Abstract
eIF3 binds to 40S ribosomal subunits and stimulates recruitment of Met-tRNAiMet and mRNA to the pre-initiation complex. Saccharomyces cerevisiae contains an ortholog of human eIF3 subunit p35, HCR1, whose interactions with yeast eIF3 are not well defined. We found that HCR1 has a dual function in translation initiation: it binds to, and stabilizes, the eIF3–eIF5– eIF1–eIF2 multifactor complex and is required for the normal level of 40S ribosomes. The RNA recognition motif (RRM) of eIF3 subunit PRT1 interacted simultaneously with HCR1 and with an internal domain of eIF3 subunit TIF32 that has sequence and functional similarity to HCR1. PRT1, HCR1 and TIF32 were also functionally linked by genetic suppressor analysis. We propose that HCR1 stabilizes or modulates interaction between TIF32 and the PRT1 RRM. Removal of the PRT1 RRM resulted in dissociation of TIF32, NIP1, HCR1 and eIF5 from eIF3 in vivo, and destroyed 40S ribosome binding by the residual PRT1–TIF34–TIF35 subcomplex. Hence, the PRT1 RRM is crucial for the integrity and ribosome-binding activity of eIF3.
Keywords: eIF3/HCR1/PRT1 RRM/40S ribosome/translation initiation
Introduction
Translation initiation in eukaryotes is a complex series of reactions leading to the formation of an 80S ribosomal complex containing methionyl initiator tRNA (Met-tRNAiMet) base paired with the initiation codon in mRNA. These reactions require the participation of numerous protein factors called initiation factors (eIFs), several of which operate at multiple steps in the process. eIF3 and eIF1A stimulate dissociation of 80S ribosomes into 40S and 60S subunits, and also promote binding of a ternary complex consisting of Met-tRNAiMet, eIF2 and GTP to the small subunit, forming the 43S pre-initiation complex (reviewed in Merrick and Hershey, 1996). eIF3 remains bound to the 43S complex and prevents its disruption by 60S ribosomal subunits (Chaudhuri et al., 1999). The 43S complex interacts with the m7G cap structure at the 5′ end of mRNA, forming the 48S complex, in a reaction stimulated by multiple mRNA-binding factors [eIF4F, eIF4A, eIF4B and poly(A)-binding protein (PAB1)] and eIF3 (Merrick and Hershey, 1996; Sachs and Varani, 2000). The 48S complex scans the mRNA until an AUG start codon is encountered, facilitated in this process by eIF1 and eIF1A (Pestova et al., 1998). Upon base-pairing between the anticodon of Met-tRNAiMet and the AUG start codon, eIF5 stimulates the hydrolysis of GTP bound to eIF2, followed by release of the eIF2–GDP binary complex and other initiation factors. The 60S ribosome then joins with the 48S pre-initiation complex, dependent on eIF5B, and the entire process is completed when eIF5B is released from the 80S initiation complex (Pestova et al., 2000).
Mammalian eIF3 has the most complex structure of the initiation factors, containing 11 non-identical subunits (Vornlocher et al., 1999). An eIF3 complex has been isolated from the yeast Saccharomyces cerevisiae by affinity purification and gel filtration that contains only five subunits, TIF32/RPG1, NIP1, PRT1, TIF34 and TIF35 (Danaie et al., 1995; Asano et al., 1998; Phan et al., 1998), which are homologs of human eIF3 subunits p170, p110, p116, p36 and p44, respectively (Asano et al., 1997). A mutation in PRT1 or depletion of NIP1 both led to defects in Met-tRNAiMet binding to 40S ribosomes in yeast cell extracts that could be complemented with the purified yeast eIF3 complex. Thus, the yeast five-subunit factor can associate with 40S subunits and stimulate Met-tRNAiMet binding, two of the key activities ascribed to mammalian eIF3 (Phan et al., 1998).
We demonstrated recently that yeast eIF3, eIF1, eIF5 and the ternary complex reside in a multifactor complex (MFC) that is an important intermediate in translation initiation (Asano et al., 2000). eIF1 and eIF5 are physically associated with eIF3 via the NIP1 subunit (homolog of mammalian eIF3-p110) (Naranda et al., 1996; Asano et al., 1998; Phan et al., 1998), whereas the Met-tRNAiMet⋅eIF2⋅GTP–eIF3 interaction is bridged by eIF5 (Asano et al., 1999, 2000). Some of these interactions have been confirmed for the corresponding mammalian factors (Bandyopadhyay and Maitra, 1999; Fletcher et al., 1999). In yeast, eIF5 and eIF1 were shown to promote stringent selection of AUG as the start codon (Huang et al., 1997); hence, eIF3 may play a structural role in AUG recognition and GTP hydrolysis in addition to promoting initiation complex assembly.
Only one additional human eIF3 subunit (p35) has an ortholog encoded in the S.cerevisiae genome. The gene encoding this protein, HCR1, was identified as a high-copy suppressor of the temperature-sensitive (Ts–) phenotype of the rpg1-1 allele of TIF32/RPG1 (Valášek et al., 1998, 1999). The hcr1Δ mutant was viable, but showed a reduction in growth rate. Combining the hcr1Δ allele with rpg1-1 exacerbated the growth defect conferred by rpg1-1. Moreover, the HCR1 and TIF32 gene products were co-immunoprecipitated from yeast cell extracts. Despite the fact that purified yeast eIF3 preparations did not contain HCR1 (Phan et al., 1998), these findings suggested that HCR1 is physically associated with yeast eIF3 in vivo.
We show here that a portion of HCR1 resides in the MFC containing eIF3, eIF5, eIF1 and the ternary complex. Importantly, the hcr1Δ mutation decreased the abundance of the MFC, implicating HCR1 in pre-initiation complex assembly. We also found that recombinant HCR1 and the internal segment of TIF32, which is 25% identical to HCR1, bound concurrently to the RNA recognition motif (RRM) of PRT1, indicating a network of physical interactions involving HCR1, TIF32 and the PRT1 RRM. A mutant PRT1 protein lacking the RRM was shown previously to interfere with translation in otherwise wild-type cells and to reside in defective complexes incapable of stable binding to 40S ribosomes (Evans et al., 1995). In agreement with our in vitro binding data, we found that these defective complexes contained TIF34 and TIF35 but lacked TIF32, HCR1 and NIP1. We propose that HCR1 serves to stabilize or modulate the interaction between TIF32 and the PRT1 RRM, which is crucial for integrity of the MFC and its association with 40S ribosomes. Unexpectedly, deletion of HCR1 reduced the abundance of 40S ribosomes, indicating a novel function for this eIF3-associated factor in ribosome biogenesis or stability.
Results
HCR1 is an eIF3-associated protein that interacts directly with PRT1 and TIF32/RPG1
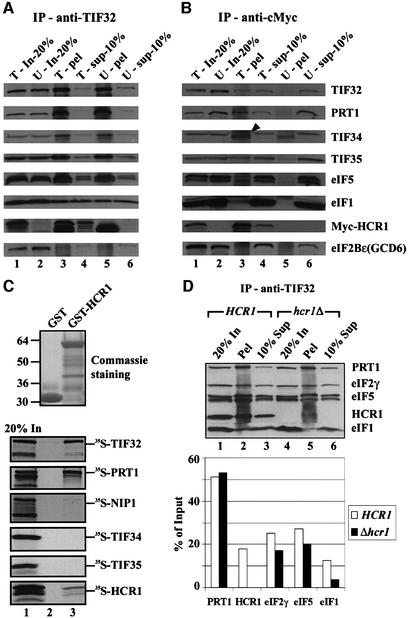
To determine whether the previously described HCR1–TIF32 complex (Valášek et al., 1999) also contained the other subunits of eIF3, we prepared whole-cell extracts (WCEs) from the hcr1Δ strain transformed with a plasmid encoding c-Myc-tagged HCR1 (Myc-HCR1) or an empty vector. These extracts were immunoprecipitated with antibodies against TIF32 or c-Myc and the immune complexes were probed with antibodies against eIF3 subunits, eIF1 and eIF5. As expected, the anti-TIF32 antibodies immunoprecipitated a large fraction (∼60%) of TIF32, PRT1, TIF34 and TIF35 (all eIF3 subunits), ∼20% of the eIF5 and ∼10% of the eIF1 from both extracts. Additionally, ∼20% of the Myc-HCR1 co-immunoprecipitated with TIF32 from the HCR1-Myc extract. The ε subunit of eIF2B (GCD6), which does not interact with eIF3 (Cigan et al., 1991), was not immunoprecipitated by the TIF32 antibodies from either extract (Figure 1A, lanes 3 and 5). The TIF32–HCR1 complexes detected above were judged to be part of eIF3 because the c-Myc antibodies immunoprecipitated ∼10–20% of the other eIF3 subunits, 1–3% of eIF1 and eIF5, and no GCD6, along with ∼60% of the Myc-HCR1 (Figure 1B, lane 3). None of these proteins was immunoprecipitated with c-Myc antibodies from the wild-type untagged extract (Figure 1B, lane 5). These results demonstrate that HCR1 is physically associated with the eIF3–eIF1–eIF5 complex.
Fig. 1. HCR1 interacts with the eIF3–eIF2–eIF1–eIF5 multifactor complex in vivo and binds in vitro to the PRT1 and TIF32/RPG1 subunits of eIF3. (A and B) Co-immunoprecipitation of HCR1 with the eIF3 complex from cell extracts. The hcr1Δ strain YLVH13 was transformed with single-copy plasmid YCpLVHM-T bearing c-myc-tagged HCR1 (T, for tagged) or with empty vector YCplac22 (U, for untagged). Aliquots containing 200 µg of WCE from each transformant were immunoprecipitated with mouse monoclonal anti-TIF32 antibodies (A) or with hybridoma supernatant anti-c-Myc antibodies (B). Immune complexes were analyzed by western blotting using antibodies against the proteins indicated to the right of (B). In-20%, 20% of the input WCE used for immunoprecipitation (lanes 1 and 2); pel, the entire immunoprecipitated fraction (lanes 3 and 5); sup-10%, 10% of the supernatant fractions from the immunoprecipitations (lanes 4 and 6). The arrowhead indicates the position of TIF34; the band beneath it was precipitated non-specifically. (C) Binding of TIF32 and PRT1 to GST–HCR1 in vitro. Full-length HCR1 fused to GST (lane 3) or GST alone (lane 2) was expressed in E.coli, immobilized on glutathione–Sepharose beads and incubated with 10 µl of 35S-labeled full-length TIF32, PRT1, NIP1, TIF34, TIF35 and HCR1 at 4°C for 2 h. The beads were washed three times with 1 ml of PBS and bound proteins separated by SDS–PAGE. Gels were first stained with Gelcode Blue Stain Reagent (Pierce) (top panel) followed by autoradiography (bottom panels). Lane 1 shows 20% of the input amounts of in vitro translated proteins added to each reaction (20% In). (D) Deletion of HCR1 reduces the amounts of eIFs 2, 5 and 1 associated with eIF3. Upper panel: the MFC was co-immunoprecipitated from either W303 (HCR1) or YLVH13 (hcr1Δ) strains essentially as in (A). Bottom panel: quantification of various factors was carried out using the NIH Image program (version beta 3b, NIH, USA). White bars show the percentage of each factor out of the total precipitated from the W303 wild-type strain; black bars correspond to the percentage of the total precipitated from the hcr1Δ strain.
The relatively small proportions of eIF3 subunits, eIF5 and eIF1 that co-immunoprecipitated with Myc-HCR1 compared with the larger amounts of these proteins that co-immunoprecipitated with TIF32 suggested that HCR1 might be a substoichiometric component of eIF3. To investigate this possibility, we compared the amounts of the c-Myc-tagged forms of these two proteins expressed in the wild-type strain W303 by western blot analysis of WCEs using antibodies against the c-Myc epitope. The results suggested that Myc-HCR1 was present at a quarter of the level of Myc-PRT1 (data not shown). Using polyclonal antibodies against HCR1 and PRT1, we found that ratios of Myc-HCR1 to total HCR1, and of Myc-PRT1 to total PRT1, were very similar in these extracts. Assuming that the specific binding activity of the c-Myc antibody was identical for the two tagged proteins, these results suggest that no more than a quarter of the total eIF3 complexes contain HCR1. We also noted that a smaller proportion of the total HCR1 was immunoprecipitated with TIF32 antibodies than was observed for other eIF3 subunits. Thus, in addition to its relatively low abundance, it appears that HCR1 is less tightly associated with eIF3 than are the five core subunits.
We next asked whether HCR1 can interact directly with an individual subunit of eIF3. A glutathione S-transferase (GST)–HCR1 fusion protein was incubated with various 35S-labeled eIF3 subunits synthesized in reticulocyte lysate. Large fractions of the radiolabeled PRT1 (∼40%) and TIF32 (∼20%) were bound to the immobilized GST–HCR1 (Figure 1C). Much lower amounts of [35S]HCR1 (∼1–2%) and no detectable radiolabeled NIP1, TIF34 or TIF35 bound to GST–HCR1. These data suggest that HCR1 binds directly to eIF3 subunits TIF32 and PRT1. It was reported previously that p170 is the binding partner in human eIF3 for p35, the homolog of HCR1 (Block et al., 1998). Thus, the association of HCR1/p35 with the largest subunit of eIF3 is evolutionarily conserved between yeast and humans.
HCR1 enhances formation of the MFC, the abundance of 40S ribosomes and the rate of translation initiation in vivo
It was of interest to determine whether deleting HCR1 would destabilize the MFC (Asano et al., 2000). In the first approach to addressing this issue, we immunoprecipitated the MFC from HCR1 and hcr1Δ extracts using TIF32 antibodies and quantified the amounts of the individual components (Figure 1D, bottom panel). We found that deletion of HCR1 reduced the amounts of eIF2 and eIF5 (by ∼30%) and the amount of eIF1 (by ∼75%) that co-immunoprecipitated with eIF3 (Figure 1D, lanes 2 and 4). Considering that HCR1 is less abundant than eIF3, this represents a significant effect on the formation or stability of the MFC.
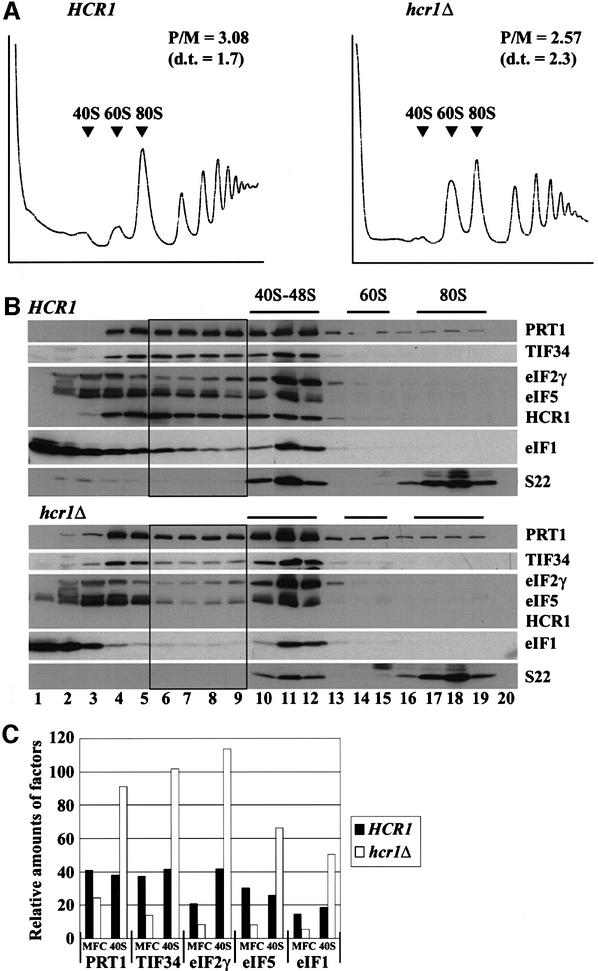
In the second approach, we separated the MFC from its individual constituents by resolving WCEs from HCR1 and hcr1Δ strains on a sucrose gradient by velocity sedimentation. At the same time, we examined the polysome profiles in these extracts to determine whether the slow growth phenotype of the hcr1Δ strain resulted from diminished translation initiation. As shown in Figure 2A, deletion of HCR1 led to only a slight reduction in the polysome/monosome ratio (2.57 in the hcr1Δ mutant versus 3.08 in the wild type). However, there was an obvious decrease in the proportion of large polysomes in the mutant, indicating a reduced rate of translation initiation. Deletion of HCR1 also led to a striking reduction in the abundance of free 40S subunits and a corresponding increase in free 60S subunits. The 60S/40S ratio for the hcr1Δ strain was 4.32 compared with 1.19 for the wild type. This last defect suggests that HCR1 is required for the biogenesis or stability of 40S ribosomes (Kressler et al., 1999) in addition to its function in eIF3.
Fig. 2. Deletion of HCR1 reduces the levels of free 40S ribosomes and destabilizes the multifactor complex. (A) Analysis of polysome profiles in isogenic HCR1 and hcr1Δ strains. Yeast strains W303 and YLVH13 were grown in YPD to an OD600 of ∼1.0, and 50 µg/ml cycloheximide was added 5 min prior to harvesting. WCEs were prepared and resolved by velocity sedimentation on 5–45% sucrose gradients for 2.5 h at 39 000 r.p.m. Fractions were collected while scanning at A254. The positions of different ribosomal species are indicated. P/M, ratio of A254 in the combined polysome fractions to that in the 80S peak; d.t., cell doubling time in hours. (B) Western blot analysis of the sucrose gradient fractions. W303 and YLVH13 strains were grown as in (A), and WCEs were separated on 7.5–30% sucrose gradients by centrifuging for 5 h at 41 000 r.p.m. Gradient fractions were collected and resolved by SDS–PAGE, followed by immunoblot analysis using antibodies against the proteins indicated on the right of each panel. The presence of 40S and 60S ribosomes was revealed by the A254 profile (not shown) and by probing for the 40S subunit protein S22. Black rectangles highlight the gradient fractions in which the distributions of eIFs 3, 2, 5 and 1 (MFC) differ substantially between the gradients in hcr1Δ versus HCR1. (C) The results in (B) were quantified using the NIH Image program for the five proteins indicated on the x-axis. The proportions of the total signals present in fractions 6–9 (MFC) or 10–12 (40S) were plotted after normalizing the values obtained for fractions 10–12 for the amounts of 40S ribosomes. Black and white bars show the values obtained for the HCR1 wild-type and hcr1Δ strains, respectively.
Western blot analysis of the gradient fractions (Figure 2B) and quantification of these data (Figure 2C) revealed that eIFs 1, 2, 3 and 5 were depleted specifically in fractions 6–9 containing the MFC (Asano et al., 2000) in the hcr1Δ mutant versus the wild type. There was a corresponding increase in the proportions of eIFs in fractions 10–12 containing 43–48S initiation complexes in the hcr1Δ mutant (Figure 2B and C). Probing for the 40S subunit ribosomal protein S22 revealed the anticipated ∼2-fold reduction in the amount of free 40S subunits in the mutant (Figure 2B). Hence, in Figure 2C, the calculated proportions of each factor in the 40–48S region were normalized for the amounts of 40S ribosomes present in the relevant fractions. These findings suggest that HCR1 promotes the formation or stability of the MFC, and that inactivation of HCR1 impedes the conversion of 43S to 48S, or 48S to 80S, initiation complexes. The results in Figure 2B (top panel) also provide additional evidence that HCR1 is a component of the MFC and is recruited to 43–48S initiation complexes along with the other constituents of the MFC.
Considering that the hcr1Δ mutation led to reduced amounts of 40S subunits, we asked whether the decreased amount of the MFC or the reduced rate of converting 43S/48S to 80S complexes seen in the mutant could be a secondary consequence of diminished 40S subunits. Inconsistent with this possibility, we found that deletion of the 40S ribosomal protein gene RP51A did not alter the distribution of eIFs between the MFC and 43–48S initiation complexes (data not shown). Importantly, rp51aΔ leads to an even greater reduction in the pool of 40S subunits and growth rate than does hcr1Δ (Foiani et al., 1991).
High-copy HCR1 is an allele-specific suppressor of prt1 mutations
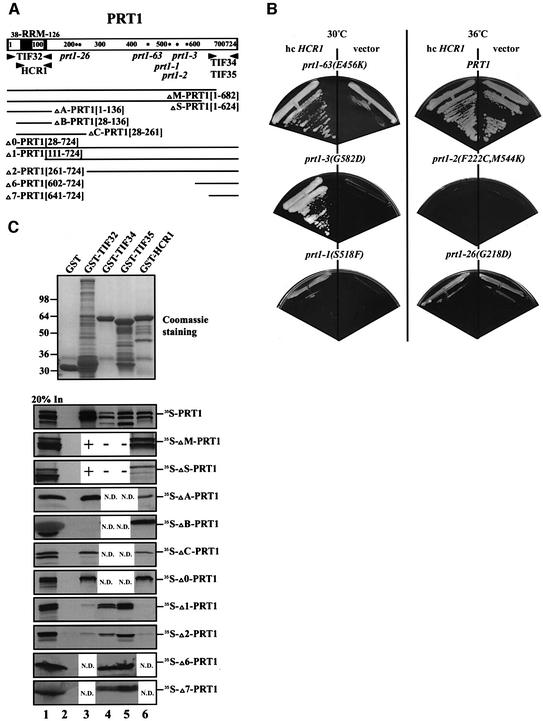
To obtain evidence that HCR1 interacts with PRT1 in vivo, we asked whether high-copy HCR1 could suppress the Ts– phenotypes of various prt1 mutations. As shown in Figure 3B, high-copy HCR1 strongly enhanced growth at 36°C in the prt1-63 and prt1-3 mutants, and weakly stimulated growth of the prt1-1 strain. The Ts– phenotypes of the prt1-2 and prt1-26 mutants were not suppressed by high-copy HCR1, even though the latter mutant has a less severe phenotype than does the prt1-63 mutant, which was strongly suppressed by high-copy HCR1 (Figure 3B). The allele-specific suppression of different prt1 mutations by high-copy HCR1 is consistent with a direct interaction between PRT1 and HCR1 in vivo.
Fig. 3. The RRM of PRT1 is sufficient and necessary for its interaction with TIF32/RPG1 and HCR1. (A) Schematic of the PRT1 sequence and location of the RRM, with arrowheads delimiting the boundaries of the minimal segments required for interactions with the indicated proteins, based on the results shown in (C). Asterisks indicate the positions of the single amino acid substitutions made by the prt1 mutations indicated beneath the asterisks. The two dots in the sequence indicate the two substitutions made by the prt1-2 mutation. The lines at the bottom depict the 35S-labeled segments of PRT1 used for the binding assays shown in (C), designated by clone name and the PRT1 amino acids present (in brackets). (B) Allele-specific suppression of the Ts– phenotypes of prt1 mutants by overexpression of HCR1. Mutant strains containing prt1-63 (TC-26-3), prt1-3 (TP13-1-2), prt1-1 (TP11-4-1), PRT1 (21R), prt1-2 (TP12-7-0) and prt1-26 (TDE/16A) were transformed with a high-copy-number plasmid (hc) carrying HCR1 (YEpLVHCR1) or with empty vector YEplac181 and examined for growth at 36°C on YPD medium. For each strain, designated above by its prt1 allele, growth of the hc HCR1 and vector transformants is shown in the left- and right-hand sectors, respectively. (C) In vitro binding of different GST fusion proteins (labeled across the top of the upper panel) to the various 35S-labeled segments of PRT1 labeled to the right of the lower panels. Binding experiments were conducted as described in Figure 1C. Lanes 2, 3, 4, 5 and 6 show binding to GST alone, GST–TIF32, GST–TIF34, GST–TIF35 and GST–HCR1, respectively. Lane 1 shows 20% of the input amounts of in vitro translated PRT1 proteins added to each reaction (20% In). + or – in lanes 3–5 for the ΔM-PRT1 and ΔS-PRT1 constructs indicates results previously published (Asano et al., 1998). N.D., not determined.
The RRM of PRT1 is necessary and sufficient for interaction with TIF32 and HCR1 in vitro
We showed previously that PRT1 could interact with all core eIF3 subunits except NIP1 (Asano et al., 1998). Having found that HCR1 also interacts with PRT1, we set out to locate precisely the binding domains in PRT1 for HCR1 and other eIF3 subunits. A battery of N- or C-terminal truncations of [35S]PRT1 (depicted in Figure 3A) was tested for in vitro binding to GST fusions containing full-length HCR1 or other eIF3 subunits. Only the first 136 residues of PRT1 (in fragment ΔA-PRT1), encompassing the RRM, were sufficient for binding to GST–TIF32 and GST–HCR1 (Figure 3C, lanes 3 and 6). Consistently, deletion of 42 or 100 residues from the C-terminus of PRT1 (ΔM-PRT1 and ΔS-PRT1) did not reduce its interaction with GST–HCR1. It was shown previously that these deletions disrupted interactions of PRT1 with GST–TIF34 and GST–TIF35, but not with GST–TIF32 (Asano et al., 1998). Removal of the first 27 residues from the PRT1 segment (yielding ΔB-PRT1) abolished its interaction with GST–TIF32 without affecting its binding to GST–HCR1. Binding to GST–TIF32 in the absence of the first 27 residues of PRT1 was restored, however, by including an additional 125 residues C-terminal to the RRM (yielding ΔC-PRT1). Deletion of the N-terminal 110 residues including most of the RRM (yielding fragment Δ1-PRT1), or residues 1–260 (Δ2-PRT1), abolished binding of PRT1 to GST–HCR1 and GST–TIF32 (Figure 3C). Thus, the RRM in PRT1 is necessary and sufficient for binding HCR1, whereas TIF32 binding requires additional PRT1 residues immediately flanking the RRM.
Previously, we showed that the C-terminal 83 residues of PRT1 were sufficient for interaction with TIF34 and TIF35 in yeast two-hybrid assays, and we confirmed this finding for TIF34 by showing that [35S]TIF34 synthesized in vitro bound to the GST–PRT1[641–724] fusion containing the C-terminal 83 residues of PRT1 (Asano et al., 1998). Additionally, deletion of residues 625–724 or 683–724 from [35S]PRT1 (ΔM-PRT1 and ΔS-PRT1) abolished its interaction with GST–TIF34 and GST–TIF35 (Asano et al., 1998). In the same study, however, [35S]TIF35 failed to interact with GST– PRT1[641–724], suggesting that the binding domain for TIF35 extends further towards the N-terminus of PRT1 than that for TIF34. However, we found here that GST–TIF35 bound strongly to [35S]PRT1 segments containing only the C-terminal 123 or 83 residues of the protein (Figure 3C, Δ6-PRT1 and Δ7-PRT1). Presumably, fusion of the C-terminal fragment of PRT1[641–724] to GST masked its TIF35-binding site without affecting that of TIF34. We conclude that HCR1 and TIF32 bind to the N-terminal segment of PRT1 harboring the RRM, whereas TIF34 and TIF35 interact with the extreme C-terminus of PRT1 (Figure 3A).
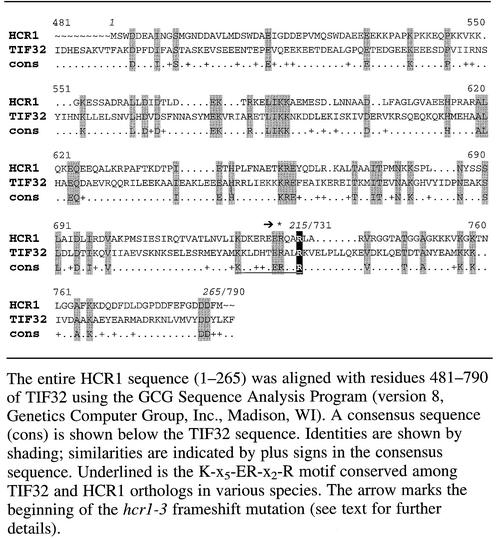
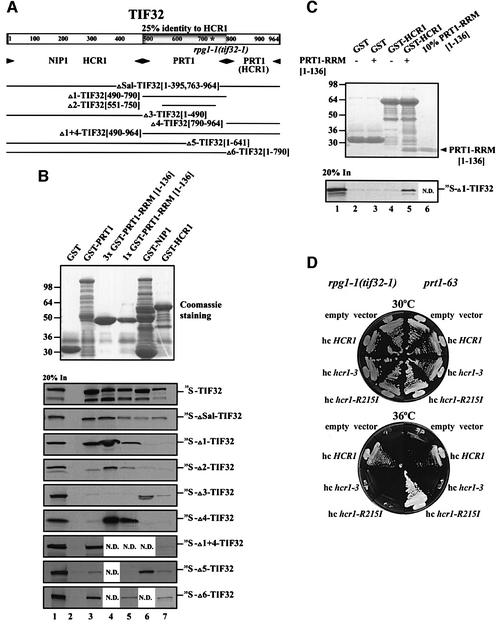
An internal segment of TIF32 similar in sequence to HCR1 is sufficient for interaction with the RRM of PRT1
Prompted by our finding that HCR1 and TIF32 both interacted with the RRM in PRT1, we aligned the HCR1 and TIF32 sequences and discovered that residues 490–790 in TIF32 are 25% identical to the entire sequence of HCR1 (Table I). Accordingly, we tested whether the HCR1-like domain (HLD) in TIF32 binds to the PRT1 RRM. A 35S-labeled fragment comprising the HLD of TIF32 (Δ1-TIF32, Figure 4A) was sufficient for strong binding to GST fusions containing either full-length PRT1 or the N-terminal 136 residues of PRT1 (GST–PRT1 RRM[1–136]) (Figure 4B). Simultaneous removal of 61 and 40 residues from the N- and C-terminus, respectively, of this internal TIF32 fragment (yielding Δ2-TIF32) reduced its binding to GST–PRT1 RRM[1–136]. TIF32 fragments Δ1 and Δ2 showed no binding to GST–NIP1 or GST–HCR1 (Figure 4B), indicating the specificity of their interaction with the PRT1 RRM.
Table I. Sequence comparison between full-length HCR1 and the middle region of TIF32/RPG1.
Fig. 4. Physical and functional interactions between homologous domains in TIF32/RPG1 and HCR1 and the PRT1 RRM. (A) Schematic of the TIF32 amino acid sequence, with the domain showing 25% identity to HCR1 (HLD) shaded and the asterisk indicating the single amino acid substitution made by rpg1-1. Arrowheads beneath the schematic delimit boundaries of the minimal segments required for interactions with the indicated proteins, based on the results shown in (B). The lines beneath the arrowheads depict the 35S-labeled segments of TIF32 used for the binding assays shown in (B), designated by clone name and the TIF32 amino acids present (in brackets). (B) Homologous domains in HCR1 and TIF32 bind to the PRT1 RRM in vitro. Binding of different GST fusion proteins (labeled across the top of the upper panel) to the various 35S-labeled segments of TIF32 labeled to the right of the lower panels. Binding experiments were conducted as described in Figure 1C. Lanes 2, 3, 4, 5, 6 and 7 show binding to GST alone, GST–PRT1, 3× GST–PRT1 RRM[1–136], 1× GST–PRT1 RRM[1–136], GST–NIP1 and GST–HCR1, respectively. Lane 3 contains three times (3×) the arbitrary amount of GST–PRT1 RRM[1–136] loaded in lane 4 (1×). Lane 1 shows 20% of the input amounts of in vitro translated proteins added to each reaction (20% In). (C) The PRT1 RRM can bridge interaction between HCR1 and TIF32 in vitro. A 10 µg aliquot of GST alone or GST–HCR1 was mixed with 10 µl of [35S]Δ1-TIF32 (lanes 2–4) in the presence (10 µg) (lanes 3 and 5) or absence (lanes 2 and 4) of PRT1 RRM[1–136] at 4°C for 2 h, essentially as described in Figure 1C. The PRT1 RRM[1–136] fragment was cleaved from the GST moiety in GST–PRT1 RRM[1–136] using factor Xa (Pharmacia) as recommended by the vendor. Lane 1 shows 20% of the input amount of in vitro translated [35S]Δ1-TIF32 added to each reaction (20% In). Lane 6 shows 10% (∼1 µg) of the PRT1 RRM[1–136] fragment (indicated by an arrowhead) purified away from the GST moiety and used in this experiment. (D) Allele-specific suppression of the Ts– phenotypes of rpg1-1 and prt1-63 strains by different HCR1 alleles in high copy number. Transformants of rpg1-1(tif32-1) strain YLV314U (left-hand sectors) and prt1-63 strain TC-26-3 (right-hand sectors) containing high-copy-number plasmids (hc) carrying empty vector (YEplac181), HCR1 (YEpLVHCR1), hcr1-R215I (YEpLVhcr1-R215I) or hcr1-3 (YEpLVhcr1-3) were examined for growth at 30°C (upper block) and 36°C (lower block) on YPD medium.
An internal deletion in TIF32 removing most of the HLD (ΔSal-TIF32) significantly reduced, but did not abolish, binding to the GST–PRT1 fusions. Thus, the HLD of TIF32 is sufficient, but not absolutely required, for TIF32 binding to the RRM domain in PRT1. Moreover, the C-terminal domain of TIF32 (Δ4-TIF32) bound to GST–PRT1 RRM[1–136] but not to GST–NIP1 or GST–HCR1, identifying a second region in TIF32 sufficient for binding to the PRT1 RRM. Deletion of both the C-terminal domain and the HLD (Δ3-TIF32 and Δ5-TIF32) abolished interaction of TIF32 with GST–PRT1 and GST–PRT1 RRM (Figure 4B). These findings suggest that TIF32 can bind to the RRM domain of PRT1 through its HLD or C-terminus. The fact that full-length GST–PRT1 did not bind to the C-terminal segment of TIF32, whereas GST–PRT1 RRM did (Figure 4B), may indicate that intramolecular interactions in PRT1 compete for interaction between the PRT1 RRM and C-terminal domain of TIF32.
The TIF32 fragments containing the N-terminal half of the protein (ΔSal-, Δ3- and Δ5-TIF32) bound to GST–NIP1 and GST–HCR1, whereas the C-terminal fragments (Δ1-, Δ2-, Δ4- and Δ1+4-TIF32) did not (Figure 4B). These data suggest that NIP1 and HCR1 interact primarily with the N-terminal half of TIF32 (Figure 4A). However, considering that none of the N-terminal fragments of TIF32 bound as strongly as full-length TIF32 to GST–HCR1, the C-terminus of TIF32 may make additional contacts with HCR1, or it may stabilize the conformation of the N-terminus of TIF32 needed for strong interaction.
We showed above that PRT1 RRM[1–136] binds both to the HLD in TIF32 and to HCR1 itself, and wished to determine whether these interactions could occur simultaneously. Having found that the HLD of TIF32 (in [35S]Δ1-TIF32) did not bind to GST–HCR1 (Figure 4B), we asked whether the PRT1 RRM could bridge an interaction between these two polypeptides. The PRT1 RRM[1–136] fragment used in these experiments was produced by factor Xa digestion of the GST–PRT1 RRM[1–136] fusion and purified from the GST moiety. As shown in Figure 4C (lane 5), the purified PRT1 RRM fragment bound to GST–HCR1 and bridged an interaction between GST–HCR1 and [35S]Δ1-TIF32. As expected, the PRT1 RRM fragment did not mediate binding of [35S]Δ1-TIF32 to GST alone (lane 3). Thus, the PRT1 RRM can interact simultaneously with HCR1 and the HLD of TIF32.
Genetic evidence for the functional significance of sequence similarity between TIF32 and HCR1
The Ts– phenotype of the rpg1-1 allele of TIF32 is conferred by a single amino acid substitution of arginine to isoleucine at position 731 (Valášek et al., 1998) in the HLD of TIF32. This arginine residue is conserved in HCR1 at position 215 (Table I) and corresponds to the C-terminal residue in a K-x5-ER-x2-R motif that is completely conserved among S.cerevisiae HCR1 and all known TIF32 orthologs (data not shown). A similar motif (K-x3-EK-x2-K) occurs in human eIF3-p35. To explore the physiological significance of the sequence similarity between HCR1 and TIF32, we asked whether substitution of Arg215 in HCR1 with isoleucine (hcr1-R215I) would alter the ability of HCR1 to function in vivo as a dosage suppressor of rpg1-1 and prt1 mutations. In parallel, we substituted the second conserved arginine present in the K-x5-ER-x2-R motif located three residues upstream of Arg215 (producing hcr1-R212I), and separately introduced a frameshift mutation that replaced the last 57 residues of HCR1 (including the K-x5-ER-x2-R motif) with 42 novel amino acids encoded in a different reading frame (hcr1-3).
The hcr1-3 frameshift mutation destroyed the ability of high-copy HCR1 to suppress rpg1-1, prt1-63, prt1-3 and prt1-1, whereas the R212I substitution had no effect on suppression of these mutations by high-copy HCR1. Interestingly, the R215I substitution eliminated suppression of rpg1-1, enhanced suppression of prt1-63, and had no effect on suppression of prt1-3 and prt1-1 by high-copy HCR1 (Figure 4D and data not shown). Furthermore, HCR1-R215I on a low-copy-number plasmid fully complemented the slow growth phenotype of hcr1Δ strain YLVH13, whereas low-copy hcr1-3 did not (data not shown). This last finding indicates that HCR1-R215I is not a null allele. We conclude that the Ile215 substitution alters the function of HCR1 in a manner that influences its ability to compensate for defects in PRT1 and TIF32 when overexpressed. The fact that mutating the equivalent amino acids in HCR1 and the HLD of TIF32 altered the functions of both proteins in vivo supports the physiological relevance of sequence similarity between HCR1 and TIF32. The different phenotypes observed when high-copy HCR1-R215I was combined with rpg1-1 or various prt1 alleles provides additional evidence for functional interactions between HCR1 and the TIF32 and PRT1 subunits of eIF3.
Interaction of TIF32 with the PRT1 RRM is required for eIF3 integrity and binding of eIF3 to the 40S ribosome
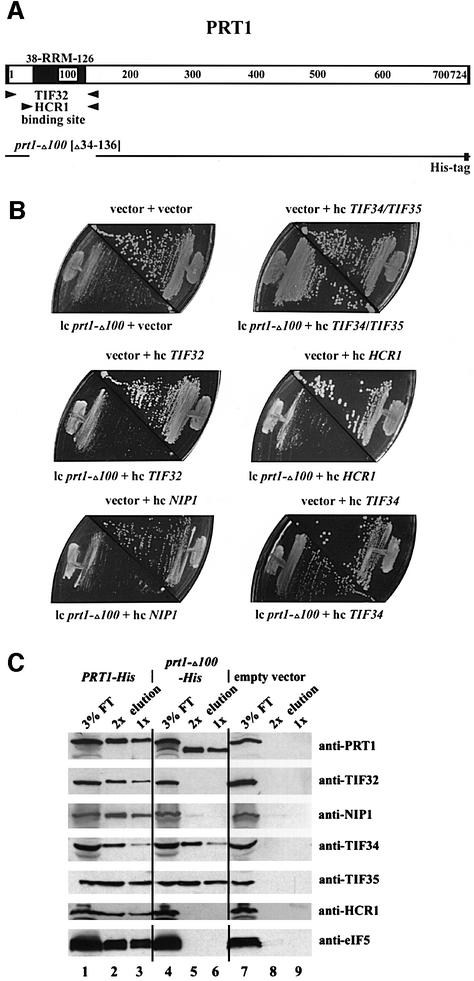
It was shown previously that deletion of the N-terminal 100 amino acids of PRT1 was lethal in vivo and that expressing the mutant protein (encoded by prt1-Δ100) in otherwise wild-type cells inhibited translation initiation through formation of PRT1-containing complexes incapable of binding to 40S ribosomes (Evans et al., 1995). Having found that the PRT1 RRM is critical for its interaction with TIF32 and HCR1 in vitro (Figure 3B), we predicted that expression of prt1-Δ100 from a low-copy plasmid (low-copy prt1-Δ100) in PRT1 cells would lead to the formation of incomplete eIF3 complexes containing TIF34 and TIF35 bound to the C-terminal domain of prt1-Δ100 but lacking TIF32, HCR1 and NIP1. As a consequence, the pool of TIF34 and TIF35 available for association with wild-type PRT1 would be decreased, leading to reduced amounts of intact eIF3.
We reasoned that if this interpretation is correct, it should be possible to restore high levels of intact eIF3 in cells containing prt1-Δ100 by overexpressing TIF34 and TIF35. Indeed, TIF34 and TIF35 present together on a high-copy plasmid fully suppressed the dominant Slg– phenotype of low-copy prt1-Δ100 in a PRT1 strain, whereas high-copy-number TIF32, NIP1 or HCR1 alone did not (Figure 5B). As high-copy TIF34 alone partially suppressed the Slg– of prt1-Δ100, whereas high-copy TIF35 did not (Figure 5B and not shown), it seems that TIF34 is the limiting factor in PRT1 cells containing the prt1-Δ100 product.
Fig. 5. The RRM of PRT1 is required for its interaction with TIF32/RPG1, HCR1 and NIP1 in vivo. (A) Schematic of the PRT1 sequence showing the locations of the RRM and minimal TIF32- and HCR1-binding sites, as in Figure 3. The line beneath the schematic depicts the amino acids present in the polyhistidine-tagged prt1-Δ100 product (prt1-Δ100-His), which lacks residues 34–136 and contains a His tag at the C-terminus. (B) The dominant Slg– phenotype of prt1-Δ100 can be suppressed by simultaneous overexpression of TIF34 and TIF35. Transformants of strain GDE303-88 containing prt1-Δ100 on low-copy-number (lc) plasmid pprt1-Δ100, or the corresponding empty vector YCplac11, were transformed additionally with one of the following high-copy-number (hc) plasmids bearing the indicated genes or with the corresponding empty vector YEp112, YEpTIF34/35T (hc TIF34/TIF35), YEpTIF32T (hc TIF32), YEpLVHCR1-1 (hc HCR1), YEpNIP1T (hc NIP1) and YEpTIF34T (hc TIF34). The resulting transformants were tested for growth at 30°C on SD medium supplemented with adenine. The plasmid combination present in each transformant is indicated above or below the appropriate plate sector. (C) Affinity purification of an eIF3 subcomplex containing prt1-Δ100-His, TIF34 and TIF35 but lacking TIF32 and NIP1. WCE was prepared in a low salt buffer (100 mM KCl) from transformants of yeast strain F353 bearing plasmid pLP101 containing PRT1-His encoding His-tagged PRT1 (lanes 1–3), pprt1-Δ100-His encoding prt1-Δ100-His (lanes 4–6) or empty vector YCplac11 (lanes 5–7). Extracts were incubated overnight with Ni2+-NTA–silica resin and the bound proteins were eluted and subjected to SDS–PAGE. Proteins were transferred to nitrocellulose membranes and probed with the antibodies indicated on the right. Lanes 1, 4 and 7 each contained 3% of the flow-through fractions from Ni2+-NTA–silica binding (3% FT); lanes 2, 3, 5, 6, 8 and 9 contained 5 µg (2×) or 2.5 µg (1×) of the corresponding eluted fractions, as indicated above the lanes.
To demonstrate directly that the prt1-Δ100 product is defective for interaction with TIF32 but retains the ability to bind TIF34 and TIF35, we inserted polyhistidine tags at the C-termini of prt1-Δ100 and wild-type PRT1, and introduced the tagged alleles on high-copy plasmids into a wild-type strain. The tagged proteins were purified on nickel–agarose resin and analyzed by western blotting. As expected, His-tagged wild-type PRT1 (PRT1-His) co-purified with all four eIF3 subunits, HCR1 and eIF5 (Figure 5C, lanes 2 and 3), whereas none of these proteins was recovered from a control strain containing only untagged PRT1 (Figure 5C, lanes 8 and 9). Importantly, TIF34 and TIF35 co-purified with prt1-Δ100-His to the same extent observed for PRT1-His, whereas no detectable TIF32, NIP1, HCR1 or eIF5 co-purified with the tagged mutant protein (Figure 5C, lanes 5 and 6). Thus, the RRM domain in PRT1 is essential for association of HCR1 and TIF32 (and consequently of NIP1 and eIF5) with eIF3 in vivo, but dispensable for the formation of a PRT1–TIF34–TIF35 subcomplex.
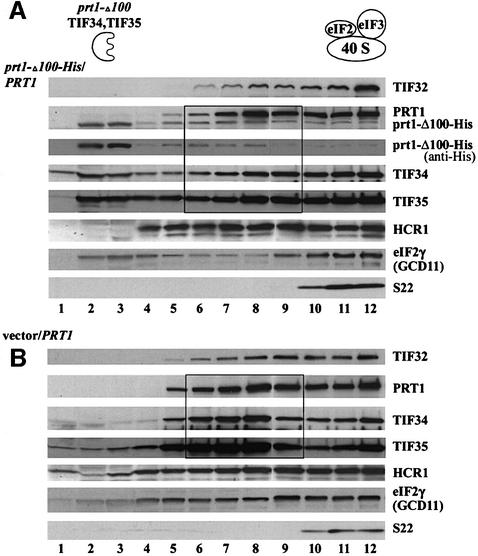
We wished to confirm that the stable subcomplexes containing TIF34, TIF35 and prt1-Δ100 were unable to bind to 40S ribosomes in vivo. We prepared extracts from PRT1 strains containing low-copy prt1-Δ100-His or an empty vector, resolved them on sucrose density gradients and probed for eIF3 subunits by western blotting. In both strains, a significant fraction of the core eIF3 subunits, HCR1 and the γ subunit of eIF2 (GCD11) were in fractions 10–12 containing the 43–48S initiation complexes (Figure 6A and B). In the wild-type extract, most of the remainder of these proteins was in fractions 6–9, corresponding to the MFC (Figure 6B). In contrast, nearly all of the prt1-Δ100-His in the mutant extract was at the top of the gradient in fractions 2–4 (Figure 6A, anti-His) along with a higher proportion of the total TIF34 and TIF35 than was seen in the corresponding fractions for the wild-type extract (Figure 6B). Interestingly, there was a decreased proportion of eIF2 in fractions 6–9 containing the MFC and an increased proportion of eIF2 at the top of the gradient (fractions 1–4) in the mutant extract (Figure 6). Together, these data are consistent with the idea that prt1-Δ100-His sequesters a fraction of TIF34 and TIF35 in defective subcomplexes that cannot form the MFC or bind to 40S ribosomes. The fact that no TIF32, HCR1 or full-length PRT1 was present at the top of the gradient in the mutant extract (Figure 6A) could indicate that the pools of these proteins expected to occur free of TIF34 and TIF35 when prt1-Δ100-His is being expressed are either degraded or form a subcomplex that binds to 40S subunits.
Fig. 6. Evidence that the eIF3 subcomplex containing prt1-Δ100, TIF34 and TIF35 is defective for binding to 40S ribosomes. Trans formants of wild-type strain W303 containing pprt1-Δ100-His bearing prt1-Δ100-His (A) or empty vector YCplac11 (B) were grown in SD medium supplemented with only the required amino acids to an OD600 of ∼1.5, and 50 µg/ml cycloheximide was added 5 min prior to harvesting. WCEs were prepared and separated by velocity sedimentation on 7.5–30% sucrose gradients for 5 h at 41 000 r.p.m. Gradient fractions were collected and resolved by SDS–PAGE, followed by immunoblot analysis using antibodies against the proteins indicated on the right of each panel. The membrane in (A) probed with anti-PRT1 antibodies was stripped and re-probed with anti-His antibody (Santa Cruz Biotechnology) to detect only the prt1-Δ100-His product. The presence of 40S ribosomes was revealed by the A254 profile (not shown) and by probing for the 40S subunit protein S22. The locations of the aberrant prt1-Δ100-His–TIF34–TIF35 subcomplex and of wild-type eIF3 and eIF2 in 40S initiation complexes are indicated schematically above (A). Black rectangles highlight the gradient fractions in which the distributions of PRT1, TIF34 and TIF35 differ substantially between the gradients in (A) versus (B).
Discussion
HCR1 associates with eIF3 and stabilizes the multifactor complex
HCR1 is a non-essential yeast protein similar in sequence to human eIF3 subunit p35. Previously, it was shown that HCR1 interacted genetically and physically with TIF32 (Valášek et al., 1999), but it was unclear whether this interaction occurred in the context of eIF3. Here we showed that a proportion of the entire eIF3 complex, and also fractions of eIF5 and eIF1, were co-immunoprecipitated with HCR1 (Figure 1). In addition, we provided evidence that HCR1 resides in the MFC (Figure 2), which is an important translation initiation intermediate in vivo (Asano et al., 2000). HCR1 was detected in 43S or 48S initiation complexes, but not in 80S ribosomes, consistent with a direct role in the initiation phase of translation. In vitro binding assays with recombinant proteins indicated that HCR1 binds directly to eIF3 subunits TIF32 and PRT1. However, it appears that HCR1 is less abundant and less tightly bound to the eIF3 complex than are the five core subunits we identified previously (Phan et al., 1998).
Deletion of HCR1 led to a slow growth phenotype and produced abnormalities in the polysome profile, indicating a reduced rate of translation initiation. Coincidentally, we observed a sharp decrease in the association of eIF1 with eIF3, and a moderate decrease in the amount of eIF5 and eIF2 associated with eIF3 in the hcr1Δ mutant (Figure 1D). The latter phenotype suggests that the abundance of the MFC is diminished in hcr1Δ cells, which we confirmed by comparing the amounts of MFC resolved on sucrose gradients between mutant and wild-type extracts (Figure 2B). Considering that both eIF1 and eIF5 bind to eIF3-NIP1, and that eIF5 bridges the interaction between eIF2 and eIF3 in the MFC (Asano et al., 2000), these defects suggest that HCR1 promotes the known interactions between NIP1 and eIFs 1 and 5. Although HCR1 did not interact with NIP1 in our binding experiments, we recently obtained evidence that NIP1 enhances the binding of HCR1 to PRT1 and TIF32 (L.Phan, L.Schoenfeld, L.Valášek and A.G.Hinnebusch, unpublished observations).
We detected an accumulation of the eIFs comprising the MFC in 43–48S complexes in hcr1Δ cells (Figure 2B and C), suggesting that recruitment of these factors to 40S ribosomes is not rate limiting in this mutant. Rather, it appears that conversion of 43S to 48S, or 48S to 80S, initiation complexes occurs inefficiently in the absence of HCR1. Recently, we observed a defect in the conversion of 48S to 80S complexes resulting from a mutation in eIF5 (tif5-7A) that also destabilized the MFC. It appears that AUG recognition or GTP hydrolysis by eIF2 is impaired when the integrity of the MFC complex is disrupted by the tif5-7A mutation (K.Asano, L.Phan, L.Valášek and A.G.Hinnebusch, unpublished observations). Similarly, the impaired interaction of eIFs 1 and 5 with eIF3 produced by hcr1Δ may have a deleterious effect on the scanning process. This would be consistent with the known roles of eIF1 and eIF5 in selection of the start codon by scanning ribosomes (Huang et al., 1997; Pestova et al., 1998).
Unexpectedly, deletion of HCR1 decreased the steady-state level of 40S subunits and produced a concomitant increase in free 60S subunits. This reduction in 40S subunits probably contributes to the diminished polysome size and slow growth phenotype of the hcr1Δ strain. It is unlikely to account for the decreased amounts of the MFC or the accumulation of 43S or 48S complexes observed in hcr1Δ cells, as these defects were not observed in the rp51aΔ mutant with an even greater depletion of 40S subunits. Hence, we conclude that HCR1 has a dual function in translation initiation: enhancing the activity of eIF3 and its associated factors, and promoting the biosynthesis or stability of 40S subunits. An attractive hypothesis is that both functions depend on the ability of HCR1 to bind to a specific site on the 40S ribosome. In this way, HCR1 could interact with factors involved in 40S biogenesis and also promote the correct orientation of the MFC on the ribosome in 43–48S initiation complexes. Its dual interactions with 40S ribosomes and eIF3 might help explain why HCR1 is less tightly associated with eIF3 than are the five core subunits. Interestingly, eIF6 may be another protein with a dual function in ribosome biogenesis and translation initiation. It was isolated originally as an anti-association factor that binds to 60S ribosomes (Merrick and Hershey, 1996) but was implicated more recently in promoting the stability or biogenesis of 60S subunits (Si and Maitra, 1999).
A network of interactions linking HCR1, TIF32 and the RRM domain at the N-terminus of PRT1
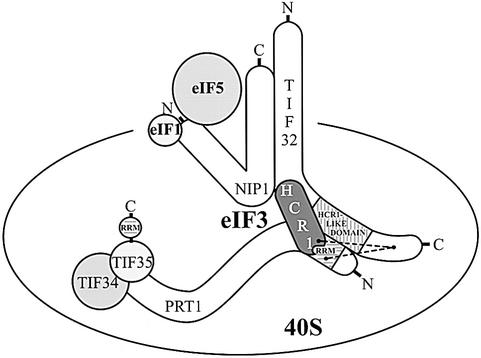
We noted that HCR1 is related in sequence to a functionally important segment of TIF32, and showed that HCR1 and the HLD in TIF32 both interact with the N-terminal segment of PRT1 containing an RRM motif. As these interactions can occur simultaneously in vitro, we propose that HCR1 stabilizes or modulates the interaction between TIF32 and the PRT1 RRM (Figure 7). HCR1 additionally interacted with TIF32 in vitro. It is unlikely that the interactions of TIF32 and HCR1 with the PRT1 RRM domain were mediated by RNA, because RNase A treatment did not reduce these interactions in GST pull-down experiments of the type shown in Figure 3 (data not shown). The interaction between TIF32 and the RRM domain of PRT1 is conserved in the human homologs of TIF32 and PRT1 (Methot et al., 1997). PRT1 also contains binding domains for TIF34 and TIF35, and thus seems to function at least partly as a scaffold in the eIF3 complex. Only the extreme C-terminus of PRT1 (residues 641–724) was required for its interactions with TIF34 and TIF35 in vitro (Figure 7).
Fig. 7. Summary of protein–protein interactions within the yeast eIF3 complex. The eIF3 subunits as well as eIF5 and eIF1 factors are shown as various shapes with sizes roughly proportional to their molecular weights. Horizontally hatched boxes depict the RRMs in TIF35 and PRT1, and the vertically hatched box indicates the HCR1-like domain of TIF32. Points of overlap between the various shapes indicate sites of known protein–protein interaction. The dashed lines depict an interaction between the PRT1 RRM, HCR1 and the C-terminal segment of TIF32 in addition to that involving the HLD of TIF32 and PRT1 RRM and the N-terminal half of TIF32 and HCR1 (see text for details). The interactions of TIF34 with the extreme C-terminus of PRT1 (Asano et al., 1998), of the N-terminal end of NIP1 with eIF1 and eIF5 (Asano et al., 2000) and between the C-terminal two-thirds of NIP1 and TIF32 (L.Valášek and A.G.Hinnebusch, unpublished observations) were identified independently of this study.
In keeping with our in vitro binding data, the prt1-Δ100 product lacking the RRM domain formed a stable subcomplex in vivo containing TIF34 and TIF35 but lacking HCR1, TIF32, NIP1 and eIF5 (Figure 5C). These results provide in vivo evidence that the PRT1 RRM is required for incorporation of TIF32 into eIF3. Since NIP1 seems to interact only with TIF32 (Asano et al., 1998), and contains the binding domain for eIF5 (Phan et al., 1998) (Figure 7), deleting the RRM from PRT1 should lead simultaneously to loss of TIF32, NIP1 and eIF5 from the complex, as we observed.
Role of the PRT1 RRM domain in 40S subunit binding
We showed that expression of prt1-Δ100 sequesters a fraction of TIF34 and TIF35 in defective subcomplexes lacking TIF32 and NIP1, which fail to bind eIF5 and HCR1 (Figure 5C) and cannot interact tightly with 40S ribosomes. Consistently, expression of these prt1-Δ100–TIF34–TIF35 subcomplexes reduced formation of the multifactor complex containing eIF2 (Figure 6A). Our conclusion that the decreased level of intact eIF3 accounts for the growth defect conferred by prt1-Δ100 is supported by the fact that this phenotype was suppressed by overexpressing TIF34 and TIF35 (Figure 5B). Presumably, providing excess amounts of these two subunits restored the concentration of intact eIF3 containing full-length PRT1 to nearly wild-type levels.
The sequestration of TIF34 and TIF35 in defective eIF3 subcomplexes incapable of forming the MFC or binding to 40S ribosomes might be expected to decrease the formation of functional 43–48S complexes. Ostensibly at odds with this expectation, we did not observe a reduced proportion of eIF3 subunits or other eIFs in the 43–48S region of the gradient in the mutant extract containing prt1-Δ100 (Figure 6). However, the predicted decrease in the rate of 43–48S complex formation in the mutant could be masked if binding of the MFC complex to 40S subunits continued after cell lysis while conversion of 48S to 80S initiation complexes occurred slowly in the extracts. In this event, there would be similar amounts of 48S complexes in mutant and wild-type extracts plus an excess of free MFCs in the wild-type extract, as we observed (Figure 6). This explanation is consistent with the recent finding that in vitro binding of the ternary complex to the 40S ribosome is ∼3-fold faster than conversion of 48S to 80S complexes (Lorsch and Herschlag, 1999). This phenomenon may also have influenced the results we obtained for the hcr1Δ extract in Figure 2B, but there we observed an excess of 43–48S complexes in the mutant, suggesting a defect at a subsequent step of the pathway.
It is possible that the ribosome-binding defect of the eIF3 subcomplex containing prt1-Δ100, TIF34 and TIF35 results from loss of interaction between the PRT1 RRM and 18S rRNA. However, the human PRT1 homolog failed to bind 18S rRNA in vitro (Methot et al., 1997; Block et al., 1998) and yeast PRT1 also showed no RNA-binding activity in vitro (Naranda et al., 1994). It is known that RRM2 of PAB1 is required for protein–protein interaction with eIF4G (Kessler and Sachs, 1998). Thus, an alternative hypothesis would be that the PRT1 RRM is the binding domain for TIF32, and TIF32 or NIP1 interacts directly with the 40S ribosome. Consistent with this last possibility, hTIF32 (p170) interacted with both 18S rRNA and β-globin mRNA (Block et al., 1998). Another interesting possibility, for which there is precedent (Scherly et al., 1990; Kessler et al., 1997), would be that binding of TIF32 to the PRT1 RRM enhances interaction of the RRM with 18S rRNA. TIF35 also contains an RRM that interacted with both 18S rRNA and β-globin mRNA in vitro (Hanachi et al., 1999). Our results indicate that the TIF35 RRM was insufficient to mediate strong ribosome association of the prt1-Δ100–TIF34–TIF35 subcomplex. Consistently, deletion of the TIF35 RRM conferred a Slg– phenotype, but was not lethal in yeast (Hanachi et al., 1999). It will be important to determine which subunits of eIF3 bind directly to the 40S ribosome to mediate recruitment of the MFC to the pre-initiation complex.
Materials and methods
Yeast strains
Details of the yeast strains used in this study are given in Table II.
Table II. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype or description | Source or reference |
|---|---|---|
| YLVH13 | MATa hcr1Δ::LEU2 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 | Valášek et al. (1999) |
| 21R | MATa ade1 leu2-3 leu2-112 ura3-52 PRT1 | G.Johnston |
| TC-26-3 | MATa leu2-3 leu2-112 ura3-52 prt1-63 cdc 63-1 | G.Johnston |
| TP11-4-1 | MATa ade1 leu2-3 leu2-112 ura3-52 prt1-1 | Evans et al. (1995) |
| TP12-7-0 | MATα prt1-2 ade1 leu2-3 leu2-112 ura1 ura3-52 his6 | Evans et al. (1995) |
| TDE/16A | MATα prt1-26 ade2 leu2-3 leu2-112 ura3-11 ura3-15 trp1-1 | Evans et al. (1995) |
| TP13-1-2 | MATa prt1-3 ade1 leu2-3 leu2-112 ura1 ura3-52 his6 | Evans et al. (1995) |
| F665 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 | A.Hopper |
| YLV314U | MATa ura3::URA3::rpg1-1 trp1-1::TRP1::rpg1-Δ2 ade2-1 can1-100 leu2-3,112 his3-11,15 | Valášek et al. (1999) |
| GDE808-33 | MATα prt1::URA3 ade leu2-3 leu2-112 ura3 his3-11 his3-15 trp1-1 pDE31 (PRT1 HIS3) | Evans et al. (1995) |
| F353 (BJ5464) | MATα ura3-52 trp1 leu2-Δ1 his3-Δ200 pep4::HIS3 prb1-Δ1.6R can1 | E.Jones |
| H1402 | MATα leu2-3 leu2-112 ura3-52 ino1 (HIS4-lacZ at ura3-52) | Foiani et al. (1991) |
| H1654 | MATα leu2-3 leu2-112 ura3-52 ino1 rp51a::LEU2 (HIS4-lacZ at ura3-52) | Foiani et al. (1991) |
Plasmid constructions and site-directed mutagenesis of HCR1
Table III contains brief descriptions of all plasmids employed in this study. Details of their construction are available as supplementary data at The EMBO Journal Online.
Table III. Plasmids used in this study.
| Plasmid | Description | Source |
|---|---|---|
| YCpLVHM-T | c-Myc-tagged HCR1, TRP1 plasmid from YCplac22 | this study |
| YCplac22 | single-copy cloning vector, TRP1 | Gietz and Sugino (1988) |
| YCpLVPRT1-Myc | c-Myc-tagged HCR1, URA3 plasmid from YCplac33 | Valášek et al. (1998) |
| YCpLVHCR1-Myc | c-Myc-tagged HCR1, LEU2 plasmid from YCplac11 | Valášek et al. (1999) |
| YCplac33 | single-copy cloning vector, URA3 | Gietz and Sugino (1988) |
| pGEX-5X-3 | cloning vector for GST fusions | Smith and Johnson (1988) |
| pGEX-HCR1 | GST–HCR1 fusion plasmid from pGEX-5X-3 | this study |
| pT7-7 | cloning vector with T7 promoter | Tabor and Richardson (1987) |
| pT7-TIF32 | TIF32 ORF cloned under T7 promoter | Asano et al. (1998) |
| pT7-PRT1 | PRT1 ORF cloned under T7 promoter | Asano et al. (1998) |
| pT7-PRT1ΔS | PRT1[1–624] ORF cloned under T7 promoter | Asano et al. (1998) |
| pT7-PRT1ΔM | PRT1[1–682] ORF cloned under T7 promoter | Asano et al. (1998) |
| pT7-NIP1 | NIP1 ORF cloned under T7 promoter | Asano et al. (1998) |
| pT7-TIF34 | TIF34 ORF cloned under T7 promoter | Asano et al. (1998) |
| pT7-TIF35 | TIF35 ORF cloned under T7 promoter | Asano et al. (1998) |
| pT7-HCR1 | HCR1 ORF cloned under T7 promoter | this study |
| pGEX-PRT1/A1 | GST–PRT1/A1[641–724] fusion plasmid from pGEX-4T-1 | Asano et al. (1998) |
| YEpLVHCR1 | high-copy HCR1, LEU2 plasmid from YEplac181 | Valášek et al. (1999) |
| YEplac181 | high-copy cloning vector, LEU2 | Gietz and Sugino (1988) |
| pGEX-TIF32 | GST–TIF32 fusion plasmid from pGEX-5X-3 | Asano et al. (1998) |
| pGEX-TIF34 | GST–TIF34 fusion plasmid from pGEX-4T-1 | Asano et al. (1998) |
| pGEX-TIF35 | GST–TIF35 fusion plasmid from pGEX-4T-1 | Asano et al. (1998) |
| pT7-ΔA-PRT1 | PRT1[1–136] ORF cloned under T7 promoter | this study |
| pT7-ΔB-PRT1 | PRT1[28–136] ORF cloned under T7 promoter | this study |
| pT7-ΔC-PRT1 | PRT1[28–261] ORF cloned under T7 promoter | this study |
| pT7-Δ0-PRT1 | PRT1[28–724] ORF cloned under T7 promoter | this study |
| pT7-Δ1-PRT1 | PRT1[111–724] ORF cloned under T7 promoter | this study |
| pT7-Δ2-PRT1 | PRT1[261–724] ORF cloned under T7 promoter | this study |
| pT7-Δ6-PRT1 | PRT1[602–724] ORF cloned under T7 promoter | this study |
| pT7-Δ7-PRT1 | PRT1[641–724] ORF cloned under T7 promoter | this study |
| pGEX-PRT1 | GST–PRT1 fusion plasmid from pGEX-5X-3 | this study |
| pGEX-PRT1 RRM | GST–PRT1 [1–136] fusion plasmid from pGEX-5X-3 | this study |
| pGEX-NIP1 | GST–NIP1 fusion plasmid from pGEX-4T-1 | Asano et al. (1998) |
| pT7-Δ1-TIF32 | TIF32[490–790] ORF cloned under T7 promoter | this study |
| pT7-Δ2-TIF32 | TIF32[551–750] ORF cloned under T7 promoter | this study |
| pT7-Δ3-TIF32 | TIF32[1–490] ORF cloned under T7 promoter | this study |
| pT7-Δ4-TIF32 | TIF32[790–964] ORF cloned under T7 promoter | this study |
| pT7-Δ1+4-TIF32 | TIF32[490–964] ORF cloned under T7 promoter | this study |
| pT7-Δ5 TIF32 | TIF32[1–640] ORF cloned under T7 promoter | this study |
| pT7-Δ6-TIF32 | TIF32[1–790] ORF cloned under T7 promoter | this study |
| YEpLVhcr1-R212I | high-copy hcr1-R212I, LEU2 plasmid from YEplac181 | this study |
| YEpLVhcr1-R215I | high-copy hcr1-R212I, LEU2 plasmid from YEplac181 | this study |
| YEpLVhcr1-3 | high-copy hcr1-R212I, LEU2 plasmid from YEplac181 | this study |
| pLP101 | His-tagged PRT1, URA3 plasmid from pRS316 | Phan et al. (1998) |
| pprt1-Δ100/pDE-1E | single-copy prt1-Δ100, LEU2 plasmid | Evans et al. (1995) |
| pprt1-Δ100-His | His-tagged prt1-Δ100, LEU2 plasmid from pDE-1E | this study |
| YCplac11 | single-copy cloning vector, LEU2 | Gietz and Sugino (1988) |
| YEp112 | high-copy cloning vector, TRP1 | Gietz and Sugino (1988) |
| YEpTIF34/35T | high-copy TIF34, TIF35, TRP1 plasmid from YEplac112 | this study |
| YEpTIF34T | high-copy TIF34, TRP1 plasmid from YEplac112 | this study |
| YEpTIF35T | high-copy TIF35, TRP1 plasmid from YEplac112 | this study |
| YEpTIF32T | high-copy TIF32, TRP1 plasmid from YEplac112 | this study |
| YEpLVHCR1-1 | high-copy HCR1, TRP1 plasmid from YEplac112 | Valášek et al. (1999) |
| YEpNIP1T | high-copy NIP1, TRP1 plasmid from YEplac112 | this study |
WCE preparation and immunoprecipitation
Yeast cells of strains W303 and YLVH13 alone or YLVH13 (hcr1Δ) transformed with either YCpLVHM-T or YCplac22 were grown in 100 ml of YPD or SC medium lacking tryptophan, respectively, to an A260 of ∼1.0. Yeast WCEs were prepared and immunoprecipitations were conducted as described previously (Asano et al., 1999) except that a different breaking buffer was used (Valášek et al., 1998) for immunoprecipitations using c-Myc antibodies. Our modifications of this protocol are available as supplementary data at The EMBO Journal online.
Polysome profile analysis
Preparation of WCEs and subsequent polysome analysis were conducted essentially as described previously (Foiani et al., 1991). Our modifications of this protocol are available as supplementary data at The EMBO Journal online.
GST pull-down experiments
Preparation of recombinant proteins, in vitro translation and in vitro binding experiments were carried out as described previously (Asano et al., 1998).
Ni2+ affinity purification of eIF3 complexes containing His-tagged PRT1
Preparation of WCEs and subsequent Ni2+ affinity purification were conducted essentially as described previously (Phan et al., 1998). Our modifications of this protocol are available as supplementary data at The EMBO Journal online.
Preparation of antibodies against HCR1
The GST–HCR1 fusion protein encoded by pGEX-HCR1 was expressed in Escherichia coli and purified from the WCE by incubation with glutathione–Sepharose 4B beads (Pharmacia). The isolated protein was resolved by SDS–PAGE (4–20% gels), excised from the gel and washed with 1× phosphate-buffered saline (PBS). Rabbits were injected with the purified protein and serum containing polyclonal antibodies against HCR1 was obtained commercially from Covance Research Products (Denver, PA).
Antibodies against TIF32 (Valášek et al., 1998), PRT1, NIP1, TIF34, eIF1, eIF5, GCD6, GCD11 (Phan et al., 1998) and TIF35 (L.Phan, L.W.Schoenfeld, L.Valášek, K.Nielsen and A.G.Hinnebusch, submitted) used in this study were described previously. Antibodies against S22 were kindly provided by Jan van’t Riet.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are indebted to Chris Barnes, Harald Nierlich, Jiri Hašek, Gerard Griffioen, Vladimir Reiser and Jan van’t Riet for sending us yeast strains, plasmids and other materials used in this study. We are grateful to Thomas Dever, Beatriz Castilho and Katsura Asano for critical reading of the manuscript, and to the members of the Hinnebusch and Dever laboratories for helpful discussions and suggestions during the course of this work.
REFERENCES
- Asano K., Vornlocher,H.-P., Richter-Cook,N.J., Merrick,W.C., Hinnebusch,A.G. and Hershey,J.W.B. (1997) Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits: possible roles in RNA binding and macromolecular assembly. J. Biol. Chem., 272, 27042–27052. [DOI] [PubMed] [Google Scholar]
- Asano K., Phan,L., Anderson,J. and Hinnebusch,A.G. (1998) Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem., 273, 18573–18585. [DOI] [PubMed] [Google Scholar]
- Asano K., Krishnamoorthy,T., Phan,L., Pavitt,G.D. and Hinnebusch, A.G. (1999) Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J., 18, 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Clayton,J., Shalev,A. and Hinnebusch,A.G. (2000) A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5 and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev., 14, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A. and Maitra,U. (1999) Cloning and characterization of the p42 subunit of mammalian translation initiation factor 3 (eIF3): demonstration that eIF3 interacts with eIF5 in mammalian cells. Nucleic Acids Res., 27, 1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block K.L., Vornlocher,H.P. and Hershey,J.W.B. (1998) Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3. J. Biol. Chem., 273, 31901–31908. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Chowdhury,D. and Maitra,U. (1999) Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40S ribosomal preinitiation complex. J. Biol. Chem., 274, 17975–17980. [DOI] [PubMed] [Google Scholar]
- Cigan A.M., Foiani,M., Hannig,E.M. and Hinnebusch,A.G. (1991) Complex formation by positive and negative translational regulators of GCN4. Mol. Cell. Biol., 11, 3217–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaie P., Wittmer,B., Altmann,M. and Trachsel,H. (1995) Isolation of a protein complex containing translation initiation factor Prt1 from Saccharomyces cerevisiae. J. Biol. Chem., 270, 4288–4292. [DOI] [PubMed] [Google Scholar]
- Evans D.R.H., Rasmussen,C., Hanic-Joyce,P.J., Johnston,G.C., Singer, R.A. and Barnes,C.A. (1995) Mutational analysis of the Prt1 protein subunit of yeast translation initiation factor 3. Mol. Cell. Biol., 15, 4525–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C.M., Pestova,T.V., Hellen,C.U.T. and Wagner,G. (1999) Structure and interactions of the translation initiation factor eIF1. EMBO J., 18, 2631–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Cigan,A.M., Paddon,C.J., Harashima,S. and Hinnebusch,A.G. (1991) GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 3203–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Hanachi P., Hershey,J.W.B. and Vornlocher,H.P. (1999) Characterization of the p33 subunit of eukaryotic translation initiation factor-3 from Saccharomyces cerevisiae. J. Biol. Chem., 274, 8546–8553. [DOI] [PubMed] [Google Scholar]
- Huang H., Yoon,H., Hannig,E.M. and Donahue,T.F. (1997) GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev., 11, 2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S.H. and Sachs,A.B. (1998) RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol., 18, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M.M., Henry,M.F., Shen,E., Zhao,J., Gross,S., Silver,P.A. and Moore,C.L. (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev., 11, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Linder,P. and de La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch J.R. and Herschlag,D. (1999) Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J., 18, 6705–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick W.C. and Hershey,J.W.B. (1996) The pathway and mechanism of eukaryotic protein synthesis. In Hershey,J.W.B., Matthews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 31–69.
- Methot N., Rom,E., Olsen,H. and Sonenberg,N. (1997) The human homologue of the yeast Prt1 protein is an integral part of the eukaryotic initiation factor 3 complex and interacts with p170. J. Biol. Chem., 272, 1110–1116. [DOI] [PubMed] [Google Scholar]
- Naranda T., MacMillan,S.E. and Hershey,J.W.B. (1994) Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J. Biol. Chem., 269, 32286–32292. [PubMed] [Google Scholar]
- Naranda T., MacMillan,S.E., Donahue,T.F. and Hershey,J.W. (1996) SUI1/p16 is required for the activity of eukaryotic translation initiation factor 3 in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2307–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov,S.I. and Hellen,C.U.T. (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature, 394, 854–859. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin,I.B., Lee,J.H., Choi,S.K., Dever,T.E. and Hellen,C.U.T. (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature, 403, 332–335. [DOI] [PubMed] [Google Scholar]
- Phan L., Zhang,X., Asano,K., Anderson,J., Vornlocher,H.P., Greenberg,J.R., Qin,J. and Hinnebusch,A.G. (1998) Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol., 18, 4935–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B. and Varani,G. (2000) Eukaryotic translation initiation: there are (at least) two sides to every story. Nature Struct. Biol., 7, 356–361. [DOI] [PubMed] [Google Scholar]
- Scherly D., Boelens,W., Dathan,N.A., van Venrooij,W.J. and Mattaj,I.W. (1990) Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B” and their cognate RNAs. Nature, 345, 502–506. [DOI] [PubMed] [Google Scholar]
- Si K. and Maitra,U. (1999) The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell. Biol., 19, 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.B. and Johnson,K.S. (1988) Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene, 67, 31–40. [DOI] [PubMed] [Google Scholar]
- Tabor S. and Richardson,C.C. (1987) DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl Acad. Sci. USA, 84, 4767–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Trachsel,H., Hašek,J. and Ruis,H. (1998) Rpg1, the Saccharomyces cerevisiae homologue of the largest subunit of mammalian translation initiation factor 3, is required for translational activity. J. Biol. Chem., 273, 21253–21260. [DOI] [PubMed] [Google Scholar]
- Valášek L., Hašek,J., Trachsel,H., Imre,E.M. and Ruis,H. (1999) The Saccharomyces cerevisiae HCRI gene encoding a homologue of the p35 subunit of human translation eukaryotic initiation factor 3 (eIF3) is a high copy suppressor of a temperature-sensitive mutation in the Rpg1p subunit of yeast eIF3. J. Biol. Chem., 274, 27567–27572. [DOI] [PubMed] [Google Scholar]
- Vornlocher H.P., Hanachi,P., Ribeiro,S. and Hershey,J.W.B. (1999) A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J. Biol. Chem., 274, 16802–16812. [DOI] [PubMed] [Google Scholar]