Abstract
The major virulence factors of toxigenic Vibrio cholerae are cholera toxin (CT), which is encoded by a lysogenic bacteriophage (CTXΦ), and toxin-coregulated pilus (TCP), an essential colonization factor which is also the receptor for CTXΦ. The genes for the biosynthesis of TCP are part of a larger genetic element known as the TCP pathogenicity island. To assess their pathogenic potential, we analyzed environmental strains of V. cholerae carrying genetic variants of the TCP pathogenicity island for colonization of infant mice, susceptibility to CTXΦ, and diarrheagenicity in adult rabbits. Analysis of 14 environmental strains, including 3 strains carrying a new allele of the tcpA gene, 9 strains carrying a new allele of the toxT gene, and 2 strains carrying conventional tcpA and toxT genes, showed that all strains colonized infant mice with various efficiencies in competition with a control El Tor biotype strain of V. cholerae O1. Five of the 14 strains were susceptible to CTXΦ, and these transductants produced CT and caused diarrhea in adult rabbits. These results suggested that the new alleles of the tcpA and toxT genes found in environmental strains of V. cholerae encode biologically active gene products. Detection of functional homologs of the TCP island genes in environmental strains may have implications for understanding the origin and evolution of virulence genes of V. cholerae.
Cholera caused by toxigenic Vibrio cholerae is an acute watery diarrhea which can occur as spreading epidemics (15). The pathogenesis of cholera depends on the synergistic effect of a number of factors produced by toxigenic V. cholerae. Profuse watery diarrhea is caused by an enterotoxin, cholera toxin (CT), produced by V. cholerae when it colonizes the small intestine (15, 31). The ctxAB operon, which encodes the A and B subunits of CT, resides in the genome of CTXΦ, a lysogenic filamentous bacteriophage (37). In addition to genes encoding CT, all strains capable of causing cholera carry genes for a colonization factor known as toxin-coregulated pilus (TCP), the expression of which is coordinately regulated with CT (21, 36). Although the major structural subunit of TCP is encoded by the tcpA gene, the formation and function of the pilus assembly require the products of a number of other genes located on a larger genetic region referred to as the TCP pathogenicity island (10, 15, 23, 27). Expression of CT and TCP are coregulated by the ToxR regulatory system, which includes the ToxT protein (9). The TCP gene cluster comprises at least 15 open reading frames, including the tcpA and toxT genes as well as a number of other regulatory genes. It has been suggested that regulators such as TcpI act downstream of the toxR and toxT genes to fine-tune the expression of TCP throughout the pathogenic cycle of V. cholerae (19). Other genes of the TCP island, including tcpP and tcpH, have also been suggested to have a role in the transcriptional activation of the toxT promoter (20). A notable example of evolutionary coadaptation is that the CTXΦ virion uses TCP as its receptor for infecting V. cholerae cells (37), whereas the toxT gene, which is located in the TCP island, encodes a transcriptional regulator which controls the expression of both TCP and CT genes in response to particular host or environmental conditions (9, 19, 20, 23).
Recently, new variants of the TcpA protein have been found in several V. cholerae non-O1, non-O139 strains (3). Although clinical isolates of V. cholerae are normally expected to carry virulence-associated genes, recent studies have also identified environmental V. cholerae strains which possess virulence genes or their homologs, including genetic variants of the TCP pathogenicity island (4, 30). These environmental strains were found to carry new alleles of the tcpA, toxT, and tcpF genes or variant forms of a regulatory sequence upstream of toxT (30). The present study was undertaken to further analyze these V. cholerae strains for pathogenic potential by using animal models and for toxigenic conversion by CTXΦ. This has implications for understanding the emergence and evolution of new pathogenic strains of V. cholerae.
A total of 14 V. cholerae strains initially cultured from three different freshwater lakes and ponds in the eastern part of Calcutta, India (4), were included in the study. These strains were previously shown to carry genetic variants of the TCP pathogenicity island with new alleles of several TCP island genes (30). Details of the strains analyzed in this study are listed in Table 1. Relevant characteristics of reference bacterial strains and properties of phages and plasmids used in this study are presented in Table 2.
TABLE 1.
Presence of major virulence-associated genes among V. cholerae non-O1, non-O139 strains isolated from environmental surface water, and susceptibility of the strains to a genetically marked derivative of CTXΦ
| Strain | Serogroup | Presence ofa:
|
Susceptibility to CTXΦ (frequency) | Competitive index of colonization | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| tcpAb | toxTb | toxR | lg1 | rstR | RstC | ctxA | ||||
| SCE4 | O8 | C | + | + | − | − | − | − | 6.0 × 10−5 | 0.35 |
| SCE188 | O44 | Env | + | + | + | + | + | + | 6.3 × 10−4 | 0.80 |
| SCE223 | O27 | Env | + | + | + | + | + | + | 0 | 0.86 |
| SCE225 | O35 | C | Env | + | − | + | + | − | 0 | 0.31 |
| SCE226 | O35 | C | Env | + | − | + | + | − | 0 | 0.53 |
| SCE227 | O35 | C | Env | + | − | + | + | − | 0 | 0.20 |
| SCE256 | O42 | C | Env | + | − | + | + | − | 0 | 0.28 |
| SCE258 | O42 | C | Env | + | − | − | − | − | 0 | 0.28 |
| SCE263 | O10 | C | Env | + | + | + | + | − | 0 | 0.33 |
| SCE264 | O42 | C | Env | + | + | + | + | − | 0 | 0.42 |
| SCE340 | O69 | E | Env | + | − | − | − | − | 3.9 × 10−4 | 1.26 |
| SCE341 | O69 | E | Env | + | − | − | − | − | 4.5 × 10−4 | 0.29 |
| SCE354 | O27 | Env | + | + | + | + | + | + | 0 | 2.78 |
| SCE359 | O8 | C | + | + | − | − | − | − | 5.6 × 10−4 | 0.82 |
Presence of genes was detected by using DNA probes and PCR assays.
Env refers to a new allele of the tcpA or toxT genes, different from those normally found in epidemic strains (30). C and E, classical and El Tor type tcpA genes, respectively.
TABLE 2.
Characteristics of V. cholerae reference strains, plasmids, and phages used in the study
| Strain | Relevant characteristic(s) | Reference |
|---|---|---|
| MSF8.2Φ | Derivative of CTX-KmΦ in which the ctxAB operon was reinstated. This construct carries both a functional ctxAB operon and a kanamycin resistance cassette | 16 |
| O395 | Classical Ogawa streptomycin-resistant strain | Laboratory collection |
| O395 (pMSF8.2) | Strain O395 carrying the RF of MSF8.2Φ | 16 |
| RV508 | Derivative of classical biotype strain 569B that constitutively expresses CT, TCP pili, and other toxR-regulated gene products | 36 |
| Bah-2 | Derivative of El Tor strain E7946 in which the entire CTX element as well as the attachment sequence attRS was deleted | 25 |
| TCP-2 | Derivative of strain O395 which carries deletions in the tcpA and ctxA genes | 35 |
| SA-317 | TCP-positive nontoxigenic V. cholerae O1 strain | 12 |
| P-27459 | Toxigenic clinical V. cholerae O1 El Tor strain | Laboratory collection |
| MO10 | Toxigenic V. cholerae O139 strain | Laboratory collection |
The genetically marked phage MSF8.2Φ used in this study was a derivative of an El Tor type CTXΦ which carried a functional ctxAB operon as well as a kanamycin resistance (Kmr) determinant (16). MSF8.2Φ was prepared for the present study from a culture of V. cholerae O395 carrying RF of the phage pMSF8.2. Aliquots of the culture supernatants were sterilized by filtration through 0.22-μm-pore-size filters (Millipore Corporation, Bedford, Mass.). The filtrate was titrated for infectious phage particles by incubating aliquots of the supernatants for 30 min at 30°C with the classical biotype strain RV508, which constitutively expresses TCP, and then selecting for colonies resistant to kanamycin.
The presence of virulence-associated genes was determined by using specific DNA probes or PCR assays. The gene probes used in this study included a 0.5-kb EcoRI fragment of pCVD27 (22) containing part of the ctxA gene and a 2.1-kb SphI-XbaI fragment of pCTX-Km (37) containing the entire zot and ace genes and part of orfU. The toxR gene probe was a 2.4-kb BamHI fragment of pVM7 (29). The rstRET probe was a SacI-XbaI fragment of pHK1 (26). Presence of the rstC gene was determined by a PCR assay with the two primers 5′ATGAGTTTGAAACCATACACTTT and 5′TTACAGTGATGGATCAGTCAAT, as described previously (13). Presence of the tcpA and acfB genes were also tested by PCR assays described previously (14, 24). Colony blots or Southern blots were prepared by using nylon filters (Hybond; Amersham Biosciences, Uppsala, Sweden) and were processed by standard methods (28, 33). The probes were labeled by random priming (17) by using a random-primers DNA labeling kit (Invitrogen Corporation, Carlsbad, Calif.) and [α-32P]dCTP (3,000 Ci/mmol; Amersham). Southern blots and colony blots were hybridized with the labeled probes, and autoradiographs were developed as described previously (14).
The susceptibility of V. cholerae strains to the genetically marked derivative of CTXΦ was assayed under laboratory conditions and inside the intestines of infant mice by previously described methods (11, 12). Representative infected colonies were grown overnight in Luria-Bertani (LB) broth containing kanamycin (50 μg/ml), and cells were precipitated by centrifugation. The supernatant fluids of the cultures were titrated for the presence of MSF8.2Φ particles by using strain RV508 as the recipient. Total DNA or plasmids were extracted from bacterial pellets by standard methods (28) and purified by using microcentrifuge filter units (Ultrafree-Probind; Sigma). The presence of the phage genome was verified by comparative Southern blot analysis of total DNA and plasmid preparations from the phage-infected strains and the corresponding native strains.
Colonization of infant mice by the environmental strains in competition with a reference strain was assayed as described previously (1, 18). Briefly, each V. cholerae test strain and the reference strain were grown to stationary phase at 30°C in LB broth. The test strain and the reference strain were mixed at a 1:1 ratio and diluted 1:1,000 in LB broth. Approximately 105 CFU of the bacterial mix in a 50-μl suspension was used to intragastrically inoculate groups of 5-day-old Swiss Albino mice. Each strain was inoculated in at least six mice, and the infections were allowed to proceed for 20 h. The mice were then sacrificed, and bacteria were recovered from the small intestines by homogenization in phosphate-buffered saline (pH 7.4). Serial dilutions of the homogenates were plated on appropriate antibiotic plates and on plates devoid of the antibiotic to determine the ratio of the number of organisms of the test strain to that of the reference strains. Competitive indices were calculated by dividing the output ratios by the inoculum input ratio of the test and reference strains. An in vitro analysis of the inoculum was also done to determine the precise ratio of test strain to reference strain and to determine the competitive indices in vitro. This was determined for each inoculum by plating serial dilutions, as described above, before and after 20 h of growth under in vitro laboratory conditions without antibiotic.
Production of CT by V. cholerae strains was determined by the GM1 ganglioside-dependent enzyme-linked immunosorbent assay (GM1-ELISA) and the rabbit ileal loop assay, as described previously (8, 32). A toxigenic strain, P27459, and a nontoxigenic strain, SA-317, were included as positive and negative control strains in each round of assay.
Diarrheal response to the V. cholerae strains was assayed in adult rabbits by using the removable intestinal tie-adult rabbit diarrhea (RITARD) model (34) with New Zealand White rabbits, as described previously (16). Each strain was inoculated in at least five different rabbits. Rabbits were observed for overt diarrhea and for death, and stools or rectal swabs were cultured on gelatin agar plates and a duplicate plate containing kanamycin (50 μg/ml) when appropriate to monitor shedding of the challenge organisms. Observations were made at 6-h intervals during the 7 days following inoculation; the number of rabbits developing moderate to severe diarrhea arbitrarily was scored, and the number of deaths was recorded.
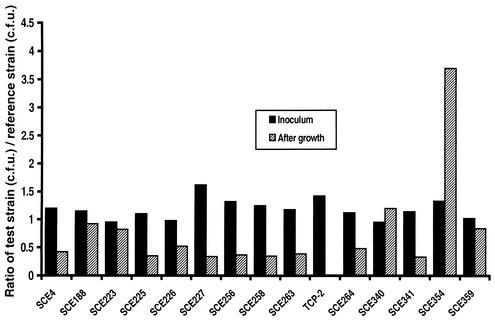
Of 14 environmental strains included in the present study, 12 strains carried one or more new alleles of the TCP island genes (30). This included the tcpA gene, which encodes the major structural subunit of TCP, and the major virulence regulatory gene toxT, which controls the expression of both CT and TCP. Other genetic variation was found in the tcpF genes or in a regulatory sequence upstream of the toxT gene (30). Since TCP is the major colonization factor of V. cholerae and a crucial factor for successful infection, we tested the ability of the strains to infect infant mice in competition with a known TCP-positive, CT-negative strain, Bah-2 (25). The reason for choosing the CT-negative strain was that most of the environmental strains tested were also negative for CT. It may be mentioned that previous studies have indicated that CT-positive strains are more efficient colonizers than the corresponding CT-negative mutants (2, 15). The reference strain Bah-2 was resistant to streptomycin, and this allowed us conveniently to differentiate the reference strain from the test strains. The proportions of test strain and reference strain recovered from the mouse intestine 20 h after intragastric inoculation of groups of infant mice with a mixture of the two strains are shown in Fig. 1. While the inoculum contained the two strains at an approximately 1:1 ratio, the ratio in the recovered samples varied between 0.33 and 3.7. In the in vitro assay, no major change was observed in the ratio of the two strains. The competitive index of colonization varied from 0.28 to 2.78 (Table 1). The TCP-negative control strain TCP-2 included in the study, however, was completely outcompeted by the reference TCP-positive strain (competition index, 0.007). This suggested that the test strains competed with the reference strain with various efficiencies for infecting infant mice. All strains included in the study carried the tcpA, toxT, and acfB genes or their homologs (Table 1) and, presumably, the entire TCP pathogenicity island. Hence, the observed moderate to high colonization efficiency compared to that of the reference strain may be attributed to the production of TCP, unless these strains produce any previously undiscovered colonization factors. It may be mentioned that at least one of these strains has previously been found to produce a pilus (4), and most of the strains demonstrated autoagglutination, a property attributed to the production of TCP by V. cholerae. The results of the present study indicate that the TCP produced by these environmental V. cholerae strains is a biologically active pilus and can contribute to colonization of the mouse intestine.
FIG. 1.
Colonization of infant mice by environmental V. cholerae strains in competition with a reference TCP-positive, nontoxigenic V. cholerae strain, Bah-2. The TCP-negative strain TCP-2 was used as a negative control.
TCP is also used by CTXΦ as its receptor for invading V. cholerae cells. We therefore tested the susceptibility of the environmental V. cholerae strains to CTXΦ. This was done by using a genetically marked phage which carried a Kmr determinant as well as a functional ctxAB operon (16). The infected strains were initially selected by their resistance to kanamycin and were later tested for the presence of CTXΦ-specific genes. The results showed that 5 of 14 environmental strains tested were infected by the phage. These five strains included two strains carrying classical type tcpA gene, two strains with El Tor type tcpA gene, and one strain with a new variant (Env type) tcpA gene (Table 1). We tested the susceptibilities of the strains both under in vitro laboratory conditions and inside the intestines of infant mice. Previous studies with CTXΦ showed that the efficiency of transduction was considerably higher in vivo, and this was attributed to more adequate expression of the phage receptor TCP in vivo than under laboratory conditions (11, 12). In the present study, we did not detect any Kmr transductants of the environmental strains in the in vitro assay, and infection with MSF8.2Φ was detectable only in the infant mouse assay. This suggested that infection of these V. cholerae strains by CTXΦ was possibly TCP dependent. Susceptibility of the environmental strains was low (the mean frequency of infection was between 5.5 × 10−5 and 6.5 × 10−4) compared to that of the control CTXΦ-negative V. cholerae O1 strain SA-317 that was included in the study. The susceptibility of this O1 strain varied between 2.1 × 10−2 and 6.3 × 10−2 in vitro and between 8.6 × 10−2 and 19.2 × 10−2 in the in vivo assays. Nevertheless, this is the first demonstration of environmental V. cholerae strains carrying new alleles of the TCP island genes being infected by CTXΦ.
Previous studies have described heteroimmunity among CTX phages mediated by widely diverse CTXΦ repressors encoded by different rstR genes. (7, 26). The existence of at least three different rstR genes carried by different CTX phages, namely, phages CTXΦET, CTXΦclass, and CTXΦCalc, has been recognized (7). More recently, some of the environmental strains included in the present study have been reported to carry novel rstR homologs (30). The genetically marked phage MSF8.2Φ used in our study carried an El Tor type rstR gene (rstRET). To investigate the reasons for the resistance of nine environmental strains to the phage, we analyzed the strains for the possible presence of an rstRET gene. This showed that all strains which were resistant to CTXΦ infection in the present study carried one or more copies of the rstRET gene, although all except one strain (SCE188) were nontoxigenic. It is interesting that nine strains carried the rstR gene independently of the other CTXΦ genes (core genes). We have previously demonstrated the horizontal transfer of the RS1 element of V. cholerae as a filamentous phage which also carries the rstR gene (13). To examine whether these environmental strains carried the RS1 element, we analyzed these strains for the RS1-specific gene rstC, which is not present in the related genetic element RS2, an integral part of the CTXΦ genome (38). PCR analysis of the environmental V. cholerae strains for rstC confirmed the findings of a previous investigation (30) that strains which were positive for rstR were also positive for rstC. This suggested that the environmental strains harbored the RS1 element carrying an rstRET gene, which accounted for the immunity of these nine strains to further infection by an El Tor type CTXΦ.
Production of CT was initially studied in vitro by GM1-ELISA by using an antibody against the B subunit of CT (Table 3). Of the 14 strains tested, strain SCE 188 was toxigenic and produced CT, whereas the remaining 13 strains were nontoxigenic. However, environmental strains which were infected by CTXΦ in our study produced CT after transduction by the phage. To further ascertain whether the toxin produced by these transductants was biologically active, we used the ligated ileal loop assay in rabbits and observed fluid accumulation. All culture supernatants which were positive for CT in the ELISA also caused fluid accumulation in the ileal loops of rabbits, confirming that these strains produced biologically active CT. The pathogenic potentials of these strains were further assessed by using the adult rabbit diarrhea model. While rabbits challenged with the CTX-negative strains did not show a diarrheal response (Table 3), V. cholerae strains carrying the MSF8.2Φ genome, as well as the native toxigenic strain SCE188 and the reference virulent strains P-27459 and MO10, produced severe diarrhea in rabbits. The environmental strains were less virulent than the control strains, since none of these strains caused fatal diarrhea. However, they caused mild to moderate diarrhea, and the test strains were excreted by challenged rabbits for 2 to 9 days following inoculation. The prolonged shedding of the challenge organisms suggested that the strains colonized the intestines of the rabbits, and the data supported the results of the mouse colonization assay.
TABLE 3.
Production of cholera toxin by environmental V. cholerae strains and their derivatives (carrying the genome of a genetically marked CTX phage, MSF8.2Φ), and diarrheal response of adult rabbits to these strains
| Strain | Cholera toxin production by:
|
Diarrheal response of adult rabbits by the RITARD model
|
||||
|---|---|---|---|---|---|---|
| GM1-ELISAa | Fluid accumulation in rabbit ileal loops (ml/cm of loops)b | No. of animals challenged | No. responding with fatal diarrheac | No. developing nonfatal diarrhead | Mean duration of shedding of strain in stools (days) | |
| SCE4 | UD | 0 | 5 | 0 | 0 | 3.6 ± 1.3 |
| SCE4 (MSF8.2) | 2.19 ± 0.61 | 2.06 ± 0.46 | 6 | 0 | 5 | 4.8 ± 1.7 |
| SCE188 | 2.92 ± 0.56 | 2.27 ± 0.82 | 5 | 1 | 4 | 4.8 ± 0.8 |
| SCE188 (MSF8.2) | 3.96 ± 0.79 | 2.95 ± 0.78 | 6 | 2 | 4 | 5.3 ± 1.5 |
| SCE340 | UD | 0 | 5 | 0 | 0 | 4.6 ± 1.6 |
| SCE340(MSF8.2) | 2.65 ± 0.45 | 2.09 ± 0.51 | 4 | 0 | 4 | 6.4 ± 0.8 |
| SCE341 | UD | 0 | 5 | 0 | 0 | 3.8 ± 0.8 |
| SCE341 (MSF8.2) | 2.71 ± 0.56 | 2.13 ± 0.64 | 6 | 0 | 5 | 3.6 ± 1.2 |
| SCE359 | UD | 0 | 5 | 0 | 4 | 6.0 ± 1.3 |
| SCE359 (MSF8.2) | 2.93 ± 0.51 | 2.12 ± 0.73 | 5 | 2 | 2 | 7.2 ± 1.6 |
| SA-317 | UD | 0 | 5 | 0 | 1 | 5.6 ± 2.1 |
| P-27459 | 2.97 ± 0.81 | 2.55 ± 0.79 | 6 | 3 | 2 | 6.3 ± 1.6 |
| MO10 | 3.16 ± 0.73 | 2.32 ± 0.65 | 6 | 4 | 2 | 6.1 ± 1.4 |
Toxin amounts are expressed in micrograms per unit of optical density of the culture at 600 nm. Values represent the averages of results of five independent observations. UD, undetectable (toxin amounts were less than 0.01 μg/ml, which was the lowest concentration of purified toxin used as control).
Values represent the averages of results of five independent observations made in different rabbits.
Rabbits developing fatal diarrhea died within 12 h after the initiation of diarrhea.
The duration of diarrhea was between 4 and 6 days for rabbits challenged with strains carrying the CTXΦ genome.
However, it is not clear from this study why the environmental strains appeared to be less virulent in the rabbit assay than the V. cholerae O1 and O139 strains used as positive controls. Remarkably, all environmental strains carrying a combination of the classical type tcpA gene and the new allele of the toxT gene were completely resistant to CTXΦ. The competitive indices of colonization of mice by these strains were also lower than those of most other strains (Table 1). If this difference in colonization was due to inadequate expression of TCP, this would mean that optimum expression of classical type TCP requires the epidemic type ToxT. Hence, the epidemic type toxT gene may have evolved from an environmental toxT driven by a need to upregulate the expression of TCP, given that the classical biotype of V. cholerae O1 was responsible for the sixth pandemic, and possibly the earlier pandemics, of cholera (15).
Demonstration of the presence of the TCP pathogenicity island in environmental strains, and particularly the genetic diversity of TCP island genes, can provide clues about the origin of the TCP pathogenicity island. Recent studies have shown that virulence genes or their homologs are dispersed among environmental V. cholerae strains belonging to diverse serogroups, whereas most previous studies suggested that virulence genes, such as the TCP island genes, are carried only by clinical isolates. This assumption was made because the studies overlooked the possibility of genetic variation within the virulence genes to the extent that the variants might escape detection with PCR or probes that were designed strictly based on the sequences of the corresponding genes found in clinical strains. For example, in a previous study, some of the same environmental strains were reported to be negative for TCP genes (4), whereas a more recent study (30) confirmed that these strains carried the TCP island, but the sequence of some of the genes differed from previously reported sequences of these genes.
In the present study, we have demonstrated that virulence gene homologs carried by environmental strains are functional genes, and such strains are potential pathogens, although their virulence potential may be somewhat lower than that of epidemic strains. The results reported here indicate that functional virulence genes possessed by clinical strains may have evolved from environmental genes. Thus, new insight is provided into the origin and evolution of virulence genes in V. cholerae. Characterization of these genes will contribute to the understanding of the ecological significance of the occurrence of virulence gene homologs in environmental strains and their relationship with virulence-associated functions. It has been suggested that virulence factors, including colonization factors and CT, may have a crucial function in the symbiotic and/or commensal association between V. cholerae and specific aquatic organisms (5, 6). It is possible that gene products described as virulence factors in the context of human infection may have additional roles while V. cholerae persists in the environment. Nonetheless, results of the present study have revealed that the environmental strains of V. cholerae not only carry variants of the TCP island genes, but also that these gene variants are functional and capable of producing TCP. Further studies will be done to understand more definitively the role of these virulence-associated factors in the natural environment as well as the environmental selection pressures for V. cholerae to carry virulence genes or their homologs.
Acknowledgments
This research was funded in part by the United States Agency for International Development (USAID) under grant HRN-5986-A-00-6005-00 with the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B) and by the United States National Institutes of Health under grant no. RO1 AI39129-01A1 with the Department of International Health, Johns Hopkins University, and ICDDR,B. The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries. Current donors providing unrestricted support include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, Japan, Kingdom of Saudi Arabia, The Netherlands, Sweden, Sri Lanka, Switzerland, and the United States of America.
We thank Matthew Waldor, New England Medical Center, Boston, Mass., for the rstR gene probes and Afjal Hossain for secretarial assistance.
Editor: A. D. O'Brien
REFERENCES
- 1.Angelichio, M. J., J. Spector, M. K. Waldor, and A. Camilli. 1999. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 67:3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baselski, V. S., R. A. Medina, and C. D. Parker. 1979. In vivo and in vitro characterization of virulence-deficient mutants of Vibrio cholerae. Infect. Immun. 24:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, E. F., and M. K. Waldor. 2002. Evolutionary and functional analyses of variants of the toxin-coregulated pilus protein TcpA from toxigenic Vibrio cholerae non-O1/non-O139 serogroup isolates. Microbiology 148:1655-1666. [DOI] [PubMed] [Google Scholar]
- 4.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colwell, R. R., and A. Huq. 1994. Vibrios in the environment: viable but nonculturable Vibrio cholerae, p. 117-133. In I. K. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington, D.C.
- 6.Colwell, R. R., and W. M. Spira. 1992. The ecology of Vibrio cholerae, p. 107-127. In D. Barua and W. B. Greenough III (ed.), Cholera. Plenum Medical Book Co., New York, N.Y.
- 7.Davis, B. M., K. E. Moyer, E. F. Boyd, and M. K. Waldor. 2000. CTX prophages in classical biotype Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J. Bacteriol. 182:6992-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De, S. N., and D. N. Chatterje. 1953. An experimental study of the mechanisms of action of Vibrio cholerae on the intestinal mucous membrane. J. Pathol. Bacteriol. 46:559-562. [DOI] [PubMed] [Google Scholar]
- 9.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascades controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everiss, K. D., K. J. Hughes, M. E. Kovach, and K. M. Peterson. 1994. The Vibrio cholerae acfB colonization determinant encodes an inner membrane protein that is related to a family of signal-transducing proteins. Infect. Immun. 62:3289-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque, S. M., Asadulghani, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic V. cholerae O1 and O139. Infect. Immun. 66:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faruque, S. M., Asadulghani, M. N. Saha, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for the origination of new strains with epidemic potential. Infect. Immun. 66:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M., Asadulghani, M. Kamruzzaman, R. K. Nandi, A. N. Ghosh, G. B. Nair, J. J. Mekalanos, and D. A. Sack. 2002. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXΦ. Infect. Immun. 70:163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque, S. M., A. K. Siddique, M. N. Saha, Asadulghani, M. M. Rahman, K. Zaman, M. J. Albert, D. A. Sack, and R. B. Sack. 1999. Molecular characterization of a new ribotype of Vibrio cholerae O139 Bengal associated with an outbreak of cholera in Bangladesh. J. Clin. Microbiol. 37:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faruque, S. M., M. M. Rahman, A. K. M. Hasan, G. B. Nair, J. J. Mekalanos, and D. A. Sack. 2001. Diminished diarrheal response to Vibrio cholerae strains carrying the replicative form of the CTXΦ genome instead of CTXΦ lysogens in adult rabbits. Infect. Immun. 69:6084-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg, A., and B. Vogelstein. 1984. A technique for radio labeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 18.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harkey, C. W., K. D. Everiss, and K. M. Peterson. 1994. The Vibrio cholerae toxin-coregulated pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect. Immun. 62:2669-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hase, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili and ToxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaper, J. B., J. G. Morris, Jr., and M. Nishibuchi. 1988. DNA probes for pathogenic Vibrio species, p. 65-77. In F. C. Tenover (ed.), DNA probes for infectious disease. CRC press, Inc., Boca Raton, Fla.
- 23.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keasler, S. P., and R. H. Hall. 1993. Detection and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 341:1661.. [DOI] [PubMed] [Google Scholar]
- 25.Kenner, J. R., T. S. Coster, D. N. Taylor, A. F. Trofa, M. Barrera-Oro, T. Hyman, J. M. Adams, D. T. Beattie, K. P. Killeen, D. R. Spriggs, J. J. Mekalanos, and J. C. Sadoff. 1995. Peru-15, an improved live attenuated oral vaccine candidate for Vibrio cholerae O1. J. Infect. Dis. 172:1126-1129. [DOI] [PubMed] [Google Scholar]
- 26.Kimsey, H. H., and M. K. Waldor. 1998. CTXΦ immunity: application in the development of cholera vaccines. Proc. Natl. Acad. Sci. USA 95:7035-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovach, M. E., M. D. Shaffer, and K. M. Peterson. 1996. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology 142:2165-2174. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Miller, V. L., and J. J. Mekalanos. 1984. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc. Natl. Acad. Sci. USA 81:3471-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay, A. K., S. Chakraborty, Y. Takeda, G. B. Nair, and D. E. Berg. 2001. Characterization of VPI pathogenicity island and CTXφ prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabbani, G. H., and W. B. Greenough. 1990. Cholera, p. 233-253. In E. Lebenthal and M. Duffy (ed.), Textbook of secretory diarrhea. Raven Press Ltd., New York, N.Y.
- 32.Sack, D. A., S. Huda, P. K. B. Neogi, R. R. Daniel, and W. M. Spira. 1980. Microtiter ganglioside enzyme-linked immunosorbent assay for Vibrio and Escherichia coli heat-labile enterotoxins and antitoxins. J. Clin. Microbiol. 11:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 34.Spira, W. M., R. B. Sack, and J. L. Froehlich. 1981. Simple adult rabbit model for Vibrio cholerae and enterotoxigenic E. coli diarrhea. Infect. Immun. 32:739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, R., C. Shaw, K. Peterson, P. Spears, and J. Mekalanos. 1988. Safe, live Vibrio cholerae vaccines? Vaccine 6:151-154. [DOI] [PubMed] [Google Scholar]
- 36.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 38.Waldor, M. K., E. J. Rubin, D. N. Gregory, H. H. Kimsey, and J. J. Mekalanos. 1997. Regulation, replication and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol. Microbiol. 24:917-926. [DOI] [PubMed] [Google Scholar]