Abstract
YopM is a leucine-rich repeat (LRR) virulence protein that is delivered into host cells when any of the three human-pathogenic species of Yersinia binds to mammalian cells. It exhibits heterogeneity of size and sequence among the yersiniae, but the functional consequences of this variability are not yet known. Yersinia pestis YopM was previously shown to accumulate in the nuclei of infected HeLa cells by a mechanism that requires vesicular trafficking. In this study, we characterized the trafficking of Y. pestis YopM in a Saccharomyces cerevisiae model previously found to support nuclear localization of YopM from an enteropathogenic Yersinia strain (C. F. Lesser and S. I. Miller, EMBO J. 20:1840-1849, 2001). Y. pestis YopM was N-terminally fused to the yeast enhanced green fluorescent protein (yEGFP) and inducibly expressed in the cytoplasm. yEGFP-YopM localized to the yeast nucleus, showing that this property is conserved for YopMs so far tested and that infection and the presence of other Yops are not required for its trafficking. When expressed in S. cerevisiae that is temperature sensitive for vesicular transport, YopM failed to accumulate in the nucleus at the nonpermissive temperature but did accumulate when the permissive temperature was restored. This shows that vesicular trafficking also is required in yeast for normal localization of YopM. YopM consists of a 71-residue leader sequence, 15 LRRs, and a 32-residue tail. Deletion analysis revealed that the leader sequence or tail is alone insufficient to direct YopM to the nucleus, showing that the LRR structure is required. Both the N-terminal and C-terminal halves of YopM localized to the nucleus, indicating the possible presence of two nuclear localization signals (NLSs) in YopM or domains in YopM where an NLS-containing protein might bind; this fits with the presence of two highly conserved regions among Yersinia YopMs. yEGFP-YopM lacking LRRs 4 to 7 or 7 to 10 accumulated in the nucleus in yeast, and YopM lacking these LRRs concentrated normally in the HeLa cell nucleus after delivery by Yersinia infection, showing that these LRRs are not essential for YopM trafficking in eucaryotic cells. However, because Y. pestis carrying either of these YopMs is strongly compromised in virulence in mice, these findings revealed that LRRs 4 to 10 map a region of YopM or support a conformation of YopM that is necessary for a pathogenic effect.
YopM of the human-pathogenic Yersinia species is one of six virulence proteins, termed Yops, that are delivered to the host cell cytoplasm from surface-adherent bacteria via a contact-activated secretion mechanism (type III secretion system) (see, e.g., references 8 and 9). Of these six, YopM is the only one for which neither a target nor an activity has been identified. It is essential for full virulence of yersiniae in mice (15, 23, 26): a YopM mutant of Y. pestis is decreased in virulence by 4 orders of magnitude in a systemic plague model, although the major consequence of the mutation is not seen during the first 3 days of infection (23). Accordingly, YopM may function primarily to counteract a secondary wave of innate defenses.
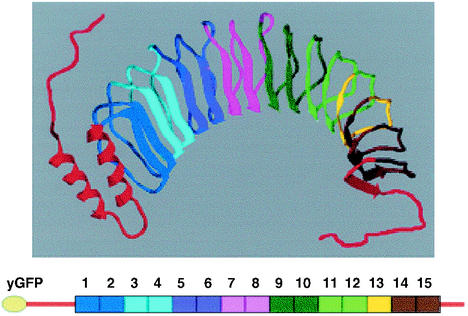
Y. pestis YopM is a very acidic 46-kDa protein that belongs to the leucine-rich repeat (LRR) structural family of proteins (6, 18). Unlike the other Yops, which are highly conserved among the human-pathogenic yersiniae, YopM shows considerable heterogeneity in numbers of LRRs and in the sequence of LRRs other than the first three and last two LRRs. LRR proteins have no common enzymatic function attributed to them so far, other than the pattern that they interact with other proteins (6). Y. pestis YopM has recently been crystallized (12). The molecule has a slightly twisted horseshoe shape [Fig. 1, top]. Each LRR contributes a parallel beta-sheet element and a kinky loop that is linked to the next LRR. The highly conserved residues that make up the LRR consensus are involved in interactions that stabilize this structure. Among LRR proteins, the Yersinia YopM proteins are unusual in that they consist almost entirely of LRRs. Y. pestis YopM contains 15 LRRs of 20 or 22 residues, a 71-residue leader sequence that is important for secretion by the type III secretion system (3), and a 32-residue C-terminal tail. Almost all other proteins in the LRR family have at least one major non-LRR domain that could provide an enzymatic activity. LRR domains can function in binding, e.g., Listeria InlA binding to E-cadherin (25), RNase inhibitor binding to RNase (20), and U2A′ binding to small nuclear RNA (29). Y. pestis YopM presents a considerably hydrophilic convex surface to its environment. However, it has a hydrophobic band around its convex surface, as well as hydrophobic residues within its concave face (12), and thus it has the potential to participate in a wide variety of interactions. It is not yet known how the heterogeneity among the Yersinia YopM proteins may translate into functional differences.
FIG. 1.
Structure of YopM and cartoon of yEGFP-YopM. (Top) Ribbon model of YopM based on PDB structure 1JL5 (12) modified by adding free-form lines to indicate residues at the beginning of the leader domain (red, at left) and at the end of the tail domain (red, at right) which were not resolved in the crystal structure. Coloring has been added to provide a visual aid for locating regions deleted in the various yEGFP-YopM molecules tested in this study (Table 1). (Bottom) Cartoon of yEGFP-YopM with numbered LRRs and the same coloring as in the ribbon model. The figure was generated using Swiss PBD Viewer and rendered with PovRay before final construction in Microsoft Powerpoint. It was printed from Adobe PhotoShop 6.0.
Y. pestis YopM was shown to bind thrombin and inhibit thrombin-elicited platelet aggregation (15, 23, 31, 33), prompting the hypothesis that YopM might function extracellularly to sequester thrombin at foci of infection and thereby play an anti-inflammatory role. However, YopM was not protective against plague in mice, by either active or passive immunization, as might be expected if it indeed played an extracellular role (27). Moreover, Y. pestis expressing a YopM variant that bound thrombin poorly had the same intermediate virulence as a strain expressing a variant that bound thrombin better than does native YopM (15). Hence, although some YopM is found in the surrounding medium when Y. pestis infects mammalian cells (34), the main function of YopM, at least in systemic plague, is likely to be within host cells after its delivery by the type III secretion system.
Within host cells, Y. pestis YopM traffics to the nucleus, and efficient nuclear accumulation requires functional vesicular trafficking (34). However, since there is no known vesicular delivery to the nucleus, we hypothesized that after its vesicular association, YopM must meet a chaperone or carrier that takes it through the nuclear pores and into the nucleus (34). YopM does not possess a canonical nuclear localization signal (NLS); hence, either it has a novel NLS or it may be carried into the nucleus by an NLS-containing protein to which it binds. These findings indicate that YopM probably has multiple interactions within the host cell: after delivery into the cytoplasm by the type III secretion system, it may interact with a component(s) on a vesicle, with a hypothetical protein that chaperones it into the nucleus, and presumably with a molecular target within the nucleus. Accordingly, YopM could have multiple pathogenic effects, one of which could be in the nucleus to modulate host cell gene expression to the benefit of the pathogen.
Recently, Lesser and Miller (22) demonstrated that YopM from an enteropathogenic Yersinia strain entered the yeast nucleus when expressed as a fusion to green fluorescent protein (GFP). This showed that that YopM did not require delivery by Yersinia or the presence of other Yops to localize to the nucleus. However, it was not determined if nuclear localization in yeast had the same requirements as did localization to the mammalian nucleus. In the present study, we determined that Y. pestis YopM also localizes in the yeast nucleus. Furthermore, we show that vesicular trafficking is necessary for this localization to be efficient. Tests with YopM domains and truncations showed that YopM may have two NLSs or binding sites for a carrier protein. Normal nuclear localization of Y. pestis YopM proteins lacking certain internal LRRs revealed a region of YopM that is dispensable for trafficking; however, these YopM proteins are defective in pathogenic function.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain DH5α (GIBCO-BRL) was used as a host for amplification and storage of plasmids. It was grown in Luria-Bertani broth (24) or on tryptose blood agar at 37°C. All Y. pestis strains were Pgm− (39). The YopM− Y. pestis strain KIM8-3233 (pPCP1− pCD1[yopM::lacZYA]) (34), also lacking the native plasmid pPCP1 encoding the surface-located protease Pla, was grown at 28°C on tryptose blood agar plates or in heart infusion broth (DIFCO Laboratories). Tests for secretion of YopM derivatives were made in the defined TMH medium lacking added Ca2+ as previously described (36). If necessary, ampicillin was present in the medium at a final concentration of 100 μg/ml.
Yeast strains and growth conditions.
S. cerevisiae strain RCD224 (Ura−, His−, Trp−, Leu−, Lys− R. C. Dickson, unpublished data; obtained from R. C. Dickson, University of Kentucky) was used for expression of different forms of YopM fused to yeast-enhanced GFP protein (yEGFP) in the expression plasmid pYES2 (Invitrogen). The protease-deficient y604pep4 S. cerevisiae (MATa ade2-1 bar1Δ can1-100 his3-11,15 leu2-3,112 pep4::HIS3 trp1-1 ura3-1 [16] [obtained from M. Mendenhall, University of Kentucky]) was initially used to determine the localization of yEGFP-YopM fusion proteins that were unstable in strain RCD224. In general, for every yEGFP-YopM ΔLRR variant, we confirmed expression of the correct-size protein in S. cerevisiae RCD224, and ones not strongly expressed or present only as degradation products were studied in S. cerevisiae y604pep4, where immunoblotting was used to show that proteins of the anticipated mass were expressed. The temperature-sensitive sec18-1 mutant S. cerevisiae MLY1854 (Matα ura3-52 his4-619 sec18-1 [obtained from M. Latterich, Salk Institute, La Jolla, Calif.]) was used to determine the effect of a block in vesicular trafficking on YopM localization.
For routine growth of yeasts, SD medium was used, with appropriate omissions of amino acids to select for plasmids carrying metabolic markers (32). All yeast strains were stored as glycerol stocks frozen at −80°C. The localization of variant YopM proteins fused to yEGFP was tested by using cells from the same culture that was used to make the glycerol stock or cells taken within 2 weeks of streaking a fresh plate from the glycerol stock. Even so, there was heterogeneity of yEGFP fluorescence intensity within the final yeast culture (for yEGFP expressed alone or for yEGFP fused to variant YopM molecules). Because we always used fresh cultures from archival stocks, this variability did not reflect a genetic shift of the population. It therefore was inherent to the yeast system. However, it did not diminish the usefulness of the system for our goal of monitoring the localization of fluorescence. For expression of YopM, yeasts were inoculated into SD medium supplemented with 2% glucose (32) and grown overnight at 30°C. They were refreshed in SD medium supplemented with 2% glucose, and when the culture reached an optical density at 600 nm of 1.0, the cells were pelleted, washed once with phosphate-buffered saline (PBS), and transferred to SD medium plus 2% sucrose for overnight growth at 30°C. The culture was diluted to an optical density at 600 nm of 0.2 to 0.3 in SD medium with 2% galactose for induction of yEGFP or yEGFP-YopM expression at 30°C. During induction, the percentage of the yeast population showing fluorescence increased over time. Only a few of the cells were positive after 4 h of induction. Overnight induction typically produced a culture in which most of the cells showed fluorescence from yEGFP or yEGFP-YopM. Unless otherwise stated, induction was done overnight. The fluorescence distribution within the cells was then determined in the majority, fluorescent subpopulation.
To test the effect of the sec18-1 mutation on the localization of YopM, sec18-1 S. cerevisiae MLY1854 carrying a plasmid encoding yEGFP or a fusion of yEGFP with YopM was grown similarly but at 25°C and the temperature was changed to the restrictive temperature (37°C) for 2 h before the yeasts were moved to SD medium with galactose. The cells were incubated further for 6 h before the localization of yEGFP-YopM was investigated. In variations of this experiment where reversibility of the sec18-1 effect was assessed, the yeasts were stepped from glucose to sucrose at 25°C as previously but were then shifted to SD medium plus galactose for 2 h to initiate YopM expression. Then they were shifted to 37°C for 5 h, at which point one set of cultures was shifted back to 25°C while the duplicate cultures remained at 37°C. After 2 h, both sets of cultures were observed for YopM localization.
DNA methods.
Cloning methods and isolation of plasmid DNA and DNA fragments were as previously described by Birnboim and Doly (2) and Maniatis et al. (24), supplemented by the use of appropriate Qiaprep spin kits or Qiaquick DNA purification kits (Qiagen Inc.). Constructs were obtained in E. coli and checked by double and single restriction endonuclease digestions; some were verified by DNA sequencing across junction areas (by the University of Kentucky Macromolecular Structure Analysis Facility or by Retrogen). Synthetic primers for PCR and DNA sequencing were made by Genosys Biotechnologies, Inc. DNA was electroporated into Y. pestis as described previously (28). Yeast were transformed with plasmid DNA by using the S. c. EasyComp transformation kit (Invitrogen) as specified by the manufacturer. After transformation into yeast, all constructs were verified by Western analysis of the expressed protein from extracts of yeast cells.
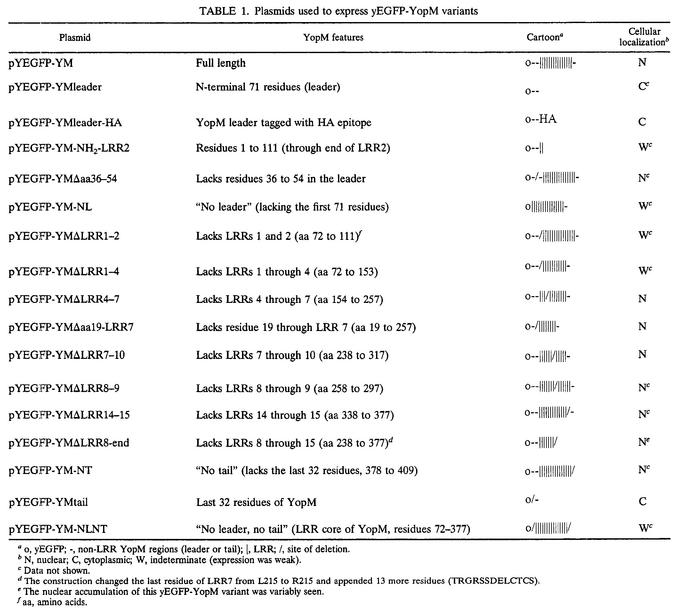
Plasmids for expression of YopM variants in yeast.
Plasmid pYM expressing YopM from the galactose-inducible promoter in pYES2 (Ura3 Apr pGal1 ColE1 ori and 2μm ori; Invitrogen) was made by amplifying yopM from the YopM-overexpressing plasmid pBS15/15 (15) and cloning into pYES2. Plasmid pYEGFP expressing the yEGFP from the galactose-inducible promoter in pYES2 was made by cloning a PCR fragment carrying the yegfp gene (7) into the EcoRI and NotI sites of pYES2. The NotI-flanked downstream primer was designed to eliminate the yegfp stop codon immediately upstream of the NotI site. The resulting yEGFP expressed from this construct accordingly had an extra 11 residues at the C terminus, 3 from the NotI site and 8 to the first stop codon in pYES2. pYEGFP was used to create a series of plasmids encoding yEGFP fused to the N terminus of YopM or YopM variants (Table 1 and Fig. 1). Deletions of sequence for LRRs 4 to 7, 8 and 9, and 7 to 10 were created previously in pBS15/15 (15) by the method of Wren et al. (41). Additional internal deletions of YopM were created similarly in pBS15/15. Then PCR amplifications were used to obtain the yopM genes, which were in frame with yegfp after NotI-XbaI digestion and ligation into NotI-XbaI-digested, phosphatase-treated pYEGFP. Constructs fusing yEGFP to the 71-amino-acid leader or the 32-amino-acid tail, the leader plus the first 2 LRRs of YopM, or YopM lacking the leader, tail, or both were made by PCR from pBS15/15 by using appropriate primers followed by cloning into NotI-XbaI-digested pYEGFP. This entire series of constructs encoded a 3-residue spacer between the C terminus of yEGFP and the first methionine of the YopM variant, due to the NotI site. One construct, encoding YopM ΔLRR8-end, was made directly in pYEGFP-YM by the PCR method of Wren et al. (41). The YopM sequence encoded by this construct ends at residue 214 (LRR 7 ends with L215) and has an additional 14 residues appended (RTRGRSSDELCTCS).
TABLE 1.
Plasmids used to express yEGFP-YopM variants
o, yEGFP; -, non-LRR YopM regions (leader or tail); |, LRR; /, site of deletion.
N, nuclear; C, cytoplasmic; W, indeterminate (expression was weak).
Data not shown.
The construction changed the last residue of LRR7 from L215 to R215 and appended 13 more residues (TRGRSSDELCTCS).
The nuclear accumulation of this yEGFP-YopM variant was variably seen.
aa, amino acids.
Plasmids for expression of YopM variants in Y. pestis.
Tests for localization of YopM within HeLa cells after delivery by infection used YopM overexpressed from its native promoter in pBluescript SK(−) (Stratagene) (pYopM) (31). Similar overexpression plasmids for YopMs lacking LRRs 4 to 7 or 7 to 10 were made by PCR using pYopMΔLRR4-7 or pYopMΔLRR7-10 (15) as templates and primers containing 5′ XhoI or XbaI sites for cloning into XhoI- and XbaI-digested pBluescript SK(−), respectively. YopM genes lacking sequence for LRRs 1 and 2 and 1 to 4 and expressed from the native yopM promoter were obtained from pBS15/15 by the method of Wren et al. (41) and then amplified by PCR and subcloned as described above into pBluescript SK(−).
Localization of YopM after delivery by Y. pestis infection.
Localization of YopM after delivery by infection with Y. pestis was carried out as described by Skrzypek et al. (34). Briefly, subconfluent HeLa cells on coverslips in RPMI 1640 supplemented with glutamine and containing 25 mM HEPES (Life Technologies no. 22400-097) were infected at a multiplicity of infection of 10 with Y. pestis that had been grown in exponential phase at 26°C in HIB. The mixed culture was incubated for 4 h at 37°C in the presence of 5% CO2 and then was fixed with 2% paraformaldehyde. The samples were permeabilized with 0.5% Triton X-100 in microtubule-stabilizing buffer (100 mM PIPES [pH 6.9], 4% [wt/vol] polyethylene glycol 6000, 1 mM EGTA). They were incubated with affinity-purified rabbit anti-YopM antibodies followed by Oregon green-conjugated secondary antibodies (Molecular Probes). The coverslips were examined with a Leica TCS laser scanning confocal microscope (DM RXE; 488-nm Ar laser) and 63× or 100× objectives. The images of optical slices ca. 1.5 μm thick were digitally recorded at 1024 by 1024 pixel resolution.
Preparation of protein extracts and immunoblot analysis.
Yeast cells were grown in SD medium as described above and washed once with water. Proteins were extracted for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the rapid method of Horvath and Riezman (17). Alternatively, proteins were isolated by using the Y-PER yeast protein extraction reagent (Pierce) as specified by the instructions enclosed in the kit. Proteins in the soluble fractions from either extraction procedure were separated by SDS-PAGE (12% [wt/vol] acrylamide), electrotransferred, and probed with polyclonal antibodies directed against the whole YopM molecule. For the hemagglutinin (HA)-tagged yEGFP-YMleader-HA, detection also used mouse monoclonal anti-HA antibodies.
Immunoblot analysis of YopM and YopM variants delivered to HeLa cells by infection with Y. pestis was performed as previously described (34). Briefly, infected monolayers were treated with trypsin to remove any surface-adsorbed YopM, and then a mixture of protease inhibitors was added. The cells were washed and then lysed in cold water containing protease inhibitors. The lysed cells were centrifuged, and the supernatant from this centrifugation (the cellular soluble fraction) was precipitated with trichloroacetic acid. Proteins recovered in this way were dissolved in SDS sample buffer (21). The pellet of the lysed cells containing bacteria and large cellular debris was dissolved in the same volume of SDS sample buffer. The culture medium from the infected culture was filtered through 0.2-μm-pore-size filters, precipitated with trichloroacetic acid, and dissolved in the same volume of SDS sample buffer. Equal volumes of each culture fraction were analyzed by immunoblotting as described above.
Localization of yEGFP-YopM derivatives in yeast and immunofluorescence colocalization tests.
Portions (1 ml) of the final induced cultures of yeast cells expressing yEGFP fusion proteins were pelleted, washed once with PBS, and resuspended in 100 μl of PBS. Yeast nuclei were stained in vivo for 20 min with Hoechst 33342 fluorescent stain (5 μg/ml; Molecular Probes). Alternatively, to obtain stronger nuclear staining, the cells were fixed and stained with 4′,6-diamidino-2-phenylindole (DAPI). The cells were prepared by a modification of a previously published method (30). Briefly, cells induced in galactose were fixed in 4% formaldehyde while still in their growth medium for 2 h at room temperature (RT). They were washed with PBS and resuspended in 1 ml of PBS containing 1.1 M sorbitol, 1% (vol/vol) β-mercaptoethanol, 110 μl of Glusulase (New England Nuclear), and 12 U of Zymolyase 20T (ICN). They were incubated at 37°C for 2 to 4 h to allow spheroplasting to occur. The spheroplasts were added to multiwell slides (Cell Point Scientific, Inc.) coated with poly-l-lysine (molecular weight, >300,000) (Sigma), allowed to settle, and then fixed sequentially in −20°C methanol (for 6 min) and −20°C acetone (for 30 s). They were air dried and blocked with 1 mg of bovine serum albumin per ml (BSA) in PBS (PBS-BSA) for at least 30 min at RT. To stain the yeast vacuole, the cells were treated overnight at 4°C with anti-yeast vacuolar carboxypeptidase Y (CPY) mouse monoclonal antibody 10A5-B5 (Molecular Probes) diluted 1:50 in PBS-BSA. The cells were washed with PBS and stained for 2 h at RT with Texas Red-X-conjugated goat anti-mouse immunoglobulin G (Molecular Probes) diluted 1:200 in PBS-BSA. Nuclei were stained with 1 μg of DAPI per ml in PBS for 12 min at RT. The cells were then mounted in Vectashield mounting medium (Vector Laboratories). All yeast samples were examined with a Zeiss Axiophot microscope with epifluorescent illumination and a 100× PLAN Achromat objective. Images were recorded through fluorescein isothiocyanate, rhodamine, or DAPI filters (for yEGFP, Texas Red-X, and Hoechst or DAPI, respectively) with a Spot charge-coupled device camera (Diagnostic Instruments Inc.), and the data were saved by using Diagnostic Instruments software (version 2.2.2).
CPY assays.
S. cerevisiae RCD224 cells containing pYM or pYEGFP-YM or vector alone (pYES2 or pYES2 expressing yEGFP) were grown overnight in SD medium with 2% galactose for standard expression of yEGFP-YopM (see above). Cultures were harvested at the mid-exponential phase for assays. Tests for an effect of YopM or yEGFP YopM on maturation of CPY employed immunoblots probed with anti-CPY monoclonal antibody 10A5-B5 at 1:1,000 dilution and alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Sigma). Assays for CPY activity in extracts of yeast expressing YopM were based on cleavage of a chromogenic substrate (N-carbobenzoxy-l-phenylalanyl-l-leucine [CBZ-Phe-Leu; Sigma], measured at 540 nm, or N-benzoyl-l-tyrosine-p-nitroanilide [BTPNA; Sigma], measured at 410 nm) compared to a standard curve with CPY (Sigma) (14, 37).
RESULTS
yEGFP-YopM localizes to the nucleus in yeast.
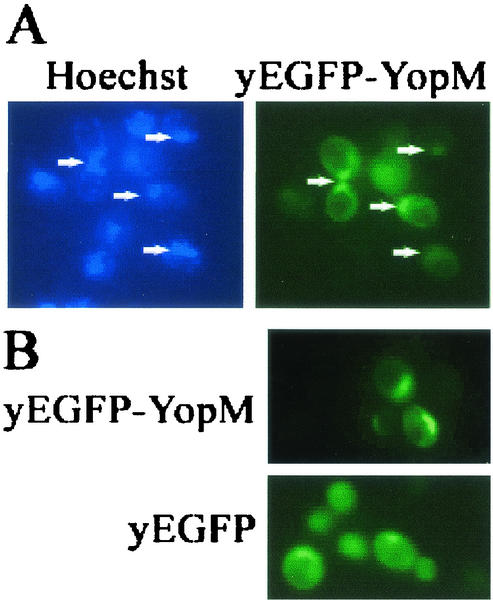
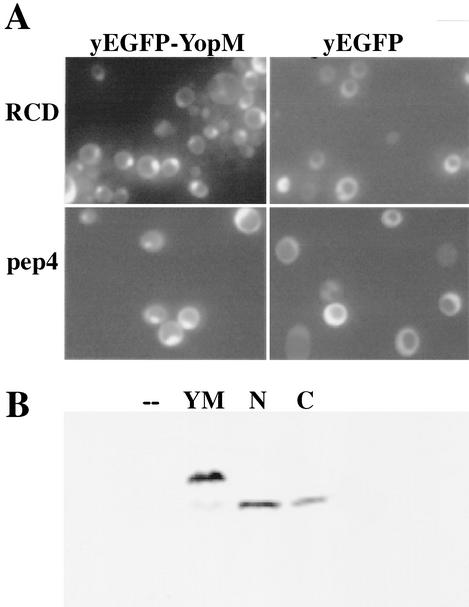
The goal of this study was to use yeast as a surrogate host cell to further characterize the requirements for trafficking of Y. pestis YopM in eucaryotic cells. We began by determining if Y. pestis YopM would traffic to the nucleus in yeast as had YopM from an enteropathogenic Yersinia strain (22). We expressed YopM tagged at its amino terminus with yEGFP from a high-copy-number expression vector (pYEGFP-YM [Table 1]) in S. cerevisiae RCD224. Figure 2 compares the distribution of yEGFP-YopM with that of yEGFP expressed from yeast containing the vector alone. yEGFP alone had a diffuse distribution, being present throughout the cytosol, in the nucleus, and sometimes in the vacuole of the cell. In contrast, yEGFP-YopM concentrated in the nucleus and colocalized with Hoechst or DAPI staining of DNA. These findings demonstrated that Y. pestis YopM does preferentially localize within the yeast nucleus after expression in the cytoplasm, showing that this property is conserved among YopM proteins tested so far. We therefore used yeast for further characterization of trafficking by Y. pestis YopM.
FIG. 2.
YopM localizes to the nucleus in yeast. S. cerevisiae RCD224 was induced for expression of yEGFP-YopM or yEGFP alone by growth in the presence of galactose. (A) The yeast DNA was stained with Hoechst 33342, and blue Hoechst fluorescence is shown in the left panel for the same cells pictured at right, where green yEGFP-YopM fluorescence is shown. Arrows point to nuclei. Note that some cells visible by Hoechst staining (lower left quadrant) are not expressing yEGFP-YopM strongly enough for detection. (B) The localization of green fluorescence is compared for S. cerevisiae RCD224 expressing yEGFP-YopM and yEGFP alone from the vector used to carry the yEGFP-YopM fusion proteins in this study. This and other figures were rendered by using Adobe Photoshop 6.0.
YopM depends on functional vesicular trafficking to localize to the nucleus in yeast.
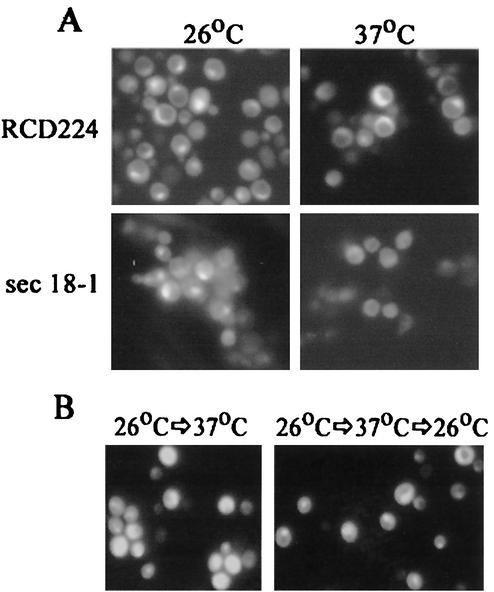
We previously showed that in HeLa cells infected by Y. pestis, YopM requires vesicular trafficking to reach the nucleus efficiently (34). This had been a surprising finding, and we wondered if the use of vesicular transport is a general feature of YopM trafficking in eucaryotic cells. To perform this test with yeast, we examined the distribution of yEGFP-YopM in the sec18-1(ts) mutant strain. Sec18p is the yeast counterpart of the mammalian NSF protein, an ATPase required for membrane fusion and vesicular transport. In yeast, Sec18p is required for most vesicular transport steps as well as for vacuolar fusion, and at the restrictive temperature (37°C), no transport occurs in the sec18-1 mutant (11, 13). We transformed the sec18-1 S. cerevisiae with pYEGFP-YM and compared the localization of yEGFP-YopM at permissive (26°C) and restrictive (37°C) temperatures. yEGFP-YopM localized to the nucleus in the wild-type strain RCD224 at both temperatures and in the sec18-1 mutant at the permissive temperature (Fig. 3A), showing that the different temperatures, per se do not affect YopM expression or distribution in a Sec18+ background. However, YopM was only weakly present in the sec18-1 mutant at the restrictive temperature, and the fluorescent signal had lost its preferentially nuclear localization. This showed that vesicular trafficking accounts for the yEGFP-YopM distribution observed and suggested that the nuclear localization of YopM in yeast requires functional vesicular trafficking. In this experiment, we had incubated the yeast for 2 h at the restrictive temperature before inducing the expression of YopM. Then we allowed a further 4 h or more of incubation (6 h was used in the experiment in Fig. 3A) to detect YopM expression by immunofluorescence. We were concerned that after this duration of inhibition of vesicular trafficking, secondary phenomena might cause poor YopM expression and that the weak, diffusely distributed fluorescence might reflect nonspecific signal. To address this issue, we began induction of YopM expression 2 h prior to the temperature shift and held the yeast at the restrictive temperature for a shorter period (5 h). Then we tested whether the normal YopM distribution could be restored after a return to the permissive temperature for a period that normally is not sufficient for visualization of de novo-synthesized yEGFP-YopM (2 h). In this protocol, the fluorescence from yEGFP-YopM was stronger in the yeast (Fig. 3B). The yeast culture that was returned to the permissive temperature showed a restored preferential localization of YopM in the nucleus, in contrast to the more diffuse YopM distribution in yeasts maintained at 37°C for the additional 2 h. This finding supports the conclusion that vesicular trafficking is necessary for YopM to reach the nucleus efficiently in yeast, as is the case for mammalian cells. Accordingly, delivery of YopM by Y. pestis or effects of other Yops are not necessary for YopM to enter a pathway to the nucleus that is promoted by vesicular trafficking. Further, the yeast system appears to provide a model for identifying a component of vesicular trafficking that promotes the nuclear localization of YopM as well as for mapping determinants on YopM that function in this process.
FIG. 3.
Efficient localization of YopM to the yeast nucleus requires functional vesicular trafficking. (A) yEGFP-YopM was expressed in S. cerevisiae RCD224 (wild type) and a temperature-sensitive sec18-1 S. cerevisiae strain. The localization of green fluorescence is compared for the two strains at the permissive temperature of 26°C, where both strains showed nuclear localization of YopM, and at the nonpermissive temperature for the sec18-1 strain (37°C), where nuclear localization occurred only in the wild-type yeast. (B) sec18-1 S. cerevisiae expressing yEGFP-YopM for 2 h was shifted to the nonpermissive temperature of 37°C for 5 h and then kept at 37°C for a further 2 h (left panel) or shifted back to the permissive temperature for 2 h (right panel) to determine if the inhibition of nuclear localization was reversible.
As previously found for a different YopM (22), expression of Y. pestis YopM causes slower growth of yeast (data not shown), and we wondered if the association of YopM with vesicular trafficking might in some way interfere with the normal handling of secreted proteins in yeast. This might represent a way in which YopM could function in host cells to interfere with the development of a timely immune response. Our colocalization tests had already indicated that YopM expression did not grossly alter the localization of the serine protease CPY in yeast (data not shown, but see Fig. 6C for an example). This enzyme is an extensively characterized substrate for the major pathway of protein sorting to the vacuole in yeast and is studied as a model for eucaryotic protein sorting in general. CPY is active only if it is secreted to the vacuole and processed there. As a more sensitive test for an effect of YopM on trafficking of CPY, we assayed CPY secretion indirectly by immunoblot analysis to detect the size variants that represent glycosylation and maturation intermediates that form during secretion. We also measured total cellular CPY enzyme activity by using chromogenic substrates. Yeasts grown overnight in medium containing galactose to induce the expression of yEGFP-YopM or YopM (from pYM) were compared to ones with vector alone in these tests, and no difference was seen in CPY maturation or activity (data not shown). This shows that YopM does not significantly affect the CPY secretion pathway in yeast. Because CPY passes through the endoplasmic reticulum and Golgi and then sorts to prevacuolar and vacuolar compartments (5, 35), these data suggest that YopM does not have a global inhibitory effect on the yeast secretory pathway.
FIG. 6.
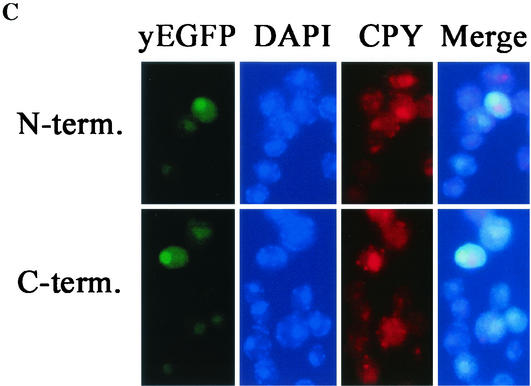
Both halves of YopM localize to the nucleus in yeast. (A) A control test showing the localization of yEGFP-YopM and yEGFP alone compared for S. cerevisiae RDC224 (wild type) (top) and the protease-deficient S. cerevisiae y604pep4 (bottom). (B) Immunoblot analysis of proteins extracted from S. cerevisiae y604pep4 after induction by galactose. Lanes: —, vector control expressing only yEGFP; YM, yEGFP-YopM; N, N-terminal half of YopM fused to yEGFP (yEGFP-YopM ΔLRR8-end); C, C-terminal half of YopM fused to yEGFP (yEGFP-YopM Δaa19-LRR7). The blots were probed with a rabbit polyclonal antibody against YopM; hence, yEGFP alone and background yeast proteins are not visualized. (C) The distribution was determined for fluorescence from yEGFP-YopM ΔLRR8-end (the N-terminal half of YopM) and yEGFP-YopM Δaa19-LRR7 (the C-terminal half of YopM) in S. cerevisiae y604pep4. From left to right, yEGFP indicates fluorescence from yEGFP-YopM proteins, DAPI indicates fluorescence from nuclei (DNA) stained with DAPI, CPY indicates fluorescence from CPY in the vacuole, and Merge indicates overlay of the green, blue, and red channels. In the Merge panel, note that the green fluorescence of yEGFP now appears turquoise due to overlap with blue DAPI fluorescence in the nucleus and that there is no overlap of yEGFP-YopM-related fluorescence with the red CPY fluorescence in the vacuole.
Tests of the possible roles of specific LRRs in nuclear localization of YopM: LRRs 4 to 7 and 7 to 10.
We next used the yeast system to determine if there is a domain that is essential for the trafficking of YopM to the nucleus. We made a series of constructs expressing truncated or internally deleted YopM variants fused at their N termini to yEGFP (Table 1; Fig. 1). These were expressed in yeast, and the localization of yEGFP fluorescence was determined.
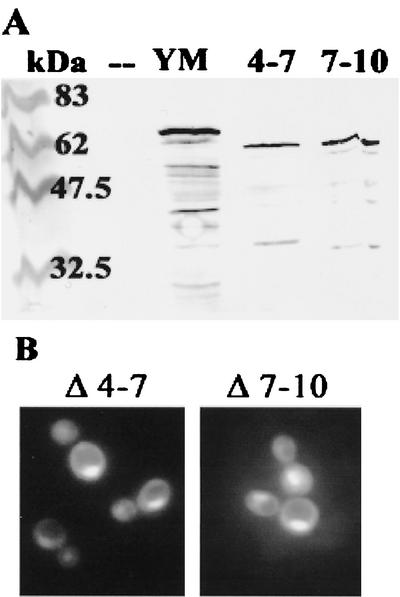
We first tested the effects of deleting LRRs 4 to 7 and 7 to 10. In a previous study, these two YopM variants had been found to differ in their ability to bind human α-thrombin, but Y. pestis strains expressing either of them instead of full-length YopM showed virulence that was decreased by 3 orders of magnitude (15). It was hypothesized that these proteins are defective in an intracellular pathogenic function. Hence, it was of interest to know if these proteins were defective in their ability to enter the nucleus. Figure 4A shows that both proteins were expressed at their expected size. Figure 4B shows that the deletions present in yEGFP-YopM ΔLRR4-7 and yEGFP-YopM ΔLRR7-10 did not prevent YopM localization to the nucleus.
FIG. 4.
Two YopM proteins deficient for pathogenic function still localize in the yeast nucleus. (A) Immunoblot analysis of proteins extracted from S. cerevisiae RCD224 after induction by galactose. Lanes: kDa, molecular mass markers; —, vector control expressing only yEGFP; YM, yEGFP-YopM; 4-7, yEGFP-YopM ΔLRR4-7; 7-10, yEGFP-YopM ΔLRR7-10. The blots were probed with a rabbit polyclonal antibody against YopM; hence, yEGFP alone and background yeast proteins are not visualized. The top bands in the other lanes represent the full-length yEGFP-YopM proteins. (B) The distribution of green fluorescence is shown for yEGFP-YopM ΔLRR4-7 (Δ 4-7) and yEGFP-YopM ΔLRR7-10 (Δ 7-10), expressed in S. cerevisiae RCD224.
yEGFP-YopM ΔLRR4-7 and yEGFP-YopM ΔLRR7-10 localize to the nucleus in HeLa cells.
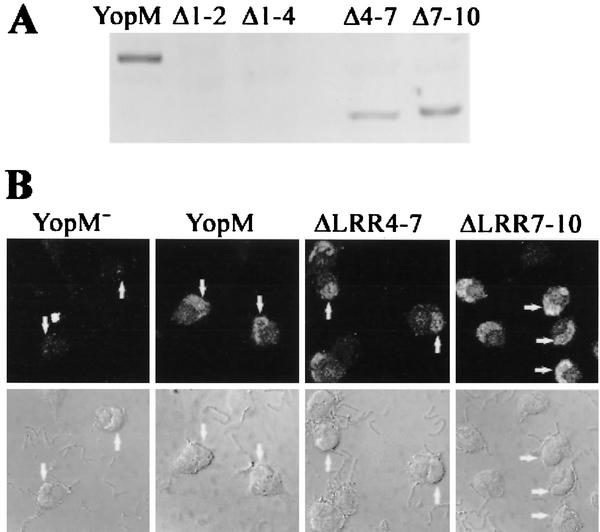
We wanted to further validate the yeast system by determining whether these YopM variants also traffic to the nucleus in mammalian cells. Accordingly, we expressed YopM ΔLRR4-7 and YopM ΔLRR7-10 as well as YopM ΔLRR1-2 and YopM ΔLRR1-4 in the YopM− Y. pestis KIM8-3233 and monitored the distribution of the mutant YopM proteins within HeLa cells after delivery by the contact-activated type III secretion system. (We were not able to test YopM proteins tagged with GFP, because an N-terminally tagged YopM would not be secreted and YopM C-terminally tagged with GFP is secretable by Y. pestis but is not detectably translocated into HeLa cells [unpublished data].) Although YopM ΔLRR1-2 and YopM ΔLRR1-4 were weakly expressed and were secreted by the bacteria in defined medium (data not shown), the expression was too weak for translocation into HeLa cells to be detectable by immunoblotting or indirect immunofluorescence (Fig. 5A and data not shown). YopM ΔLRR4-7 and YopM ΔLRR7-10 were expressed similarly to YopM and were delivered to HeLa cells by infection (Fig. 5A). Both proteins also localized to the nucleus as well as did YopM (Fig. 5B). (Note that the HeLa cells illustrated in Fig. 5 had rounded up due to the effects of other Yops also delivered by the type III secretion system.) We conclude that LRRs 4 to 10 are not essential for nuclear localization of YopM in HeLa cells, as predicted from the yeast experiments. Further, the avirulence of Y. pestis carrying these versions of YopM is not due to the inability of these proteins to traffic and enter the nucleus but, instead, is due to the absence of a region or conformation of YopM that is crucial for virulence-related activity within the host cell.
FIG. 5.
YopM ΔLRR4-7 and YopM ΔLRR7-10 also localize to the nucleus in HeLa cells after delivery by Y. pestis infection. HeLa cells were infected for 4 h with YopM− Y. pestis KIM8-3233 or Y. pestis KIM8-3233 expressing YopM, YopM ΔLRR1-2, YopM ΔLRR1-4, YopM ΔLRR4-7, or YopM ΔLRR7-10. After a brief trypsin treatment, the infected cells were lysed with water to obtain the soluble cellular fraction (cytosol). YopM proteins that had been delivered to the HeLa cytosol by the surface-adherent yersiniae were visualized by immunoblot analysis and probed with a rabbit polyclonal antibody raised against the whole YopM, which recognizes all of the YopM proteins being tested. Lanes: YopM, wild-type YopM; Δ1-2, YopM ΔLRR1-2; Δ1-4, YopM ΔLRR1-4; Δ4-7, YopM ΔLRR4-7; Δ7-10, YopM ΔLRR7-10. (B) The distribution of YopM protein in the infected HeLa cells was determined by indirect immunofluorescence. The primary antibody was the rabbit polyclonal used in panel A; the secondary antibody was conjugated to the Oregon Green fluorochrome. The upper panels show Oregon Green fluorescence from an optical slice obtained by laser scanning confocal microscopy; the lower panels show the differential interference contrast image of the same cells. From left to right, the panels illustrate two, two, five, and six infected HeLa cells. The arrows on some of the cells point to the kidney bean-shaped nucleus that lies to one side of each cell.
Two nuclear localization domains in YopM?
We assayed two constructs that would test the ability of the two halves of YopM to localize to the nucleus. yEGFP-YopM Δaa19-LRR7 had the first 18 residues of YopM fused to LRR8. This C-terminal half of YopM was unstable in S. cerevisiae RCD224, and its fluorescence was present in one or several small dots per cell (data not shown; see Discussion). We wondered if we might obtain stable expression in S. cerevisiae y604pep4, which lacks Pep4p, an aspartyl protease that functions to activate degradative enzymes in the yeast vacuole (1, 5, 40). This protease-deficient yeast showed the same cellular distributions of yEGFP and yEGFP-YopM as did S. cerevisiae RCD224 (Fig. 6A). We obtained stable expression of full-length yEGFP-YopM Δaa19-LRR7 in S. cerevisiae y604pep4 (Fig. 6B), and the protein clearly localized to the nucleus (Fig. 6C). Unless an NLS was located within the initial 18 amino acids whose codons typically contain the type III secretion system signal, this indicates the presence of a nuclear localization domain within the C-terminal half of the molecule. Our second construct, yEGFP-YopM ΔLRR8-end, contained the N-terminal half of YopM through LRR7 and was stably expressed in S. cerevisiae RCD224 as well as in S. cerevisiae y604pep4. It also localized to the nucleus (Fig. 6C; note the turquoise-colored overlap of yEGFP and DAPI fluorescence and no overlap with the red signal from CPY in the vacuole), although in some of our experiments its nuclear localization was weak and much YopM remained in the cytoplasmic compartment. This indicates that there may also be a nuclear localization domain in the N-terminal half of YopM.
The LRR structure is required for nuclear localization.
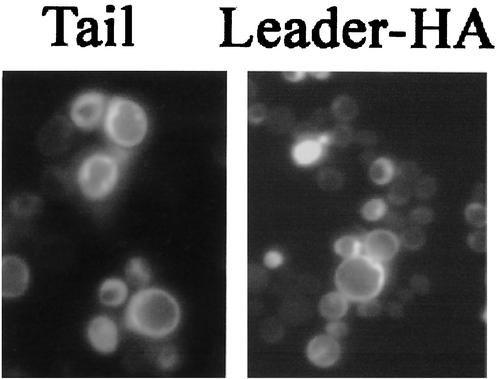
We tested whether the putative NLSs in the two halves of YopM were located in the 71-residue N-terminal leader and 32-residue C-terminal “tail” of YopM. Both of these regions are highly conserved among the YopM proteins of the three human-pathogenic Yersinia species. Interestingly, yEGFP-YopMtail was localized throughout the yeast cytoplasm as well as being present in the nuclear area (Fig. 7). Hence, it alone did not possess nuclear localization activity. YopM lacking the tail (yEGFP-YopM-NT) efficiently localized to the nucleus (data not shown), confirming that the tail was not essential for nuclear localization, as was expected from tests of the two YopM halves.
FIG. 7.
The leader and tail of YopM tested alone do not concentrate preferentially in the yeast nucleus. (Left) The ability of the 32-residue tail of YopM to conduct yEGFP into the nucleus was tested by determining the fluorescence distribution for yEGFP-YopMTail in S. cerevisiae RCD224. (Right) Fluorescence distribution from yEGFP-YopMleader-HA in S. cerevisiae y604pep4.
We tested the leader as yEGFP-YopMleader and also as yEGFP-YopMleader-HA, which contained the leader C-terminally tagged with an HA epitope, in S. cerevisiae y604pep4. Immunoblot analysis for the HA epitope verified that the full-length leader was present when this protein was expressed in S. cerevisiae y604pep4 (data not shown). However, neither leader protein localized to the nucleus (Fig. 7; data shown only for yEGFP-YopMleader-HA). Taken together, these tests with the YopM leader and tail showed that one or more LRRs are necessary for NLS function.
We attempted further mapping of the YopM NLSs by testing constructs containing the leader plus the first two or four LRRs. Although full-length fusion proteins were weakly made in S. cerevisiae y604pep4 (immunoblots not shown), too few of the yeast cells showed strong enough expression for fluorescence analysis. Tests with YopM variants lacking the leader or both the leader and the tail encountered the same roadblock.
DISCUSSION
In this study, we characterized the yeast system for trafficking of Y. pestis YopM and examined the YopM molecule for domains that confer the ability to localize to the nucleus. We found that, as in HeLa cells (34), vesicular trafficking must be functioning normally for YopM localization to the nucleus to be efficient. This property of YopM trafficking was unexpected when it was found in HeLa cells and is hard to explain by current cell biological paradigms. It is significant, therefore, that we found a requirement for vesicular trafficking for the nuclear accumulation of YopM in a completely different model system and indicated that this is a general property of the dynamics of YopM within eucaryotic cells. This suggests further that the yeast system is a robust model system to study the intracellular trafficking of YopM and ruled out the possibility that the yeast system would model only the nuclear entry step of YopM.
It is unlikely that YopM, expressed in the cytoplasm, travels to the nucleus inside a vesicle, because YopM does not have a classical signal sequence to insert it into the secretory pathway at the endoplasmic reticulum. Rather, we think that YopM may interact with proteins associated with the surfaces of vesicles. These interactions are likely to be shared between yeast and mammalian cells. Perhaps the association of YopM with a vesicle brings YopM to a region of the cell where it encounters a chaperone molecule that takes it into the nucleus. Alternatively, vesicular association of YopM may support a conformation of YopM that is conducive for its interaction with a chaperone.
We think that the nuclear accumulation of YopM is probably an active process, as opposed to passive diffusion coupled to trapping by binding to a nuclear target, because yEGFP-YopM is too large for passive entry into the nucleus, and if a trapping mechanism were operating to concentrate YopM in the nucleus, both halves of YopM would need to be able to bind the YopM target independently to account for both being accumulated in the nucleus. We do not think that YopM accumulates in the nucleus by nonspecific tight binding of acidic YopM to basic chromatin, because in ongoing studies with epithelial cells, we find that YopM localizes oppositely to condensing chromatin in the nucleus (T. Myers-Morales and S. C. Straley, unpublished data). (We could not make this determination in yeast, because yeast chromatin does not condense to the extent seen in mammalian cells.)
For two of the yEGFP-YopM derivatives, yEGFP-YopM Δaa19-LRR7 and yEGFP-YopM Δaa36-54 (Table 1), the full-length form was hard to detect by immunoblotting in extracts prepared from S. cerevisiae RCD224. However, we discovered that the degradation of such yEGFP-YopM variants could be circumvented by expressing them in the protease-deficient S. cerevisiae y604pep4. The unstable yEGFP-YopM molecules were expressed stably as full-length fusion proteins, and they showed a distribution like that of yEGFP-YopM. We then used this yeast strain to examine the distribution of other yEGFP-YopM derivatives in our study. It should be noted that this beneficial effect of the pep4 mutation was not predictable in advance. Pep4p is a protease in the yeast vacuole, responsible for activation of other degradative enzymes in the vacuole. However, we never observed an accumulation of green fluorescence within the vacuole of S. cerevisiae y604pep4, and we have no evidence suggesting that YopM normally transits the vacuole on the way to the nucleus (e.g., variants with less tight nuclear localization than full-length YopM did not accumulate in the vacuole). Transport to the vacuole is a terminal step of vesicular trafficking for degradative enzymes and for proteins to be degraded, and YopM is a cytoplasmically located protein that we think does not normally become incorporated within vesicles in its trafficking. So how could an enzyme (Pep4p) within the vacuole affect the localization of YopM? Perhaps the pep4 mutation in some way decreases or retards a process called autophagy, which is used by the yeast to nonspecifically deliver cytosolic proteins to the vacuole for degradation (5, 19). This could allow extra time for yEGFP-YopM derivatives that may be defective in some intracytoplasmic interactions to move to the nucleus, perhaps less efficiently than does full-length yEGFP-YopM. In any case, the pep4 yeast strain was valuable for our studies and may prove useful to other investigators working with yeast models in studies of bacterial virulence proteins.
We used the yeast system to ask whether we could identify a discrete domain capable of directing the entry of YopM into the nucleus. The data indicate that YopM potentially contains two independently functional NLSs, in its N-terminal and C-terminal halves, respectively, since both yEGFP-YopM Δaa19-LRR7 and yEGFP-YopM ΔLRR8-end localized to the nucleus. The C-terminal half of YopM present in yEGFP-YopM Δaa19-LRR7 still has the first 18 amino acids of the leader, and one could argue that this was responsible for the nuclear localization of that protein. However, that sequence does not resemble any reported NLSs and has no significant similarity to anything (other than YopM) in the database, including the leader of IpaH9.8 of Shigella flexneri, another LRR protein that has been shown to traffic to the nucleus (38).
YopM of various Yersinia species and serotypes is heterogeneous. The number of LRRs ranges from 13 to at least 21 (4, 15), and the internal LRRs vary in sequence, sometimes by as much as 25%. However, we have found that YopM proteins from Y. enterocolitica WA (serotype O:8) and Y. pseudotuberculosis PB1/+ (serotype I), having five or six more LRRs, respectively, than Y. pestis YopM (15; E. Skrzypek and S. C. Straley, unpublished data), both localize to the yeast nucleus when tested as yEGFP-YopM proteins (data not shown), consistent with the results of Lesser and Miller (22). We think that it is significant that in all YopM proteins for which the sequence is available, the leader plus first three LRRs and the last two LRRs plus the tail are highly conserved. Accordingly, we focused our attention on these N- and C-terminal-most parts of Y. pestis YopM in our efforts to identify domains with NLS function.
In the YopM tail, the sequence 379-VEDLRMNS-386 bears some resemblance to a sequence in the nucleoprotein of influenza virus (338-FEDLRVLS-345) that is necessary for accumulation of the protein in the nucleus (10). However, the tail alone did not sponsor preferential localization of N-terminally fused yEGFP to the yeast nucleus. It is possible that the tail and the highly conserved LRRs 14 and 15 contribute the C-terminal NLS activity in YopM; however, our tests did not prove this, because the necessity of LRRs 14 and 15 for nuclear localization was tested only in a construct that also contained the putative N-terminal NLS.
We attempted to map the N-terminal NLS beyond being within the first half of the molecule, but this effort generated equivocal findings, because constructs containing either yEGFP fused to the N-terminal leader plus only the first two or four LRRs or yEGFP-YopM lacking LRRs 1 and 2 or 1 to 4 were poorly expressed even in pep4 yeast. The point at which one defines the beginning of the first LRR is somewhat arbitrary, and in our work we set it at amino acid 72 as opposed to residue 74 (used in a recent report [12]). Hence, it is conceivable that we truncated an NLS that completely resides within a 73-amino-acid leader and that this was why the leader alone did not exhibit complete nuclear localization. However, we favor the idea that the LRR structure participates in creating the NLS. The recent crystal structure of YopM has revealed that residues 34 to 73 of the leader assume a helix-turn-helix structure that lies tightly against the first LRR and provides a hydrophilic cap over otherwise exposed hydrophobic core residues of the LRR structure (12) (Fig. 1 top). We found from testing yEGFP-YopM Δaa36-54, yEGFP-YopM Δaa19-LRR7, and yEGFP-YopM-NL that sequence within the leader is important for YopM stability. This cap makes multiple contacts with the adjacent LRR and could play a scaffolding role to promote the proper folding of the protein (12). Accordingly, the C-terminal half of the leader is not likely to be available free in solution but instead is present in complex with the first LRR. We envisage that it is this unit that constitutes the NLS. Accordingly, the highly conserved LRRs 1 to 3 might contribute to NLS function.
YopM's leader sequence is also necessary for the delivery of YopM into host cells by surface-adherent bacteria. The portion of the gene essential for secretion of the protein to the bacterial surface lies within the first 40 codons, but significant delivery to a host cell of a YopM fused at its C terminus to the adenylate cyclase domain (Cya) of Bordetella pertussis cyclolysin requires 100 amino acids (leader plus almost two LRRs) (3). Delivery was more efficient if the N-terminal 141 residues of YopM were included in the fusion protein (3). The C-terminal half of the YopM leader has some sequence similarity and predicted secondary-structure similarity to the similar-length leader in Shigella IpaH9.8, Salmonella secreted proteins SlrP, SspH2, and SspH1, and three ORFs in the Y. pestis chromosome predicted to encode LRR proteins (12). Other than IpaH9.8, it is not known at this writing whether these or other IpaH proteins localize to the nucleus. However, they are all LRR proteins and would share the need to stabilize the LRR superstructure, and the C-terminal half of the leader probably plays a role in this. How it and initial LRRs may also collaborate in directing a protein to the host cell nucleus is not yet understood.
In conclusion, our findings indicate that vesicular trafficking is a general feature of the nuclear localization of YopM in eucaryotic cells, independent of delivery by Yersinia. By studying the distribution of yEGFP-YopM variants in yeast, we have found that YopM possesses two independent NLSs or binding sites for a nuclear chaperone. Our data support the hypotheses that the NLS structure contains at least one LRR and that the process of localization to the nucleus is separable from pathogenic function. Because the NLS domain of YopM does not resemble other NLSs described so far, we predict that the nuclear entry mechanism of YopM will prove to be novel.
Acknowledgments
This study was supported by Public Health Service grant AI41668 from the National Institute of Allergy and Infectious Diseases.
We acknowledge Allison Shearer and Jason Bowles for outstanding technical assistance. We thank Pete Mirabito (Department of Biological Sciences, University of Kentucky) and Bob Dickson, and Mike Mendenhall (both from the Department of Biochemistry, University of Kentucky) for their generous gifts of yeast strains and protocols. This study benefited from the expertise and instrumentation provided by the University of Kentucky Imaging Facility.
Editor: J. T. Barbieri
REFERENCES
- 1.Ammerer, G., C. P. Hunter, J. H. Rothman, G. C. Saari, L. A. Valls, and T. H. Stevens. 1986. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Biol. 6:2490-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland, A., M.-P. Sory, M. Iriarte, C. Kerbourch, P. Wattiau, and G. R. Cornelis. 1996. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 15:5191-5201. [PMC free article] [PubMed] [Google Scholar]
- 4.Boland, A., S. Havaux, and G. R. Cornelis. 1998. Heterogeneity of the Yersinia YopM protein. Microb. Pathol. 25:343-348. [DOI] [PubMed] [Google Scholar]
- 5.Bryant, N. J., and T. H. Stevens. 1998. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev. 62:230-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchanan, S. G. St. C., and N. J. Gay. 1996. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol. 65:1-44. [DOI] [PubMed] [Google Scholar]
- 7.Cormack, B. P., G. Bertram, M. Egerton, N. A. Gow, S. Falkow, and A. J. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, G. R. 1998. The Yersinia deadly kiss. J. Bacteriol. 180:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., and H. Wolf-Watz. 1997. The Yersinia Yop virulon: a bacterial system for subverting eucaryotic cells. Mol. Microbiol. 23:861-867. [DOI] [PubMed] [Google Scholar]
- 10.Davey, J., N. J. Dimmock, and A. Colman. 1985. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell 40:667-675. [DOI] [PubMed] [Google Scholar]
- 11.Eakle, K. A., M. Bernstein, and S. D. Emr. 1988. Characterization of a component of the yeast secretion machinery: identification of the SEC18 gene product. Mol. Cell. Biol. 8:4098-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evdokimov, A. G., D. E. Anderson, K. M. Routzahn, and D. S. Waugh. 2001. Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit. J. Mol. Biol. 312:807-821. [DOI] [PubMed] [Google Scholar]
- 13.Graham, T. R., and S. D. Emr. 1991. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec 18 (NSF) mutant. J. Cell. Biol. 114:207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemmings, B. A., G. S. Zubenko, A. Hasilik, and E. W. Jones. 1981. Mutant defective in processing of an enzyme located in the lysosome-like vacuole of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:435-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hines, J., E. Skrzypek, A. V. Kajava, and S. C. Straley. 2001. Structure-function analysis of Yersinia pestis YopM's interaction with α-thrombin to rule on its significance in systemic plague and to model YopM's mechanism of binding host proteins. Microb. Pathog. 30:193-209. [DOI] [PubMed] [Google Scholar]
- 16.Hodge, A., and M. Mendenhall. 1999. The cyclin-dependent kinase inhibitory domain of the yeast Sic1 protein is contained within the C-terminal 70 amino acids. Mol. Gen. Genet. 262:55-64. [DOI] [PubMed] [Google Scholar]
- 17.Horvath, A., and H. Riezman. 1994. Rapid protein extraction from Saccharomyces cerevisiae. Yeast 10:1305-1310. [DOI] [PubMed] [Google Scholar]
- 18.Kajava, A. V. 1998. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277:519-527. [DOI] [PubMed] [Google Scholar]
- 19.Kim, J., and D. J. Klionsky. 2000. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 69:303-342. [DOI] [PubMed] [Google Scholar]
- 20.Kobe, B., and J. Deisenhofer. 1995. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374:183-186. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lesser, C. F., and S. I. Miller. 2001. Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 20:1840-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung, K. Y., B. S. Reisner, and S. C. Straley. 1990. YopM inhibits platelet aggregation and is necessary for virulence of Yersinia pestis in mice. Infect. Immun. 58:3262-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Mengaud, J., M. Lecuit, M. Lebrun, F. Nato, J.-C. Mazie, and P. Cossart. 1996. Antibodies to the leucine-rich repeat region of internalin block entry of Listeria monocytogenes into cells expressing E-cadherin. Infect. Immun. 64:5430-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulder, B., T. Michiels, M. Simonet, M.-P. Sory, and G. Cornelis. 1989. Identification of additional virulence determinants on the pYV plasmid of Yersinia enterocolitica W227. Infect. Immun. 57:2534-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemeth, J., and S. C. Straley. 1997. Effect of Yersinia pestis YopM on experimental plague. Infect. Immun. 65:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry, R. D., M. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price, S. R., P. R. Evans, and K. Nagai. 1998. U2B"-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature 394:645-650. [DOI] [PubMed] [Google Scholar]
- 30.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 31.Reisner, B. S., and S. C. Straley. 1992. Yersinia pestis YopM: thrombin binding and overexpression. Infect. Immun. 60:5242-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 33.Skrzypek, E., and S. C. Straley. 1996. Interaction between Yersinia pestis YopM protein and human α-thrombin. Thrombosis Res. 84:33-43. [DOI] [PubMed] [Google Scholar]
- 34.Skrzypek, E., C. Cowan, and S. C. Straley. 1998. Targeting of the Yersinia pestis YopM protein into HeLa cells and intracellular trafficking to the nucleus. Mol. Microbiol. 30:1051-1065. [DOI] [PubMed] [Google Scholar]
- 35.Stack, J. H., B. Horazdovsky, and S. Emr. 1995. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu. Rev. Cell Dev. Biol. 11:1-33. [DOI] [PubMed] [Google Scholar]
- 36.Straley, S. C., and W. S. Bowmer. 1986. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect. Immun. 51:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabuchi, M., O. Iwaihara, Y. Ohtani, N. Ohuchi, J.-I. Sakurai, T. Morita, S. Iwahara, and K. Takegawa. 1997. Vacuolar protein sorting in fission yeast: cloning, biosynthesis, transport, and processing of carboxypeptidase Y from Schizosaccharomyces pombe. J. Bacteriol. 179:4179-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyotome, T., T. Suzuki, A. Kuwae, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, T. Toyofuku, M. Hori, and C. Sasakawa. 2001. Shigella protein IpaH9.8 is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 276:32071-32079. [DOI] [PubMed] [Google Scholar]
- 39.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pst− phenotypes of yersiniae. Infect. Immun. 43:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woolford, C. A., L. B. Daniels, F. J. Park, E. W. Jones, J. N. Van Arsdell, and M. A. Innis. 1986. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol. Cell. Biol. 6:2500-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wren, B. W., J. Henderson, and J. M. Ketley. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16:994-996. [PubMed] [Google Scholar]