rv2604c and the two upstream genes, Rv2605c and Rv2606c, are likely in a single operon (Fig. 3A). Rv2603c is 130 bp downstream of Rv2604c and thus might also be part of the putative operon. Both Rv2603c and Rv2604c are highly conserved but without known function. Rv2605c is homologous to acyl coenzyme A thioesterase II genes in bacteria, and Rv2606c is homologous to a family of genes conserved among prokaryotes and eukaryotes that confer resistance to singlet oxygen generated by light induction of photosensitizers such as methylene blue. Consistent with the possibility that these genes have a similar function in mycobacteria, MmW04 showed a fivefold increase in sensitivity to singlet oxygen produced by light induction of methylene blue (Fig. 4A). The increased sensitivity did not result from the loss of pigment because the other white mutants were equivalent to the wild type in this assay. Unlike the other pigment mutants, MmW04 also showed reduced intracellular growth in J774 cells, with an eightfold-lower CFU count than the wild type at 4 days after infection (Fig. 4B). Thus, in addition to the lack of pigmentation, this mutant showed a general decrease in the ability to survive environmental stress. The transposon insertion in MmW04 is not sufficiently close to the known carotenoid biosynthesis genes to be linked by PCR or to be on the same BamHI fragment (∼10 kb) as the pigmentation locus, as assessed by Southern blot analysis (data not shown).
FIG. 4.
Examination of resistance to light induction of methylene blue (A) and intracellular replication within J774 macrophages (B) for M. marinum pigmentation mutants and the complemented strain. (A) Dilutions of M. marinum strains were plated onto 7H10 plates containing 25 μM methylene blue and incubated in the dark or in ambient light for 7 days. The data are percentages of CFU in the dark divided by CFU in ambient light for each strain. Data for the wild type (WT), three pigmentation mutants, and the complemented MmW04 strain are graphed. The data are means of results from two experiments under the same conditions. (B) CFU associated with J774 cells were enumerated at time zero and after 96 h of intracellular growth. Data for the same strains as those used for panel A are graphed. The means and standard deviations of results from quadruplicate experiments performed on two different days are depicted. pRv2603c-Rv2606c indicates complementation with M. tuberculosis genes encompassing Rv2603c to Rv2606c. Differences between MmW04 and the WT or the complemented strain are statistically significant (P < 0.05); the levels of growth of the WT and the complemented mutant are not different.
Complementation of the pigmentation mutants.
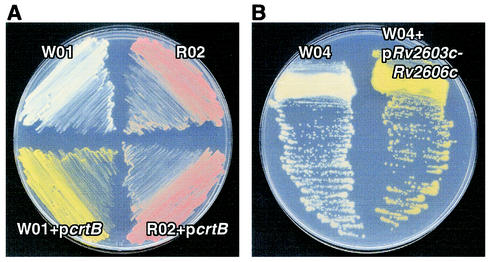
To prove that the mutant phenotypes were specifically caused by the transposon insertions, we carried out complementation studies by expressing the corresponding wild-type coding sequence within an appropriate mutant. We first developed a mycobacterial expression vector, pLYG204.Zeo, which carries a Zeor gene immediately downstream from a mycobacterial groEL promoter and multiple cloning sites for the insertion of a gene of interest. The M. marinum crtB gene coding sequence (30) was cloned into the multiple cloning site of pLYG204.Zeo and transformed into MmW01, and the Zeor colonies were examined for complementation of pigmentation. As shown in Fig. 5A, the mutant MmW01 was fully complemented by episomal expression of crtB to produce wild-type pigment. Complementation of MmW01 by crtB was specific, since the empty vector did not complement (data not shown), and the expression of crtB did not complement the MmR02 mutant (Fig. 5A).
FIG. 5.
Complementation of carotenoid biosynthesis for M. marinum mutants. (A) A plasmid expressing the M. marinum crtB gene was introduced into both MmW01 (W01) and MmR02 (R02), but it restored pigment synthesis only in MmW01. (B) A plasmid expressing M. tuberculosis genes Rv2603c to Rv2606c was used to complement the MmW04 (W04) mutant to produce wild-type pigment. Both plates were exposed to light for 48 h prior to imaging.
The homology of the insertion site in MmW04 with an M. tuberculosis locus provided an opportunity to assess the ability of M. tuberculosis genes to function in M. marinum. Because of the possibilities that (i) several of the genes in the region from Rv2603c to 2606c are likely in an operon, (ii) the direction of transcription of the transposon insertion is opposite to the direction of transcription of these genes (Fig. 3A), leading to the possibility of antisense transcripts arising from the transposon, and (iii) the insertion was in a regulatory region for one or more of these genes, we attempted to complement the MmW04 phenotypes by using an M. tuberculosis genomic fragment encompassing Rv2603c to Rv2606c. An ∼3.3-kb genomic fragment containing these genes was PCR amplified from the M. tuberculosis Erdman strain and cloned into pLYG204.Zeo. The expression plasmid was transformed into MmW04, and the phenotypes of the transformants were determined. The defects of the MmW04 mutant in pigmentation, resistance to singlet oxygen, and intracellular growth were completely restored to wild-type levels by expression of this M. tuberculosis genomic fragment (Fig. 4 and 5B). Thus, an M. tuberculosis operon was able to complement the defects of the MmW04 mutant.
DISCUSSION
We have developed a method for M4 that is efficient and simple. The mariner transposon inserts at a TA recognition site in M. marinum, without any other apparent consensus sequence requirement, as is the case in other organisms. This property allowed for the construction of a transposon mutant library containing remarkably random insertions. There are ∼70,000 possible mariner insertion sites in a typical mycobacterial chromosome. Since there are ∼4,000 genes, this suggests that there are many potential sites of transposition in each gene. Indeed, in our library of ∼15,000 individual mutants, we found mutations in multiple expected genes of M. marinum in the carotenoid biosynthesis pathway, suggesting that with M4 it will be possible to generate mutations within most genes compatible with survival in vitro. The ∼15,000 mutants that we screened were generated from a single electroporation, and a mutant library with a larger size can be easily obtained by plating out more Kanr bacteria on Kan- and Suc-containing plates, which could potentially saturate all possible TA sites. High-efficiency transposition by a slightly different mariner transposon has been reported for E. coli and several mycobacterial species (33). Together, these studies demonstrate the utility of the mariner transposon in random bacterial mutagenesis.
Our first mutant screen has enhanced the understanding of the regulation of carotenoid biosynthesis in M. marinum. We found one mutant (MmR03) that produces a red pigment in the dark, unlike wild-type M. marinum, which is white in the absence of light. Since the MmR03 mutant accumulated lycopene in the dark and produced beta carotene upon exposure to light, the normal gene product (CrtR) likely has an inhibitory activity on crtI and crtB that is released upon light induction. Consistent with this, the ORF interrupted by the transposon in MmR03 shows homology to the MarR family of transcription repressors, which bind DNA through a helix-turn-helix motif. Release of repression by MarR is thought to occur by direct binding of inducers to the repressor, leading to loss of DNA binding. Based on this, we hypothesize that CrtR is a constitutive inhibitor of crtI and crtB that is released when M. marinum is exposed to light. It will be interesting to determine the nature of the light-activated inducer of the crtI/crtB pathway in M. marinum. Expression of crtY also apparently requires induction by light, since the MmR03 mutant accumulates red, but not gold, pigment in the dark, yet it does develop a gold color when exposed to light. It is possible that there is a common inducer for both steps in carotenoid production.
Our screen also identified one nonpigmented mutant (MmW04) that had a transposon insertion outside the known carotenoid biosynthesis locus within a region that exhibits significant sequence homology to M. tuberculosis genes. Since the mutation also caused decreased resistance of M. marinum to singlet oxygen and compromised intracellular growth, the normal gene product (CrtP) is likely involved in the positive regulation of a general stress response in mycobacteria. M. tuberculosis does not produce colored pigments, yet its homologous locus complemented the defects of MmW04 in pigmentation, resistance to singlet oxygen, and intracellular growth. Thus, the role of the locus in regulating stress response is conserved among Mycobacterium species. In fact, genes homologous to those in this locus, such as SOR 1 (singlet oxygen resistance) (10), SNZ 1 (stationary-phase gene) (4), pyroA (pyridoxine biosynthesis) (25), and HEVER (Hevea ethylene responsive) (37), are conserved in distantly related organisms. All of these genes have been implicated in responses to a variety of stresses. As intracellular pathogens, mycobacteria must have mechanisms for protection from the stresses of the intraphagosomal environment; it is likely that the crtP locus (Rv2606c to Rv2603c) has a role in this response in M. tuberculosis, as its homologue does in M. marinum. In M. marinum, this locus has an additional role in regulating carotenoid biosynthesis. Since no other nonpigmented mutants have defects in intracellular survival or resistance to singlet oxygen, carotenoids apparently are not directly involved in protection against either of these stresses. This implies that the crtP locus activates stress responses in addition to pigmentation that more directly affect bacterial survival in the intracellular environment. We are currently investigating which gene(s) of the crtP locus is responsible for mediating the stress response in M. marinum.
The potential for using M. marinum as a model to study molecular mechanisms of M. tuberculosis pathogenesis has been explored systematically by Chan et al. (5) and Ramakrishnan et al. (29) as well as others (40). We have extended the potential utility of this model by showing that for genes with M. tuberculosis homologues, the M. tuberculosis genes can be used to complement the mutant M. marinum phenotypes. While this has been done to some extent in M. smegmatis, this organism does not persist in macrophages or other host cells and therefore cannot be used to study genes involved in intracellular growth and/or persistence. In contrast, M. marinum is able to replicate within macrophages, making it possible to screen for mutants defective in intracellular survival or growth by using the random transposon mutagenesis technique we describe here. Indeed, we have screened ∼1,000 mutants from this library for the ability to survive in macrophages and have identified 23 different genes, in addition to the MmW04 gene, that are involved in intracellular survival. Significantly, each of these genes has an M. tuberculosis homologue, and in all 10 mutants in which we have tried, we have successfully corrected the intracellular survival defect of the mutant with the M. tuberculosis homologue. Thus, M. marinum is a model organism well suited not only for discovery of mycobacterial genes involved in intracellular survival but also for functional analysis of M. tuberculosis genes required for this aspect of pathogenesis.
Acknowledgments
We thank Sophie Goyard and Luiz R. O. Tosi for technical assistance, J. Hiroshi Morisaki for assistance with preparing the manuscript, and Luisa M. Stamm for critical reading of the manuscript.
This work was supported by grants from the National Institutes of Health to E.J.B., S.M.B., and J.S.C. L.-Y.G. is supported by T32 AI07334.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessereau, J. L., A. Wright, D. C. Williams, K. Schuske, M. W. Davis, and E. M. Jorgensen. 2001. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413:70-74. [DOI] [PubMed] [Google Scholar]
- 4.Braun, E. L., E. K. Fuge, P. A. Padilla, and M. Werner-Washburne. 1996. A stationary-phase gene in Saccharomyces cerevisiae is a member of a novel, highly conserved gene family. J. Bacteriol. 178:6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, K., T. Knaak, L. Satkamp, O. Humbert, S. Falkow, and L. Ramakrishnan. 2002. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc. Natl. Acad. Sci. USA 99:3920-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, H. F., and C. C. Shepard. 1963. Effect of environmental temperatures on infection with Mycobacterium marinum (Balnei) of mice and a number of poikilothermic species. J. Bacteriol. 86:1057-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens, D. L., and M. A. Horwitz. 1995. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J. Exp. Med. 181:257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Dolin, P. J., M. C. Raviglione, and A. Kochi. 1994. Global tuberculosis incidence and mortality during 1990-2000. Bull. W. H. O. 72:213-220. [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrenshaft, M., P. Bilski, M. Y. Li, C. F. Chignell, and M. E. Daub. 1999. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc. Natl. Acad. Sci. USA 96:9374-9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fadool, J. M., D. L. Hartl, and J. E. Dowling. 1998. Transposition of the mariner element from Drosophila mauritiana in zebrafish. Proc. Natl. Acad. Sci. USA 95:5182-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, L. Y., and Y. A. Kwaik. 2000. The modulation of host cell apoptosis by intracellular bacterial pathogens. Trends Microbiol. 8:306-313. [DOI] [PubMed] [Google Scholar]
- 13.Gaora, P. O. 1998. Expression of genes in mycobacteria. Methods Mol. Biol. 101:261-273. [DOI] [PubMed] [Google Scholar]
- 14.Gueiros-Filho, F. J., and S. M. Beverley. 1997. Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276:1716-1719. [DOI] [PubMed] [Google Scholar]
- 15.Guilhot, C., I. Otal, I. Van Rompaey, C. Martin, and B. Gicquel. 1994. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J. Bacteriol. 176:535-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartl, D. L., A. R. Lohe, and E. R. Lozovskaya. 1997. Modern thoughts on an ancyent marinere: function, evolution, regulation. Annu. Rev. Genet. 31:337-358. [DOI] [PubMed] [Google Scholar]
- 17.Keane, J., H. G. Remold, and H. Kornfeld. 2000. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164:2016-2020. [DOI] [PubMed] [Google Scholar]
- 18.Krubasik, P., and G. Sandmann. 2000. A carotenogenic gene cluster from Brevibacterium linens with novel lycopene cyclase genes involved in the synthesis of aromatic carotenoids. Mol. Gen. Genet. 263:423-432. [DOI] [PubMed] [Google Scholar]
- 19.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 20.Manca, C., L. Tsenova, A. Bergtold, S. Freeman, M. Tovey, J. M. Musser, C. E. Barry, V. H. Freedman, and G. Kaplan. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl. Acad. Sci. USA 98:5752-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McAdam, R. A., T. R. Weisbrod, J. Martin, J. D. Scuderi, A. M. Brown, J. D. Cirillo, B. R. Bloom, and W. R. J. Jacobs. 1995. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect. Immun. 63:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendiola, M. V., C. Martin, I. Otal, and B. Gicquel. 1992. Analysis of the regions responsible for IS6110 RFLP in a single Mycobacterium tuberculosis strain. Res. Microbiol. 143:767-772. [DOI] [PubMed] [Google Scholar]
- 23.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noss, E. H., R. K. Pai, T. J. Sellati, J. D. Radolf, J. Belisle, D. T. Golenbock, W. H. Boom, and C. V. Harding. 2001. Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of Mycobacterium tuberculosis. J. Immunol. 167:910-918. [DOI] [PubMed] [Google Scholar]
- 25.Osmani, A. H., G. S. May, and S. A. Osmani. 1999. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J. Biol. Chem. 274:23565-23569. [DOI] [PubMed] [Google Scholar]
- 26.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. J. Jacobs, B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plasterk, R. H., Z. Izsvak, and Z. Ivics. 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15:326-332. [DOI] [PubMed] [Google Scholar]
- 28.Ramakrishnan, L., and S. Falkow. 1994. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect. Immun. 62:3222-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 30.Ramakrishnan, L., H. T. Tran, N. A. Federspiel, and S. Falkow. 1997. A crtB homolog essential for photochromogenicity in Mycobacterium marinum: isolation, characterization, and gene disruption via homologous recombination. J. Bacteriol. 179:5862-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramakrishnan, L., R. H. Valdivia, J. H. McKerrow, and S. Falkow. 1997. Mycobacterium marinum causes both long-term subclinical infection and acute disease in the leopard frog (Rana pipiens). Infect. Immun. 65:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson, H. M., and D. J. Lampe. 1995. Distribution of transposable elements in arthropods. Annu. Rev. Entomol. 40:333-357. [DOI] [PubMed] [Google Scholar]
- 33.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell, D. G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2:569-577. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 36.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sivasubramaniam, S., V. M. Vanniasingham, C. T. Tan, and N. H. Chua. 1995. Characterisation of HEVER, a novel stress-induced gene from Hevea brasiliensis. Plant Mol. Biol. 29:173-178. [DOI] [PubMed] [Google Scholar]
- 38.Steinmetz, M., D. Le Coq, S. Aymerich, G. Gonzy-Treboul, and P. Gay. 1985. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol. Gen. Genet. 200:220-228. [DOI] [PubMed] [Google Scholar]
- 39.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, and G. F. Hatfull. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 40.Talaat, A. M., R. Reimschuessel, S. S. Wasserman, and M. Trucksis. 1998. Goldfish, Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect. Immun. 66:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talaat, A. M., and M. Trucksis. 2000. Transformation and transposition of the genome of Mycobacterium marinum. Am. J. Vet. Res. 61:125-128. [DOI] [PubMed] [Google Scholar]
- 42.Ting, L. M., A. C. Kim, A. Cattamanchi, and J. D. Ernst. 1999. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J. Immunol. 163:3898-3906. [PubMed] [Google Scholar]
- 43.Tonjum, T., D. B. Welty, E. Jantzen, and P. L. Small. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tosi, L. R., and S. M. Beverley. 2000. cis and trans factors affecting Mos1 mariner evolution and transposition in vitro, and its potential for functional genomics. Nucleic Acids Res. 28:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viveiros, M., P. Krubasik, G. Sandmann, and M. Houssaini-Iraqui. 2000. Structural and functional analysis of the gene cluster encoding carotenoid biosynthesis in Mycobacterium aurum A+. FEMS Microbiol. Lett. 187:95-101. [DOI] [PubMed] [Google Scholar]