Abstract
Mycoplasma arthritidis causes a severe septic arthritis in rats under natural and experimental conditions. An earlier study implicated a membrane lipoprotein designated MAA1 in cytadherence of M. arthritidis. In addition, a spontaneous adherence-deficient mutant was shown to contain a nonsense mutation in the gene encoding MAA1, resulting in production of a truncated product, MAA1Δ. In the present study, a wild-type maa1 gene carried on transposon Tn4001T was introduced into the low-adherence mutant by polyethylene glycol-mediated transformation. The presence of the tranposon and the wild-type maa1 gene in the chromosome of transformants was confirmed by PCR and Southern hybridization. The latter procedure also confirmed that each transformant contained a single copy of the transposon. Western immunoblotting showed that transformants produced both wild-type MAA1 and MAA1Δ, indicating that the introduced wild-type maa1 gene was functional. This phenotype was stably maintained after multiple subcultures even in the absence of antibiotic selection. Finally, transformants were shown to adhere to rat L-2 lung cells in culture at wild-type levels, providing confirmation for an important role for MAA1 in adherence.
Mycoplasma arthritidis is a natural pathogen of rats. Experimentally induced disease manifests as a severe acute to chronic inflammatory polyarthritis that is usually self-limiting (4). Convalescent rats are protected against reinfection; resistance is antibody mediated (2) and can be induced by immunization with a variety of cell components (11).
In an earlier study, we identified two putative adhesins by using an in vitro attachment-inhibition assay and a panel of monoclonal antibodies (MAbs) raised against membrane antigens from virulent M. arthritidis strain 158p10p9 (12). Two MAbs, 7a and A9a, inhibited adherence of M. arthritidis to rat L-2 lung cells in culture. The former was directed against a 61.4-kDa size- and phase-variable membrane lipoprotein designated MAA2 (12, 14, 16). The latter was directed against an 86.5-kDa lipoprotein, MAA1, that was neither size nor phase variable (12-14). Both proteins were able to elicit protective immunity in rats. Notably, MAb A9a, directed against MAA1, was almost completely protective (15). This indicates that MAA1 may be a target for protective immunity in natural and experimental infection with M. arthritidis and suggests that MAA1 may function as a virulence factor for strain 158p10p9. As previously noted, database searches indicated that MAA1 shows no significant similarities to other known proteins (13). This may reflect the specificity of binding to particular host cell receptors.
We also isolated a spontaneous mutant from 158p10p9 with greatly diminished adherence capacity compared to the wild type. We designated this mutant the low-adherence (LA) variant (also referred to as LC in an earlier publication [12]) and showed that it contained a point mutation at nucleotide 695 in the maa1 gene that generated a premature stop codon at that site. This resulted in the production of a truncated product, MAA1Δ. Although MAA1Δ was processed by the mycoplasmal cell and expressed on the cell surface, it was apparently nonfunctional (12, 13). Although this strongly implicated MAA1 as an adhesin, we could not rule out either an accessory role for this protein or an additional mutation in the LA variant that led to the LA phenotype. The purpose of the present study was to reintroduce a wild-type version of maa1 into the LA variant and determine whether that alone could restore adherence. This would provide stronger evidence for a direct role for MAA1 in attachment of M. arthritidis to host cells.
MATERIALS AND METHODS
M. arthritidis strain and culture conditions.
The parent strain for all of the transformants and mutants used in this investigation was the virulent M. arthritidis strain 158p10p9 (3), in which MAA1 was originally identified (12). It was grown in a modified Edward-type medium (5) containing 2% (wt/vol) BBL mycoplasma broth base (Becton Dickinson Microbiology Systems, Cockeysville, Md.), 20% (vol/vol) heat-inactivated horse serum (HyClone, Logan, Utah), 0.5% (wt/vol) l-arginine HCl (Sigma-Aldrich, St. Louis, Mo.), 0.002% (wt/vol) phenol red, 0.5% (vol/vol) BBL IsoVitaleX, 0.2% (wt/vol) denatured herring sperm DNA (Sigma-Aldrich), and 50 μg of ampicillin/ml. Agar medium was prepared by adding 1.1% (wt/vol) BBL select agar to the above formula and omitting phenol red. Stock cultures were stored at −70°C.
DNA preparation.
Mycoplasmal DNA was isolated as described previously (16), except for the addition of a 2-h incubation at 55°C with 50 microliters (20 mg/ml) of proteinase K (Sigma) immediately after the sodium dodecyl sulfate (SDS) and RNase A treatments (8). Plasmid DNA was isolated from Escherichia coli JM109 by a large-scale alkaline lysis method (1). DNA was dissolved in deionized water or 10 mM Tris-HCl-1 mM EDTA (pH 8.0) buffer and then quantitated spectrophotometrically. Final chromosomal DNA concentrations ranged from 0.5 to 2 μg/μl and plasmid DNA concentrations ranged from 14 to 15 μg/μl.
Vectors, constructs, and transformation procedures.
Vector pIVT was constructed by Dybvig et al. (5) from plasmid pISM2062. pISM2062 contains the modified staphylococcal transposon Tn4001mod, which was engineered by Knudtson and Minion for use as a mycoplasmal cloning vector by placement of BamHI and SmaI restriction sites near the end of one of the IS256 arms (7). Because the bifunctional aminoglycoside resistance determinant aacA-aphD carried by Tn4001 appears not to function in M. arthritidis, Dybvig et al. further modified pISM2062 by inserting a 5-kb HincII fragment containing tetM, which encodes resistance to tetracycline, into the SmaI site, generating the modified transposon Tn4001T on vector pIVT (5). Voelker and Dybvig showed that tetM was a useful marker for M. arthritidis transformation (10), and Dybvig et al. showed that pIVT could be used to deliver Tn4001T into the M. arthritidis chromosome (5).
The maa1 gene plus its putative promoter and transcription terminator was amplified from M. arthritidis 158p10p9 chromosomal DNA by PCR (see below), placing BglII sites at each end. This fragment was inserted into the BamHI site of pIVT. BglII was used for maa1 because the gene contains an internal BamHI site (13); the two enzymes generate compatible ends. A diagram of the resulting construct, designated pIVT-maa1, showing relevant restriction sites and PCR primer locations is shown in Fig. 1. Prior to use, pIVT-maa1 was methylated with AluI methylase (New England Biolabs, Beverly, Mass.); adequate methylation was confirmed by demonstrating resistance to digestion with HindIII. This was necessary because most M. arthritidis strains, including 158p10p9, contain an AluI-like restriction-modification system (10). Transformation was carried out as described previously (5). Briefly, a 3-ml culture of M. arthritidis was grown to approximately mid-log phase. Cells were pelleted by centrifugation, resuspended in 250 μl of ice-cold 100 mM CaCl2, held on ice for 30 min, and mixed with ca. 20 μg of methylated DNA. The mixture was added to 2 ml of polyethylene glycol (PEG 8000, 40% [wt/vol] in 10 mM Tris-HCl [pH 6.5]), vortexed briefly, immediately diluted with 10 ml of 10 mM Tris-HCl (pH 6.5), and vortexed again. The mixture was centrifuged at ca. 17,000 × g for 12 min at room temperature. The cell pellet was resuspended in 1 ml of Edward broth without antibiotics. After 3 h of incubation at 37°C, cells were plated onto Edward agar plates containing 5 μg of tetracycline/ml. After 4 to 10 days of incubation, agar plugs containing individual colonies were transferred by Pasteur pipette to 1 ml of Edward broth containing 5 μg of tetracycline/ml. The 1-ml cultures were expanded to 10 ml when color changes appeared, and samples were frozen at −70°C for later analysis. All strains and transformants were reidentified as derivatives of M. arthritidis strain 158p10p9 by PCR (see below).
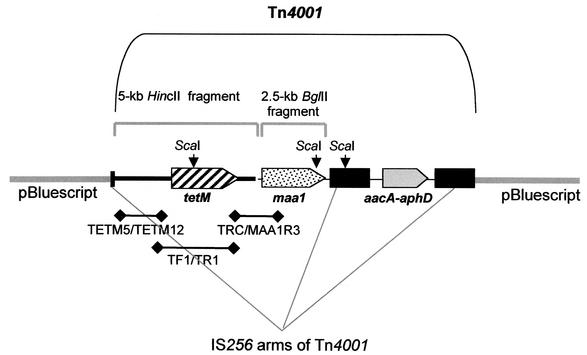
FIG. 1.
Diagram of pIVT-maa1. The portion of pIVT containing Tn4001T and its inserts is bracketed. Locations and orientations of three open reading frames are shown by shaded open arrows. The first (diagonal stripes) encodes tetracycline resistance determinant TetM and is located on a 5-kb HincII fragment inserted into the SmaI site of plasmid pISM2062 (7) by Dybvig et al. (5), generating vector pIVT. The second (stippled) encodes wild-type MAA1 plus its putative promoter and transcription terminator and is located on a 2.5-kb BglII fragment inserted into the BamHI site of pIVT in the present study. The third (shaded gray) is the bifunctional aminoglycoside resistance gene aacA-aphD, which is native to Tn4001 but does not function in M. arthritidis (5). The IS256 arms of Tn4001 are shaded in black. ScaI restriction sites are indicated by black arrows, and the approximate locations and sizes of PCR products are indicated by heavy lines below the diagram.
Plasmids pIVT and pIVT-maa1 were expanded by introduction into E. coli strain JM109 (Promega, Madison, Wis.) by CaCl2-mediated transformation (1) and electroporation (Gene Pulser II; Bio-Rad Laboratories, Hercules, Calif.), respectively. E. coli transformants and electroporants were grown in Luria broth (1) supplemented with 100 μg of ampicillin and 10 μg of tetracycline/ml.
PCR and Southern hybridization.
The maa1 gene plus its putative promoter and transcription terminator was amplified for cloning with primers F4BLG2 (5′-GTTAGATCTCATAATTGAAAAAGATGCCG-3′) and R2BGL2 (5′-GCTAGATCTGATAAAGCGCACTGTTGTGC-3′). These primers placed BglII sites at each end and were modified from previously described primers PF4 and PR2 (13).
To confirm the identity of strains and mutants used in the present study, primers F3 and HMPR, described previously (16), were used to amplify the 158p10p9 strain-specific maa2 gene.
To confirm the presence of Tn4001T in M. arthritidis transformants, an ∼1.6-kb DNA fragment upstream of the tetM gene was amplified with primers TETM5 (5′-CCCAATCCCATAGCCATACCTATC-3′) and TETM12 (5′-GGACATCCAATTATTTGTTCCCGC-3′) (Fig. 1). To confirm the presence in M. arthritidis transformants of the tetM-maa1-containing construct, a ∼1.3-kb DNA fragment extending from a region flanking the 3′ end of tetM into the 5′ end of the adjacent maa1 gene was amplified by primers TRC (5′-GCTCTTTCCGTTATGTATGG-3′) and MAA1R3 (5′-GTAGAAATTTTATGTGCGCCC-3′) (Fig. 1). Primers TF1 (5′-GAAAAGAACGGGAGTAATTGG-3′) and TR1 (5′-CCATACATAACGGAAAGAGC-3′) were used to amplify a 2.7-kb DNA fragment containing the entire tetM coding region for use as a probe in Southern hybridization assays (Fig. 1). Primer MAA1R3 was designed from the published sequence of maa1 (13); primers TRC, TF1, and TR1 were designed from the published sequence of transposon Tn916 (6).
All PCRs were carried out in 0.2-ml thin-walled tubes in 50-μl reaction mixes, each containing 0.25 U of Taq DNA polymerase (Promega), 200 μM deoxynucleoside triphosphates (Pharmacia Biotech, Piscataway, N.J.), 1× Mg2+-free reaction buffer (Promega), 1.5 mM (for primer pairs F3/HMPR and TETM5/TETM12) or 2.5 mM (for all others) MgCl2, 0.5 μM concentrations of primers, and ca. 150 to 300 ng of template DNA. The following thermal cycler settings were used for all reactions: 1 cycle of denaturation for 5 min at 94°C; 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 52°C, and polymerization for 3 min at 72°C; and final polymerization for 10 min at 72°C.
For Southern hybridization, 3 to 5 μg of chromosomal and 160 ng of plasmid DNA were digested with ScaI, electrophoresed on a 0.6% (wt/vol) agarose gel, and transferred to a nylon membrane (Hybond-N+; Amersham Pharmacia Biotech, Piscataway, N.J.) by vacuum blotting (VacuGene XL; Amersham Pharmacia) (9). The membrane was baked at 80°C, UV cross-linked for 5 min, prehybridized at 60°C for 30 min, and hybridized overnight at 60°C. The probe consisted of a 2.7-kb DNA fragment containing tetM amplified by PCR with primers TF1 and TR1 as described above and tagged by random-primed labeling with Fluorescein-11-dUTP (ECL random prime labeling and detection systems; Amersham Life Sciences). Probe labeling and signal detection were performed as described by the manufacturer.
Western immunoblotting.
Next, ∼5 μg of total mycoplasmal protein was electrophoresed on 10 or 12.5% (wt/vol) T (acrylamide) and 2.7% (wt/vol) C (N,N′-methylene-bisacrylamide) SDS-polyacrylamide gels and then transferred to nitrocellullose membranes as described previously (14). Membranes were blocked overnight by incubation at room temperature in 20 mM Tris-137 mM NaCl (pH 7.6) containing 0.1% (vol/vol) Tween 20 (TBST) and 5% (wt/vol) dry milk (TBST-milk). Blots were then incubated for 1 h at room temperature with anti-MAA1 MAb A9a diluted 1:5,000 in TBST-milk, rinsed six times in TBST, and incubated for 1 h at room temperature with peroxidase-conjugated immunoglobulin G fraction rabbit anti-mouse antibody (whole molecule specific; Organon Teknika, Cappel Division, Durham, N.C.) diluted 1:100,000 in TBST-milk. After six additional rinses, blots were incubated for 5 min in SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, Ill.) with peroxide as described by the manufacturer, wrapped in plastic wrap, and exposed to X-ray film for 30 s to 1 min.
Adherence assay.
Adherence assays were carried out as described previously (12). Briefly, rat L-2 lung cells (ATCC CCL 149) were grown to confluency in 24-well tissue culture plates in Dulbecco minimal essential medium with 10% fetal bovine serum (HyClone) and penicillin as the only antibiotic. L-2 lung cells were chosen because this is a defined cell line and representative of one of the organs targeted in M. arthritidis infection. Although arthritis is a primary manifestation of M. arthritidis disease, infection is often systemic, involving multiple organs, including the respiratory tract (4). Mycoplasma stock cultures were thawed, diluted if necessary, pelleted by centrifugation, and resuspended in 1.6 ml of Dulbecco minimal essential medium. Medium was aspirated from the tissue culture wells, and 0.5 ml of mycoplasma suspension containing ca. 106 to 107 CFU was added to each well. After 6 h of incubation at 37°C in 5% CO2, the medium was aspirated from each well, and monolayers were rinsed six times with 0.01 M phosphate-buffered 0.15 M saline (pH 7.4). L-2 cells and adherent mycoplasmas were dislodged from wells by trypsinization in a total volume of 0.5 ml per well. For enumeration of mycoplasmas remaining attached to cells after the rinsing step and for calculation of adherence efficiency, colony counts were performed on the original mycoplasma inocula and on each trypsinized suspension. An adherence index was calculated for each test sample by dividing the percentage of the original mycoplasma inoculum remaining attached to L-2 cells by the percentage calculated for wild-type 158p10p9.
Statistical analysis.
Adherence data were analyzed by using the Dunnett t test. Differences were considered statistically significant at P < 0.05.
RESULTS AND DISCUSSION
Transformation of the LA variant with pIVT-maa1 and pIVT.
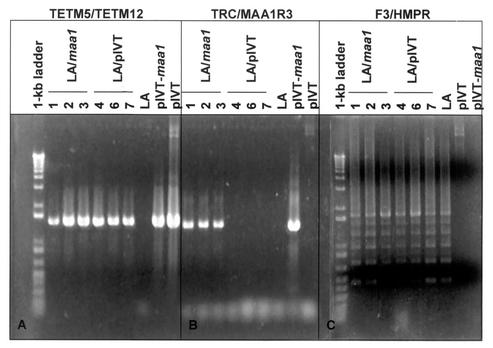
The LA variant was passaged in broth culture four to five times on consecutive days prior to transformation. Cultures were used when they had reached early log phase to mid-log phase, as estimated by the color change in the medium. After methylation, plasmid DNA was introduced into M. arthritidis by polyethylene glycol-mediated transformation. Because pIVT does not replicate in M. arthritidis, effective transformation requires the insertion of its resident transposon into the mycoplasmal chromosome. Three clones transformed with pIVT-maa1 (LA/maa1-1, -2, and -3) and three transformed with pIVT alone (LA/pIVT-4, -6, and -7) were selected for further study. The latter were included to rule out any effect the transposon itself might have either on expression of maa1 or on adherence. DNA was extracted from each clone. The presence of the appropriate inserts in the transformant chromosomes and their derivation from strain 158p10p9 were confirmed by PCR (Fig. 2). All transformants contained Tn4001T, as indicated by amplification of products with primers TETM5 and TETM12 from all six transformed clones and from both plasmids but not from the LA variant. Only the clones transformed with pIVT-maa1 contained adjacent copies of tetM and maa1, as indicated by the amplification of products with primers TRC and MAA1R3 from LA/maa1-1, -2, and -3 and from pIVT-maa1 but not from LA/pIVT-4, -6, and -7, the LA variant, or pIVT. Finally, the presence of a six-runged “ladder” pattern with primers that amplified the 158p10p9-specific maa2 gene proved that all of the DNA samples were derived from that strain and not from a contaminant.
FIG. 2.
PCR products amplified from DNA extracted from the LA variant, the transformants, pIVT, and pIVT-maa1. Locations of primers used in reactions shown in panels A and B are shown in Fig. 1. (A) Fragments amplified with primers just upstream of tetM. Products were amplified only from constructs containing the Tn4001T transposon. (B) Fragments amplified with primers designed from the 3′ end of the fragment containing tetM and the 5′ end of wild-type maa1. Products were amplified only from constructs in which tetM and maa1 were adjacent. (C) Fragments amplified with primers F3 and HMPR, which are specific for the maa2 gene. These primers typically yield a ladder pattern because maa2 undergoes rapid size variation due to loss or gain of one or more of the 264-bp tandem repeat units that comprise the bulk of the gene (15). The presence of six “rungs” indicates that the LA variant and its progeny were derived from strain 158p10p9 and did not arise from accidental contamination of cultures with another strain.
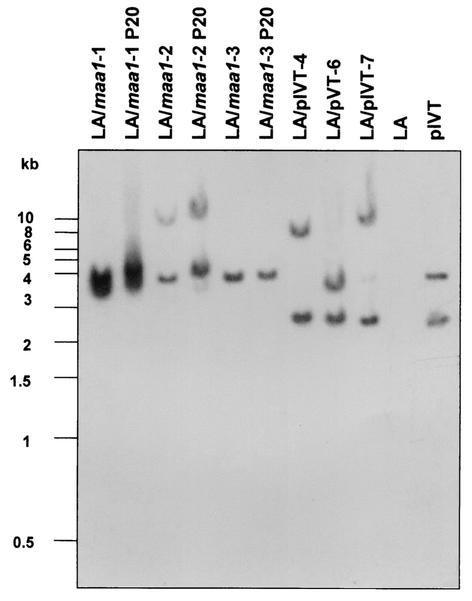
To determine whether each clone arose from a separate transformation event, ScaI-digested chromosomal DNA was subjected to Southern hybridization with the tetM probe (Fig. 3). This probe was amplified by PCR with primers TF1 and TR1 and contained the entire tetM coding region (Fig. 1). tetM, maa1, and the IS256 arms of Tn4001 each possess a single ScaI site. If each transformant contained just a single copy of the transposon, it should show two hybridizing ScaI fragments, one of which would be a fragment from within the insert (the internal fragment) and the other a fragment overlapping the insert and adjacent M. arthritidis chromosomal DNA, its size dependent upon the integration site (the variable fragment). The LA/maa1 internal fragments should be ∼4.3 kb and the LA/pIVT, ∼2.6 kb in size. Two distinct hybridizing fragments could be seen in each clone except for LA/maa1-3, which had just one, and the transposon appears to have integrated into a different site in each transformant. It is not clear why LA/maa1-3 had just the single internal ScaI fragment. It is possible that this band actually consisted of two comigrating fragments or that this particular clone contained a deletion or mutation in its DNA insert. Sequencing of the integrated fragment will be required to determine the exact cause. However, this mutation or deletion had no apparent effect on tetracycline resistance or expression of the wild-type maa1 gene (see below).
FIG. 3.
Southern hybridization of ScaI-digested chromosomal DNA from M. arthritidis strains and transformants and from vectors used in the present study with a Fluorescein-11-dUTP-labeled tetM probe (Fig. 1). Each transformant except LA/maa1-3 contained two hybridizing ScaI fragments, indicating that each contained a single copy of transposon Tn4001T. One fragment in each transformed clone was an internal ScaI fragment present on the DNA insert. For the LA/maa1 clones, that fragment is ∼4.3 kb in size and overlaps the 3′ end of tetM and the 5′ end of the adjacent wild-type maa1. For the LA/pIVT clones, that fragment is ∼2.6 kb in size and overlaps the 3′ end of tetM and the 5′ end of the adjacent IS256 arm of Tn4001T. Lanes labeled P20 contain DNA from transformed clones that had been passaged 20 times in broth medium in the absence of antibiotic selection. Identical patterns between passaged and unpassaged samples indicate that the transposon was stable in the chromosome and that there had been no secondary transposition events.
Stability of the transposons in transformant chromosomes was determined by passaging LA/maa1-1, -2, and -3 20 times on consecutive days in broth medium with or without tetracycline. DNA from the 20th passage in the absence of tetracycline was digested with ScaI and subjected to Southern hybridization. The results shown in Fig. 3 (lanes P20) indicate that the transposon remained stably integrated into the chromosomes of all three transformants even in the absence of antibiotic selection and that there were no secondary transposition events.
Expression of wild-type maa1 in LA variant transformants.
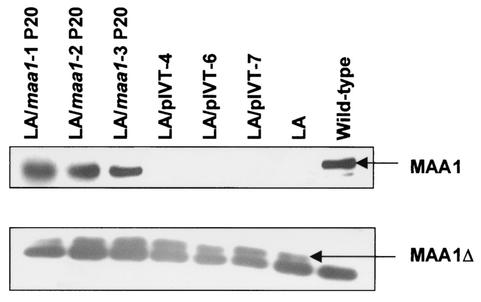
Whole-cell lysates of LA/maa1-1, -2, and -3, LA/pIVT-4, -6, and -7, the LA variant, and wild-type 158p10p9 were electrophoresed on 10 and 12.5% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with anti-MAA1 MAb A9a. All lanes contained approximately equal amounts of mycoplasmal protein. Figure 4 confirms that the LA variant and LA/pIVT-4, -6, and -7 produced only the truncated protein MAA1Δ and that the wild-type parent strain 158p10p9 produced only the full-sized protein. This indicates that the presence of Tn4001T alone had no effect on normal expression of maa1. The figure also shows that LA/maa1-1, -2, and -3 produced both truncated and full-sized products, indicating that they were, in fact, derived from the LA variant and that the wild-type maa1 gene introduced on Tn4001T was transcribed and translated to approximately wild-type levels in each clone. The LA/maa1-1, -2, and -3 antigens used here were derived from 20th passage cultures, further indicating the stability and functionality of the DNA inserts.
FIG. 4.
Western immunoblot of whole lysed mycoplasmal cells from the wild-type strain 158p10p9, the LA variant, and its transformed progeny with anti-MAA1 MAb A9a (12). Equal amounts of protein were added to each lane. (A) Full-sized MAA1 from a 10% SDS-PAGE gel. (B) MAA1Δ from a 12.5% gel. Also shown here is a dark band that migrates with the dye line and has been consistently present in all samples since this MAb has been in use (12). It probably represents degradation products of MAA1 and MAA1Δ. These figures show that the LA/maa1 transformants produced both wild-type and truncated versions of MAA1, indicating that the wild-type gene introduced on transposon Tn4001T was expressed, whereas the LA variant and clones derived from transformation with pIVT alone produced only MAA1 and the wild-type strain 158p10p9 produced only the full-sized product.
Restoration of cytoadherence by the cloned maa1 gene.
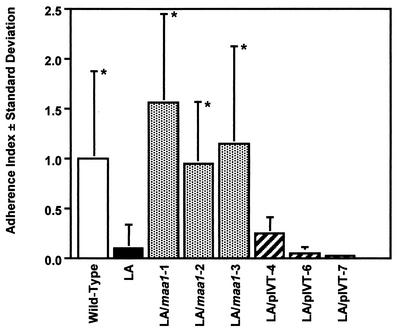
The six transformants, the LA variant, and wild-type 158p10p9 were tested for their ability to adhere to rat L-2 lung cells in culture (Fig. 5). Differences in adherence between the LA variant and LA/pIVT-4, -6, and -7 were not statistically significant, indicating that Tn4001T alone did not influence cytadherence. However, LA/maa1-1, -2, and -3 all adhered as well as or better than the wild-type 158p10p9 and significantly better than both the LA variant (P < 0.003) and the LA/pIVT transformants (P values ranged from < 0.0001 to 0.038).
FIG. 5.
Adherence of transformants compared to the LA variant and wild-type strain 158p10p9. The adherence index was calculated as the percentage of each test sample divided by the percentage of wild-type strain 158p10p9 adhering to rat L-2 lung cells in culture. Each bar represents the results from at least six replicate samples ± the standard deviation. LA/maa1-1, -2, and -3 adhered as well as or better than the wild-type strain 158p10p9 and significantly better than the LA variant (P < 0.003) and LA/pIVT-4, -6, and -7 (P ≤ 0.038). There were no statistically significant differences in adherence between the LA variant and clones LA/pIVT-4, -6, an -7. An asterisk indicates significantly greater adherence than the LA variant.
These data confirm that functional genes can be introduced into M. arthritidis by vector pIVT. They also indicate that cytadherence in 158p10p9 depends upon the presence of an intact gene encoding full-sized MAA1. They do not distinguish between a primary and a critical accessory role for MAA1, but they do demonstrate that the sole adherence-associated mutation in the LA variant was in maa1. These data plus the earlier observation that an anti-MAA1 MAb was able to confer complete protection against M. arthritidis-induced arthritis in rats provide a strong indication that MAA1 is an important player in M. arthritidis host-parasite interaction.
Acknowledgments
We thank C. T. French, University of Alabama at Birmingham (and now at Emory University), for valuable assistance and advice in preparing the constructs and conducting the transformations described in this study. We thank Richard Duman, University of South Dakota, for assistance in preparation of many of the media and reagents used in this study.
This work was supported in part by a grant to L.R.W. from the South Dakota Health Research Foundation and by the U.S. Public Health Service grant AR44252 to K.D. from the National Institutes of Health.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Cole, B. C., J. F. Cahill, B. B. Wiley, and J. R. Ward. 1969. Immunological responses of the rat to Mycoplasma arthritidis. J. Bacteriol. 98:930-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, B. C., J. R. Ward, R. S. Jones, and J. F. Cahill. 1971. Chronic proliferative arthritis of mice induced by Mycoplasma arthritidis. I. Induction of disease and histopathological characteristics. Infect. Immun. 4:344-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, B. C., L. R. Washburn, and D. Taylor-Robinson. 1985. Mycoplasma-induced arthritis, p. 108-160. In S. Razin and M. F. Barile (ed.), The mycoplasmas, vol. IV. Mycoplasma pathogenicity. Academic Press, Inc., New York, N.Y.
- 5.Dybvig, K., C. T. French, and L. L. Voelker. 2000. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J. Bacteriol. 182:4343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flannagan, S. E., L. A. Zitzow, Y. A. Su, and D. B. Clewell. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32:350-354. [DOI] [PubMed] [Google Scholar]
- 7.Knudtson, K. L., and F. C. Minion. 1993. Construction of Tn4001 lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene 137:217-222. [DOI] [PubMed] [Google Scholar]
- 8.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 9.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 10.Voelker, L. L., and K. Dybvig. 1996. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J. Bacteriol. 178:6078-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washburn, L. R., S. Hirsch, M. McKenzie, and L. L. Voelker. 1992. Vaccination of Lewis rats against Mycoplasma arthritidis-induced arthritis. Am. J. Vet. Res. 53:52-58. [PubMed] [Google Scholar]
- 12.Washburn, L. R., S. Hirsch, and L. L. Voelker. 1993. Mechanisms of attachment of Mycoplasma arthritidis to host cells in vitro. Infect. Immun. 61:2670-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Washburn, L. R., E. J. Miller, and K. E. Weaver. 2000. Molecular characterization of Mycoplasma arthritidis membrane lipoprotein MAA1. Infect. Immun. 68:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washburn, L. R., L. L. Voelker, L. J. Ehle, S. Hirsch, C. Dutenhofer, K. Olson, and B. Beck. 1995. Comparison of Mycoplasma arthritidis strains by enzyme-linked immunosorbent assay, immunoblotting, and DNA restriction analysis. J. Clin. Microbiol. 33:2271-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Washburn, L. R., and E. J. Weaver. 1997. Protection of rats against Mycoplasma arthritidis-induced arthritis by active and passive immunizations with two surface antigens. Clin. Diagn. Lab. Immunol. 4:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washburn, L. R., K. E. Weaver, E. J. Weaver, W. Donelan, and S. Al-Sheboul. 1998. Molecular characterization of Mycoplasma arthritidis variable surface protein MAA2. Infect. Immun. 66:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]