Abstract
Proteus mirabilis, a cause of complicated urinary tract infection, produces urease, an essential virulence factor for this species. UreR, a member of the AraC/XylS family of transcriptional regulators, positively activates expression of the ure gene cluster in the presence of urea. To specifically evaluate the contribution of UreR to urease activity and virulence in the urinary tract, a ureR mutation was introduced into P. mirabilis HI4320 by homologous recombination. The isogenic ureR::aphA mutant, deficient in UreR production, lacked measurable urease activity. Expression was not detected in the UreR-deficient strain by Western blotting with monoclonal antibodies raised against UreD. Urease activity and UreD expression were restored by complementation of the mutant strain with ureR expressed from a low-copy-number plasmid. Virulence was assessed by transurethral cochallenge of CBA mice with wild-type and mutant strains. The isogenic ureR::aphA mutant of HI4320 was outcompeted in the urine (P = 0.004), bladder (P = 0.016), and kidneys (P ≤ 0.001) 7 days after inoculation. Thus, UreR is required for basal urease activity in the absence of urea, for induction of urease by urea, and for virulence of P. mirabilis in the urinary tract.
Proteus mirabilis commonly infects the urinary tracts of individuals with structurally abnormal urinary tracts or long-term indwelling catheters (20). Many virulence factors have been identified from this organism, including several types of fimbriae (1, 17, 18), hemolysin (28), flagella (21), and immunoglobulin A (IgA) protease (26, 30). Importantly, P. mirabilis produces a urea-inducible urease, which hydrolyzes urea into ammonia and ultimately carbon dioxide. As a result of ammonia production, an increase in local pH causes precipitation of normally soluble calcium and magnesium ions. These salt crystals can grow to remarkable size to produce bladder and kidney stones (16), which are a hallmark of infections with Proteus spp. (6).
We have shown previously that urease is an essential virulence factor in P. mirabilis urinary tract infections (UTIs) (11, 12). A urease-negative mutant was constructed by using homologous recombination to disrupt ureC, the main urease structural subunit. The urinary tracts of mice infected with the P. mirabilis isogenic urease mutant were colonized with significantly fewer numbers than those of mice infected with the parental strain (12). This mutant also demonstrated a reduced ability to persist after a 1- or 2-week infection and did not promote bladder and kidney stone formation (11).
The P. mirabilis urease gene cluster consists of ureABC, which encode the apoenzyme structural subunits, and ureDEFG, which encode proteins that facilitate insertion of the essential nickel ions into the catalytic site (22). Urease operon expression is positively activated by UreR, a member of the AraC/XylS family of transcriptional regulators (3, 9, 23). UreR, a dimer of identical 293-amino-acid polypeptides (24), binds urea (5), causing the protein to bind avidly to both the ureR and ureD promoters within the 491-bp ureR-ureD intergenic region (27). RNA polymerase is recruited by UreR, thus activating transcription (27). To date, all of the work elucidating the functional role of UreR activation of the urease gene cluster has focused on UreR in vitro by using reporter constructs expressed in Escherichia coli. Since UreR controls expression of the entire gene cluster, we investigated how UreR function contributes to basal urease production and virulence of P. mirabilis in a mouse model of ascending UTI.
P. mirabilis HI4320, originally isolated from an elderly (>65 years old) woman with urinary catheter-associated bacteriuria, possesses urea-inducible urease activity (13, 29). E. coli DH5α was used as the host strain for transformation of plasmids other than pGP704 (19) and derivatives which require E. coli DH5αλpir. Plasmid pMID1010 carries the entire P. mirabilis gene cluster cloned into pBR322 (14). Strains were maintained on Luria broth supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml) when necessary. Nonswarming agar (per liter, 10 g of tryptone, 5 g of yeast extract, 0.5 g of NaCl, and 15 g of agar, with 0.5% glycerol) was used to prevent P. mirabilis swarming when cultured on plates (2).
Construction of the ureR::aphA mutation in P. mirabilis HI4320.
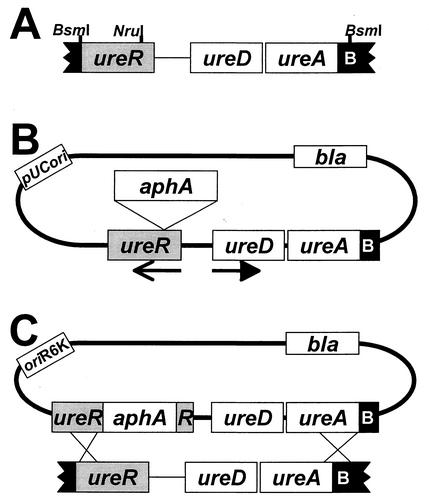
To determine the contribution of ureR to urease expression in vivo, we constructed a P. mirabilis strain deficient in UreR production according to the scheme shown in Fig. 1. The ureR open reading frame was subcloned and insertionally inactivated with a kanamycin resistance cassette. The fragment containing disrupted ureR was cloned into a pGP704-based suicide vector as described in the legend to Fig. 1. The resulting suicide construct, pJD12, was electroporated into P. mirabilis HI4320. To verify that a double crossover event had occurred, Kanr colonies were screened for ampicillin sensitivity; 4 of 525 colonies were identified as having appropriate characteristics (Fig. 1C). Homologous recombination of the wild-type ureR with ureR::aphA was confirmed by PCR by using chromosomal DNA and primers specific for ureR. A band of approximately 0.9 kb was amplified in the parent strain, while a band of approximately 2.2 kb (ureR plus Kanr cassette) was observed in the Kanr Amps mutant strain (data not shown). This mutant, designated P. mirabilis HI4320 ureR::aphA, was found to swarm normally and had a growth rate identical to that of the wild type (data not shown).
FIG. 1.
Construction of a P. mirabilis ureR mutant. (A) A 2.535-kb BsmI fragment from pMID1010 encompassing most of ureR, ureD, ureA and the first 51 nucleotides of ureB was treated with T4 DNA polymerase to fill in overhanging ends and render the fragment blunt and cloned into the SmaI site of pBluescript SK+ treated with shrimp alkaline phosphatase to create pJD10. (B) After digestion of pJD10 with NruI (corresponding to codon 25 of ureR) and shrimp alkaline phosphatase treatment, a blunt-ended kanamycin resistance cassette (aphA), excised from pUC-4κ by digestion with HincII, was inserted into pJD10. Kanamycin- and ampicillin-resistant versions of DH5α harboring the new construct, pJD11, were isolated. Arrows represent the direction of transcription. (C) A 3.8-kb XbaI-KpnI fragment containing the entire insert region of pJD11 was excised from an agarose gel (XmnI digestion was also used to remove the pBluescript vector from the desired fragment) and subcloned into pGP704 (19) to create the suicide construct pJD12. The suicide construct pJD12 was electroporated into P. mirabilis HI4320, and kanamycin-resistant transformants were selected on nonswarming agar.
Urease activity in the ureR::aphA mutant.
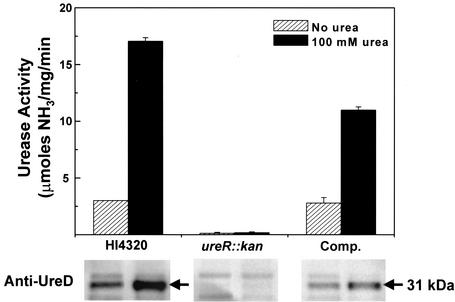
Since UreR functions as the transcriptional activator of the urease gene cluster, urease activity was quantified in vitro for P. mirabilis HI4320 and the isogenic ureR::aphA mutant (Fig. 2). The wild-type strain displayed a 5.7-fold increase in urease activity after culture in the presence of 100 mM urea. Conversely, although the level of detection for the urease assay is quite sensitive (approximately 10 nmol of ammonia), urease activity was undetectable for the mutant strain under identical experimental conditions.
FIG. 2.
P. mirabilis urease activity in wild-type HI4320, ureR::aphA mutant, and complemented strains. Urease activity (upper panel) was determined by using the phenol-hypochlorite reaction (31). Overnight cultures (5 ml) of the P. mirabilis wild-type strain HI4320, the ureR mutant (ureR::aphA), and the complemented strain (Comp.) were used to inoculate fresh medium (1:100) with or without 100 mM urea containing the appropriate antibiotics when necessary. Bacteria from mid-exponential-phase cultures (optical density at 600 nm, 0.4) (3 ml) were resuspended in 1 ml of 50 mM HEPES, pH 7.5, and lysed by passage through a French pressure cell at 20,000 lb/in2. The protein concentration of crude extract was determined by the bicinchoninic acid assay (Pierce, Rockford, Ill.). Total protein (10 μg) was diluted into 1 ml of assay buffer (50 mM HEPES, pH 7.5, and 25 mM urea) and incubated for 20 min at 37°C. Ammonia production was measured by using the reaction of phenyl nitroprusside and alkali hypochlorite (31). Optical density was measured at 625 nm by using ammonium chloride as a standard. For Western blotting (lower panel), monoclonal anti-UreD serum was prepared from a murine hybridoma cell line as previously described (8). Crude extract prepared for urease assays was assessed for production of UreD. Protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred overnight onto Immobilon-P membrane (Millipore, Inc.). Anti-UreD serum (1:100) and anti-mouse IgG conjugated with horseradish peroxidase (1:2,000) were used as primary and secondary antibodies, respectively. Protein bands were visualized on X-ray film by using chemiluminescence reagents from Amersham Biosciences (Piscataway, N.J.).
Complementation of the ureR mutation.
To determine whether urease activity could be restored to the P. mirabilis ureR mutant by complementation, ureR was cloned into the EcoRV-BamHI site of pACYC184 and was expressed from the tetracycline promoter. Following electroporation of the construct into the ureR mutant strain, urease activity was measured and the production of UreD was assayed by Western blotting (Fig. 2). Complementation by ureR in trans resulted in wild-type levels of basal urease activity as well as a fourfold induction (∼60% of that of the wild type) after culture in the presence of 100 mM urea. While ureR provided in multicopy restored urea inducibility, the level of induction was not statistically equivalent to that observed in the wild-type strain.
The data to date suggest that the genes encoding the urease structural subunits and accessory proteins (except ureG) are transcribed on a single polycistronic mRNA (4). Because UreR is a transcriptional activator, it is likely that only a few copies of the gene product are required for activation. It is therefore not surprising that UreR was undetectable on Western blots of either wild-type P. mirabilis or the isogenic ureR mutant (data not shown). To determine if expression of the urease operon occurred, we measured UreD production by using Western blotting for the parent, mutant, and complemented mutant (Fig. 2). As predicted from the presence of basal urease activity, P. mirabilis produced a small but detectable amount of UreD in the absence of urea. The amount of UreD increased significantly upon addition of 100 mM urea, which correlated with the fold increase observed for urease activity. Under identical experimental conditions, UreD expression was not detected in the P. mirabilis ureR::aphA mutant regardless of urea levels, whereas UreD expression was restored by complementation with cloned ureR expressed in trans from a plasmid. These data demonstrated that a fully functional ure gene cluster was present and that insertional inactivation of ureR did not exert polar effects on other ure genes. Importantly, it was revealed that UreR is responsible for the low basal levels of urease activity in uninduced P. mirabilis.
The P. mirabilis ureR::aphA mutant is outcompeted by the parent strain in a murine cochallenge model of UTI.
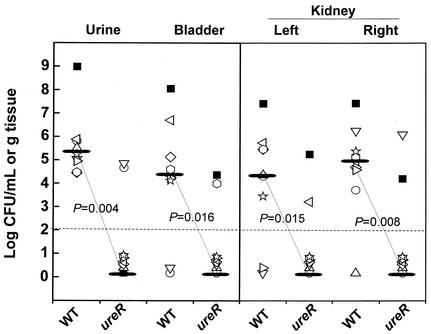
The virulence of P. mirabilis parent and mutant strains was assessed in a murine cochallenge model (15) by using a modification (10) of the procedure developed by Hagberg et al. (7). For cochallenge experiments, 10 6- to 8-week-old female CBA mice (Harlan Sprague Dawley, Indianapolis, Ind.) were infected transurethrally with an approximate 1:1 ratio (5 × 106 CFU of each strain) of the P. mirabilis parent and ureR::aphA mutant strains. Seven days after inoculation, urine was collected and mice were sacrificed. Quantitative counts from the urine, bladder, and kidneys were obtained by plating on nonswarming agar (2) to allow growth of both parent and mutant strains and on nonswarming agar with kanamycin (50 μg/ml) to detect the mutant strain alone. The limit of detection for determination of viable bacteria was 102 CFU per milliliter of urine or per gram of tissue. In these studies, the mutant was unable to colonize the bladder or kidneys in 8 of 10 mice (median value, 102 CFU, which is below the limit of detection) (Fig. 3). As a result of the inability to produce urease, the ureR::aphA mutant strain was dramatically outcompeted by the wild-type strain in the urine (P = 0.004), bladder (P = 0.016) and kidneys (P < 0.001) as well as the urinary tract overall (P < 0.001).
FIG. 3.
Cochallenge of P. mirabilis parent and ureR::aphA isogenic mutant strains in the CBA mouse model of ascending UTI. Mice were transurethrally infected with 107 CFU containing a 1:1 ratio of P. mirabilis HI4320 and ureR mutant strains. After a 7-day infection, urine, bladder, and kidney specimens were collected and the CFU per milliliter or per gram of tissue of parent and mutant strains was determined for each mouse. Each unique symbol refers to CFU for the wild type (WT) and the ureR mutant from one cochallenged mouse. The horizontal dashed line represents the lower limit of detection of viable bacteria. For ease of comparison, the median values (black bars) are connected by dotted lines. Statistical differences between the median number of log10 CFU per milliliter or per gram of tissue for P. mirabilis HI4320 and its ureR::aphA mutant were determined by using the Wilcoxon rank sum test.
UreR is required for basal urease activity in the absence of urea, for induction of urease by urea, and for virulence in the urinary tract.
Urease is an essential virulence factor for P. mirabilis colonization and persistence in UTI (11, 12). UreR, an AraC/XylS family transcriptional regulator, has been shown in vitro to positively activate ure gene cluster expression in a urea-dependent manner. To properly assess the role of UreR regulation activity in P. mirabilis in vivo, homologous recombination was used to construct a P. mirabilis mutant that was incapable of producing UreR. A kanamycin resistance gene, aphA, was inserted at the ureR locus in the P. mirabilis chromosome by homologous recombination with a pGP704-based suicide construct (Fig. 1). This mutant did not produce urease in the absence or in the presence of urea as measured by using the phenol-hypochlorite assay on mid-exponential-phase cultures (Fig. 2). We have known for many years that P. mirabilis produced basal levels of urease activity in the absence of exogenously added urea (14). Since there is a complete loss of basal urease activity in the ureR mutant, we conclude that in P. mirabilis, low levels of UreR may account for basal urease expression in the absence of urea. Urease activity was restored upon complementation with ureR expressed on a low-copy-number plasmid (Fig. 2). Interestingly, full wild-type levels of induction were not reached in the complemented mutant. It is possible that translational coupling plays a role in which newly translated UreR must bind immediately to the adjacent ureR-ureD intergenic region. UreR synthesized from the complementing plasmid would have to diffuse to the chromosomal target, an event that may be less efficient than the wild-type mechanism.
The isogenic ureR::aphA mutant of strain HI4320 was outcompeted by the parental strain in 10 of 10 CBA mice following transurethral infection (Fig. 3); indeed, in only one mouse were the numbers of the ureR mutant and parent strain similar. In our experience, as well as that of others (25), coinfection with parental and mutant strains within the same host represents a more sensitive method to test for attenuation of virulence. To our knowledge, this is the first report of a cochallenge experiment with P. mirabilis and a urease-negative mutant. We infected 10 CBA mice with a 1:1 mixture (∼5 × 106 CFU of each strain) of the HI4320 parent and ureR::aphA mutant strains. The parental strain outcompeted the mutant strain in all mice tested (Fig. 3). The low fitness and complete attenuation of the mutant strain confirms the role of urease as an essential virulence factor for P. mirabilis and demonstrates that the urease of the wild-type strain is not sufficient to sustain the urease-negative mutant in the urinary tract. Interestingly, in a previously published study (12), the wild type and the urease-negative (ureC) mutant of P. mirabilis were independently inoculated into the bladders of mice. While the mutant colonized the urine, bladder, and kidneys in ∼100-fold fewer CFU per milliliter or per gram, this mutant was nevertheless able to colonize the urinary tract to some degree in the absence of a wild-type strain. The inability of the wild-type strain to support the cocolonization of the ureR mutant suggests that the alteration of urine pH by urease is not the sole action that provides advantage to P. mirabilis. It is likely that urease, a cytoplasmic protein, provides additional advantages, such as availability of ammonia for glutamine production and protein synthesis. These latter functions could not be provided to the mutant by the wild-type strain.
Cochallenge of CBA mice with both parent and mutant strains represents a powerful method to directly compare the requirement of urease production to P. mirabilis virulence in a single mouse. High levels (107 to 108 CFU/g of tissue) of the P. mirabilis wild type were recovered from a moribund mouse (Fig. 3, black square) due to the formation of an obstructive bladder stone. Interestingly, the highest mutant counts were also observed for this mouse. Our lab has previously visualized P. mirabilis attached to the surfaces of bladder and kidney epithelium and within crevices of bladder and kidney stones (16). We hypothesize that residing close to the stone may help P. mirabilis evade the host immune response. This observation suggests that stone formation may not only contribute to P. mirabilis persistence in the urinary tract (11) but may also provide an advantage to nonureolytic pathogens as well.
To our knowledge, UreR has not been shown to affect the expression of any other operon besides the ure gene cluster. Since our data demonstrate that the absence of the UreR regulatory protein leads to inhibition of urease expression, this protein may serve as a possible target for treatment or eradication of P. mirabilis from the urinary tract. Further experimentation is required to determine if other ureR-positive Enterobacteriaceae family members will display a similar loss of urease activity following perturbation of ureR.
Acknowledgments
This work was supported by Public Health Service grants AI23328 and DK47920 from the National Institutes of Health. J.D.D. was funded by a National Research Service Award (F32 AI51078) from the NIH.
Editor: D. L. Burns
REFERENCES
- 1.Bahrani, F. K., G. Massad, C. V. Lockatell, D. E. Johnson, R. G. Russell, J. W. Warren, and H. L. Mobley. 1994. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 62:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belas, R., D. Erskine, and D. Flaherty. 1991. Transposon mutagenesis in Proteus mirabilis. J. Bacteriol. 173:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Orazio, S. E. F., and C. M. Collins. 1993. The plasmid-encoded urease gene cluster of the family Enterobacteriaceae is positively regulated by UreR, a member of the AraC family of transcriptional activators. J. Bacteriol. 175:3459-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Orazio, S. E. F., and C. M. Collins. 1995. UreR activates transcription at multiple promoters within the plasmid-encoded urease locus of the Enterobacteriaceae. Mol. Microbiol. 16:145-155. [DOI] [PubMed] [Google Scholar]
- 5.Gendlina, I., D. M. Gutman, V. Thomas, and C. M. Collins. 2002. Urea-dependent signal transduction by the virulence regulator UreR. J. Biol. Chem. 277:37349-37358. [DOI] [PubMed] [Google Scholar]
- 6.Griffith, D. P., D. M. Musher, and C. Itin. 1976. Urease: the primary cause of infection-induced urinary stones. Investig. Urol. 13:346-350. [PubMed] [Google Scholar]
- 7.Hagberg, L., I. Engberg, R. Freter, J. Lam, S. Olling, and C. Svanborg-Eden. 1983. Ascending unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heimer, S. R., and H. L. T. Mobley. 2001. Interaction of Proteus mirabilis urease apoenzyme and accessory proteins identified with yeast two-hybrid technology. J. Bacteriol. 183:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Island, M. D., and H. L. T. Mobley. 1995. Proteus mirabilis urease: operon fusion and linker insertion analysis of ure gene organization, regulation, and function. J. Bacteriol. 177:5653-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, D. E., C. V. Lockatell, M. Hall-Craigs, H. L. T. Mobley, and J. W. Warren. 1987. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J. Urol. 138:632-635. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, D. E., R. G. Russell, C. V. Lockatell, J. C. Zulty, J. W. Warren, and H. L. T. Mobley. 1993. Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect. Immun. 61:2748-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, B. D., C. V. Lockatell, D. E. Johnson, J. W. Warren, and H. L. T. Mobley. 1990. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 58:1120-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, B. D., and H. L. T. Mobley. 1987. Genetic and biochemical diversity of ureases of Proteus, Providencia, and Morganella species isolated from urinary tract infection. Infect. Immun. 55:2198-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, B. D., and H. L. T. Mobley. 1988. Proteus mirabilis urease: genetic organization, regulation, and expression of structural genes. J. Bacteriol. 170:3342-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, X., D. E. Johnson, and H. L. T. Mobley. 1999. Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis. Infect. Immun. 67:2822-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, X., H. Zhao, C. V. Lockatell, C. B. Drachenberg, D. E. Johnson, and H. L. Mobley. 2002. Visualization of Proteus mirabilis within the matrix of urease-induced bladder stones during experimental urinary tract infection. Infect. Immun. 70:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massad, G., F. K. Bahrani, and H. L. Mobley. 1994. Proteus mirabilis fimbriae: identification, isolation, and characterization of a new ambient-temperature fimbria. Infect. Immun. 62:1989-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massad, G., C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 1994. Proteus mirabilis fimbriae: construction of an isogenic pmfA mutant and analysis of virulence in a CBA mouse model of ascending urinary tract infection. Infect. Immun. 62:536-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mobley, H. L. 1996. Virulence of Proteus mirabilis, p. 245-269. In H. L. Mobley and J. W. Warren (ed.), Urinary tract infections: molecular pathogenesis and clinical management. ASM Press, Washington, D.C.
- 21.Mobley, H. L., R. Belas, V. Lockatell, G. Chippendale, A. L. Trifillis, D. E. Johnson, and J. W. Warren. 1996. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 64:5332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson, E. B., E. A. Concaugh, P. A. Foxall, M. D. Island, and H. L. Mobley. 1993. Proteus mirabilis urease: transcription and regulation by UreR. J. Bacteriol. 175:465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poor, C. A., C. Coker, J. D. Dattelbaum, and H. L. T. Mobley. 2001. Identification of domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 183:4526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect. Immun. 69:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senior, B. W., M. Albrechtsen, and M. A. Kerr. 1987. Proteus mirabilis strains of diverse type have IgA protease activity. J. Med. Microbiol. 24:175-180. [DOI] [PubMed] [Google Scholar]
- 27.Thomas, V. J., and C. M. Collins. 1999. Identification of UreR binding sites in the Enterobacteriaceae plasmid-encoded and Proteus mirabilis urease gene operons. Mol. Microbiol. 31:1417-1428. [DOI] [PubMed] [Google Scholar]
- 28.Toth, V., and L. Emody. 2000. Proteus virulence: involvement of the pore forming alpha-hemolysin (a short review). Acta Microbiol. Immunol. Hung. 47:457-470. [PubMed] [Google Scholar]
- 29.Warren, J. W., J. H. Tenney, H. L. Hoopes, H. L. Muncie, and W. C. Anthony. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 146: 719-723. [DOI] [PubMed] [Google Scholar]
- 30.Wassif, C., D. Cheek, and R. Belas. 1995. Molecular analysis of a metalloprotease from Proteus mirabilis. J. Bacteriol. 177:5790-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weatherburn, M. W. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39:971-974. [Google Scholar]