Abstract
Although light is the ultimate substrate in photosynthesis, it can also be harmful and lead to oxidative damage of the photosynthetic apparatus. The main target for light stress is the central oxygen-evolving photosystem II (PSII) and its D1 reaction centre protein. Degradation of the damaged D1 protein and its rapid replacement by a de novo synthesized copy represent the important repair mechanism of PSII crucial for plant survival under light stress conditions. Here we report the isolation of a single-copy nuclear gene from Arabidopsis thaliana, encoding a protease that performs GTP-dependent primary cleavage of the photodamaged D1 protein and hence catalysing the key step in the repair cycle in plants. This protease, designated DegP2, is a homologue of the prokaryotic Deg/Htr family of serine endopeptidases and is associated with the stromal side of the non-appressed region of the thylakoid membranes. Increased expression of DegP2 under high salt, desiccation and light stress conditions was measured at the protein level.
Keywords: D1 protein/DegP2 protease/non-appressed thylakoids/photodamage/primary cleavage
Introduction
Organisms that perform oxygenic photosynthesis are subject to inhibition of their photosynthetic function when exposed to excessive illumination. This process is referred to as photoinhibition (for reviews see Barber and Andersson, 1992; Prasil et al., 1992; Aro et al., 1993). At ambient temperatures, photoinhibition occurs primarily at the level of photosystem II (PSII) and involves reversible inactivation of PSII due to the arrest of electron transport within this complex followed by irreversible damage to the subunits of the PSII reaction centre (for reviews see Barber and Andersson, 1992; Prasil et al., 1992; Aro et al., 1993; Long et al., 1994). PSII of higher plants, algae and cyanobacteria is a multisubunit complex, comprising >25 different protein subunits (Barber and Kühlbrandt, 1999). At the core of the complex is the reaction centre consisting of two related D1 and D2 proteins, which bind all the cofactors involved in primary and secondary electron flow (Barber and Kühlbrandt, 1999). Among the PSII reaction centre proteins, the D1 protein is the main target of oxidative damage (Vass et al., 1992; Telfer et al., 1994). Rapid degradation of the photodamaged D1 protein (Mattoo et al., 1981; Depka et al., 1998) and its replacement by a de novo synthesized functional copy (Kyle et al., 1984; van Wijk et al., 1996; Melis, 1999) represent an important repair mechanism crucial for plant survival under light stress conditions.
By analogy with the L subunit of the bacterial reaction centre (Michel and Deisenhofer, 1986), the D1 protein has been predicted to have five transmembrane α-helices, which are connected by stromal and lumenal loops (Trebst, 1986). Degradation of the D1 protein proceeds via at least two steps. The primary cleavage occurs on the stromal loop connecting the membrane-spanning helices D and E, and yields N-terminal 23 kDa (Greenberg et al., 1987) and C-terminal 10 kDa (Canovas and Barber, 1993) proteolytic fragments. Although the rapid turnover of D1 protein has been known for a long time, the identity of the protease(s) involved in the primary proteolytic cleavage of the photodamaged D1 protein has remained unknown despite intensive research efforts. Biochemical analysis suggested that this step is a GTP-stimulated process (Spetea et al., 1999) mediated by an unknown serine-type protease (Virgin et al., 1990; Shipton and Barber, 1991). The second step of D1 protein degradation involves a further digestion of the primary cleavage products. Recently, the thylakoid membrane FtsH metalloprotease was proposed to be involved in the secondary proteolysis of the D1 protein (Lindahl et al., 2000). It was demonstrated that overexpressed and purified recombinant FtsH degraded the 23 kDa fragment when introduced into the isolated photoinactivated PSII complex in vitro.
In this work, we isolated an Arabidopsis DegP2 gene encoding a novel chloroplast homologue of the prokaryotic trypsin-type Deg/Htr serine proteases. We investigated the topology of DegP2 in the thylakoid membranes and showed that this enzyme is peripherally associated with the outer surface of the thylakoid membrane. The expression pattern of DegP2 was investigated under various stress conditions, and it is shown that the DegP2 protein level increased in response to a high concentration of NaCl, desiccation and illumination with high intensity light. Finally, we demonstrated that the physiological target of DegP2 is the damaged D1 protein of PSII. The DegP2 protease performed the primary cleavage of the D1 protein on the stromal D–E loop, generating the typical proteolytic fragments in a GTP-dependent manner.
Results
Isolation of a single-copy gene encoding a homologue of bacterial Deg/Htr protease in Arabidopsis thaliana
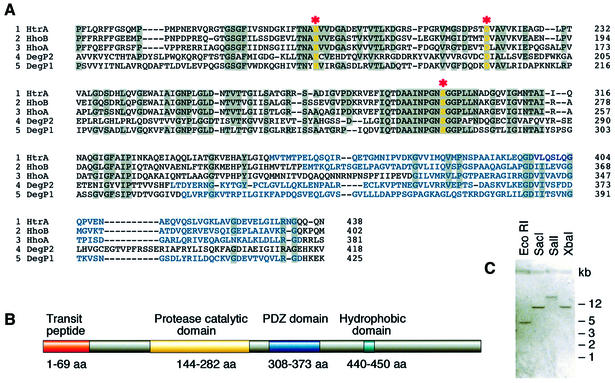
The DegP/Htr family in Prokaryota, including cyanobacteria from which chloroplasts derive, consists of three serine-type endopeptidases: DegP (also named HtrA), DegQ (also named HhoA) and DegS (also named HtrH or HhoB) (Gottesman, 1996; Pallen and Wren, 1997). Recently, a homologue of the cyanobacterial DegP protease was identified in chloroplasts of Arabidopsis and pea (this protease is designated here as DegP1) and shown to be associated with the lumenal side of the thylakoid membranes (Itzhaki et al., 1998). BLAST searches of the Arabidopsis database with the conserved amino acid region of the Deg/Htr protease family from the cyanobacteria Synechocystis sp. strain PC6803 revealed the presence of another highly conserved homologue of the DegP/HtrA protease. We named this protein DegP2 (for second DegP described from higher plants). A gene located on chromosome 2 of Arabidopsis (DDBJ/EMBL/GenBank accession No. AC005309, protein identity AAC63648) is predicted to encode the DegP2 protein.
We designed primers based on the predicted genomic sequence of DegP2 and used PCR to amplify DegP2 cDNA from an Arabidopsis cDNA library. The DegP2 cDNA sequence contained an 1821 bp open reading frame (ORF) encoding a protein composed of 607 amino acids (relative molecular mass 66.8 kDa). Alignment of the deduced amino acid sequences revealed that the overall sequence similarity of DegP2 and DegP1 to cyanobacterial Deg/Htr proteases was 44 and 48–51%, respectively. A comparison of the conserved regions of DegP2 and DegP1 from Arabidopsis and homologous proteases of the Deg/Htr family from Synechocystis is shown in Figure 1A. Three amino acids, serine (S), histidine (H) and aspartic acid (E), involved in the catalytic activity (catalytic triad) of serine proteases from the Deg/Htr family (Gottesman, 1996) are present in DegP2 at highly conserved positions. The predicted DegP2 protein possesses an N-terminal transit peptide (amino acids 1–69), typical for proteins imported into plastids, and two strongly conserved domains (Figure 1B). A catalytic domain specific for serine proteases of the trypsin type was found between amino acids 144 and 282, and a 65 amino acid residue repeat, called the PDZ domain, between amino acids 308 and 373. The PDZ domain in Eukaryota has generally been accepted as a protein–protein interaction site, and the same function was proposed for Prokaryota (Spiess et al., 1999). Hydropathy plots revealed that the mature DegP2 in Arabidopsis is predominantly a hydrophilic protein with a molecular mass of 60 kDa and has no predicted membrane-spanning domains but a short hydrophobic segment located at the C-terminus (amino acids 440–450).
Fig. 1. A single-copy nuclear DegP2 gene in A.thaliana encodes a putative serine-type protease homologous to the Deg/Htr protease family from the cyanobacteria Synechocystis sp. strain PCC 6803. (A) Sequence alignment of the conserved regions of HtrA (CyanoBase accession No. slr1204), HhoA (CyanoBase accession No. sll1679) and HhoB (CyanoBase accession No. sll1427) proteases from Synechocystis sp., DegP1 (DDBJ/EMBL/GenBank accession No. AF028842) from Arabidopsis and a novel DegP2 protease (DDBJ/EMBL/GenBank accession No. AF245171). The sequences were aligned manually with the assistance of the multiple alignment program CLUSTAL_W (Thompson et al., 1994). Identical amino acids are shown on a grey background, the PDZ domain is marked in blue, gaps are indicated by dashes, and catalytic amino acids are highlighted in yellow and marked by asterisks. (B) Schematic representation of the predicted structure of DegP2. Localization prediction and determination of the processing site were analysed by the programs PSORT (Nakai and Kanehisa, 1992), CHLOROP ver.1.1 (Emanuelsson et al., 1999) and TARGETP ver.1.01 (Emanuelsson et al., 2000). Secondary structure prediction was performed using PREDATOR (Frishman and Argos, 1996) and the Dense Alignment Surface Program (Cserzo et al., 1997). Protein pattern and motif predictions were determined using PROSITE database (Hofmann et al., 1999). (C) Southern blot analysis of genomic DNA digested with restriction enzymes and hybridized with a DegP2 cDNA probe under low stringency conditions.
To investigate the copy number of the DegP2 gene in Arabidopsis, genomic DNA was isolated, digested by restriction endonucleases and hybridized to DegP2 cDNA. Independently of the stringency of hybridization, a single band was detected (Figure 1C). This indicated that the DegP2 gene exists at a single locus in the Arabidopsis genome.
Biochemical characterization of DegP2 proteolytic activity
In order to explore the proteolytic nature of DegP2, we produced DegP2 protein in a bacterial overexpression system as a fusion protein with its C-terminus attached to a His tag. The proteolytic activity of an Escherichia coli cell extract with overexpressed DegP2 and the fractions containing recombinant DegP2 purified by affinity chromatography were tested on a non-specific protease substrate, gelatine (Figure 2A). As negative controls, an E.coli cell extract without recombinant DegP2 (Figure 2A) and an E.coli cell extract with an overexpressed non-proteolytic protein, the class 1a metallothionein (not shown), were analysed on activity gels under the same experimental conditions. The results revealed (Figure 2A, lower panel) that fractions containing overexpressed DegP2 were able to degrade gelatine in the region of DegP2 migration. A weaker proteolytic band visible in DegP2-containing fractions might represent an autodegradation product of DegP2. Although several proteolytic activities were visible in control fractions (not shown), the absence of gelatine degradation in the region corresponding to DegP2 migration (Figure 2A) proved that the measured proteolysis was mediated by recombinant DegP2 protease.
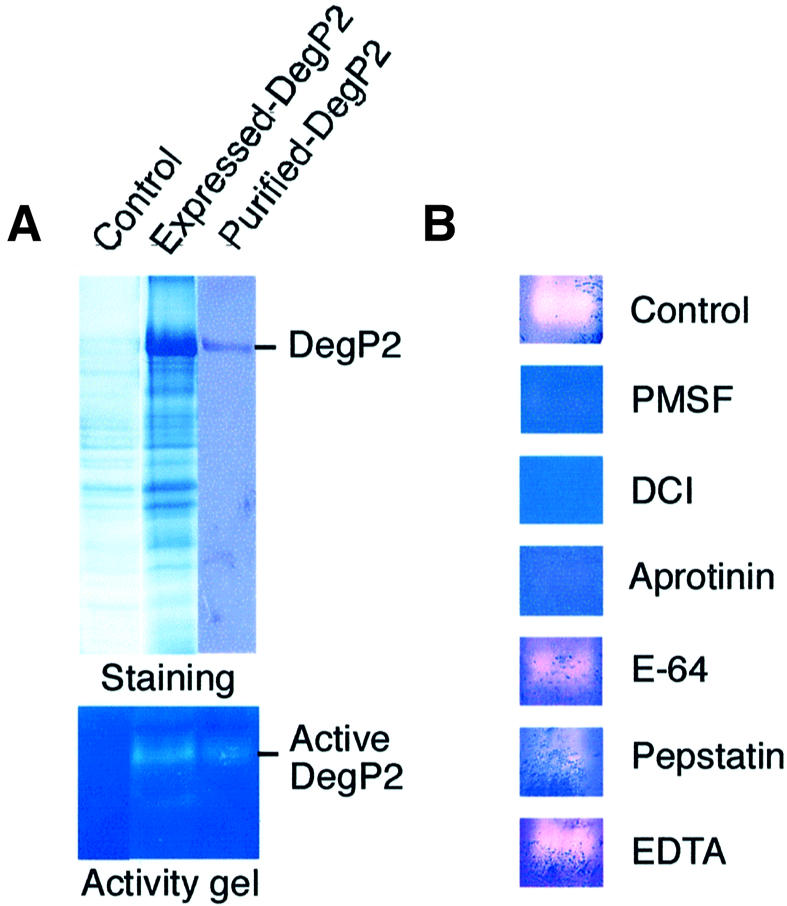
Fig. 2. DegP2 is a serine-type endopeptidase. (A) An E.coli extract without recombinant DegP2 (control) and with overexpressed DegP2, and His-tagged purified DegP2 were analysed on activity gels containing gelatine as a non-specific protease substrate. (B) To classify DegP2 protease according to its catalytic centre, activity gels were incubated in the absence (control) or presence of protease inhibitors directed against serine- (PMSF, DCI and aprotinin), cysteine- (E-64), aspartic- (pepstatin) and metallo- (EDTA) endopeptidases.
The presence of conserved catalytic amino acids in the DegP2 sequence (Figure 1A) suggested that this enzyme is a serine endopeptidase of the trypsin type. To confirm this prediction experimentally, we performed in vitro studies with protease inhibitors (Figure 2B). The degradation of gelatine in activity gels by overexpressed DegP2 was completely arrested or strongly reduced in the presence of serine protease inhibitors such as 3,4-dichloroisocoumarin (DCI), aprotinin or phenylmethylsulfonyl fluoride (PMSF). Inhibitors directed against cysteine-, aspartic- or metallo-endopeptidases did not significantly influence the proteolytic activity of DegP2.
Chloroplast localization of DegP2 and thylakoid membrane topological studies
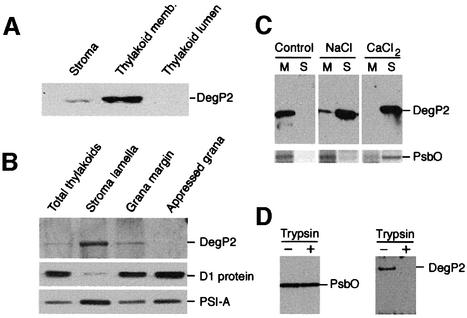
To examine the subcellular location of DegP2, we raised a polyclonal antibody against the recombinant protein. Fractionation of isolated intact chloroplasts into stroma, thylakoid membrane and thylakoid lumen, followed by immunoblot analysis of these fractions revealed that the vast majority of the DegP2 protease was associated with the thylakoid membranes and only residual amounts were detected in the stroma fraction (Figure 3A). The presence of DegP2 in the stroma might result from contamination of this fraction with small membrane fragments that did not sediment during centrifugation. These results are consistent with the location predicted from the DegP2 protein presequence. Further fractionation of the thylakoid membranes into appressed and non-appressed membrane regions using differential centrifugation and aqueous polymer two-phase partition (Andersson and Anderson, 1980) demonstrated that 80–90% of this enzyme was present in the stroma lamellae and 10–20% in fractions enriched in the non-appressed regions of grana stacks consisting of grana margins and end grana membranes (Figure 3B). The distribution of subunit A from the PSI reaction centre (PSI-A) and the D1 protein was in accordance with previously published data (Morrissey et al., 1986). Whilst the vast majority of the D1 protein was detected in the grana region, the majority of the PSI-A was found in the stroma lamellae (Figure 3B).
Fig. 3. DegP2 is an extrinsic thylakoid membrane protein located on the stromal side of stroma lamellae and in non-appressed regions of the grana stacks. (A) Isolated intact Arabidopsis chloroplasts were separated into stroma, thylakoid membrane and thylakoid lumen fractions as described in Materials and methods. Proteins in each fraction were separated by SDS–PAGE and the DegP2 location assayed by immunoblotting. (B) Localization of DegP2 in appressed and non-appressed regions of thylakoid membranes assayed by immunoblotting. As references, the distributions of the D1 protein from the PSII reaction centre and subunit-A from the PSI reaction centre (PSI-A) were visualized under the same experimental conditions. (C) The topology of DegP2 protease in the thylakoid membranes was tested by incubation of membranes in the absence (control) or presence of 0.5 M NaCl or 0.5 M CaCl2. The membrane pellet (M) and supernatant (S) containing extracted proteins were analysed by SDS–PAGE and immunoblotting. As a reference, the distribution of the 33 kDa protein from the PSII oxygen-evolving complex (PsbO) was assayed by SDS–PAGE and Coomassie Blue staining. (D) A protease protection assay was carried out after addition of trypsin (50 µg/ml) to the isolated thylakoid membranes (1 mg chlorophyll/ml) and incubation of samples at 4°C for 30 min. The protection of DegP2 against degradation by trypsin was tested by immunoblotting. As a reference, the protection of PsbO, a peripheral protein localized at the lumenal side of the membrane, is shown.
To determine whether DegP2 protease is a peripheral or an integral membrane protein, the isolated thylakoid membranes were washed with salt to release extrinsic membrane proteins. Extraction of DegP2 with 0.5 M NaCl or 0.5 M CaCl2 from the thylakoids indicated that this protease is a peripheral membrane protein (Figure 3C, upper panel). Furthermore, the presence of DegP2 in the NaCl supernatant suggested that this protein is associated with the stromal side of the membranes. As a reference, the fractionation pattern of the 33 kDa protein from the oxygen-evolving complex (PsbO) of PSII is shown (Figure 3C, lower panel). The PsbO is a peripheral membrane protein located at the lumenal side of the thylakoid membrane (Andersson et al., 1984). It was demonstrated that PsbO is easily extracted from the membrane with 1 M CaCl2 but not with 1 M NaCl (Murata and Miyao, 1985).
The topology of DegP2 within the thylakoid membranes was verified further by its susceptibility to added trypsin (Figure 3D, right panel). Incubation of the isolated thylakoid membranes with trypsin led to digestion of DegP2, demonstrating that this protein was not protected by the lipid bilayer and was thus accessible to trypsin attack. In contrast, PsbO was protected against tryptic digestion (Figure 3D, left panel).
Expression of DegP2 differs under various stress conditions.
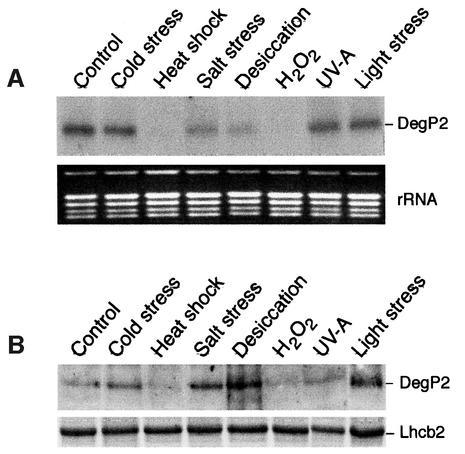
It was reported that heat shock treatment led to increased expression of E.coli DegP/HtrA (Gottesman, 1996), human HtrA2 (Gray et al., 2000) and Arabidopsis DegP1 (Itzhaki et al., 1998). To test the effect of various stresses on expression of the DegP2 gene, the leaves of Arabidopsis were exposed to cold stress, heat shock, high salt, desiccation, oxidative stress, UV-A radiation and light stress, and the level of DegP2 transcript and protein was assayed by northern (Figure 4A) or western blotting (Figure 4B), respectively. As a control, the leaves were exposed to ambient light and temperature conditions, and the amount of DegP2 transcript and protein expressed under these condition was used as a reference. Only traces of DegP2 transcript and protein were detected after exposure of leaves to elevated temperatures and treatment with H2O2. The amount of DegP2 transcript was also reduced after exposure of leaves to salt stress or desiccation (Figure 4A). In contrast, the amount of DegP2 protein in the thylakoid membranes was enhanced 2- to 4-fold under the latter two conditions as compared with the control sample. In addition, the light stress treatment promoted the accumulation of DegP2 in the thylakoid membrane (Figure 4B).
Fig. 4. Expression pattern of DegP2 under various stress conditions. (A) Arabidopsis leaves were exposed for 2 h to various stress conditions, and total RNA was isolated and used for northern blot analysis. As a reference, the rRNA pattern in the gel, visualized by staining with ethidium bromide, is shown. (B) Western blot analysis of the DegP2 level after 2 h exposure of leaves to various stress conditions. As a reference, the level of Lhcb2, the major antenna protein of the PSII, was assayed by SDS–PAGE and Coomassie Blue staining.
Physiological target of DegP2 is photodamaged D1 protein of the PSII reaction centre
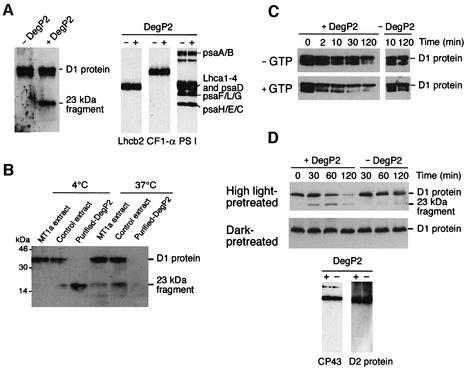
Since the degradation of the D1 protein following photoinhibition is catalysed by a membrane-bound serine-type protease (Aro et al., 1993), it was of interest to assess the physiological target of DegP2 protease. Addition of His-tagged purified DegP2 to isolated thylakoid membranes, whose endogenous proteolytic activities were inhibited by heating, resulted in the specific disappearance of a 32 kDa membrane protein, as verified by SDS–PAGE and Coomassie Blue staining (not shown). The 32 kDa protein band was not influenced when control E.coli extract was added to the reconstitution assay (not shown). To determine the identity of the 32 kDa protein, immunoblotting analyses using a polyclonal antibody against the D1 protein were performed. The band degraded during the reconstitution with purified recombinant DegP2 was identified as the D1 protein of the PSII reaction centre (Figure 5A, left panel). The immunoblotting also detected the typical N-terminal 23 kDa proteolytic fragment of the D1 protein (Greenberg et al., 1987). The corresponding C-terminal 10 kDa proteolytic fragment (Canovas and Barber, 1993) was not recognized by the D1 antibodies used. Immunoblotting with antibodies against Lhcb2, the α-subunit of the F1 ATPase and purified total PSI (Tjus and Andersson, 1991) demonstrated that the level of these proteins in the thylakoid membranes did not change significantly in the presence of DegP2 (Figure 5A, right panel). This confirms the selectivity of DegP2 proteolysis.
Fig. 5. DegP2 mediates primary cleavage of the damaged D1 protein from the PSII reaction centre in a GTP-dependent manner. (A) Isolated heat-treated thylakoid membranes were incubated in the absence (– DegP2) or presence (+ DegP2) of purified recombinant DegP2 and the degradation of D1 protein was assayed by immunoblotting after 30 min incubation at 37°C. Immunoblots showing the α-subunit of the F1 ATPase (CF1-α), several subunits of the PSI complex and the major antenna protein of the PSII complex (Lhcb2) confirm the selectivity of the DegP2 action. (B) Isolated heat-treated thylakoid membranes were reconstituted with purified recombinant DegP2 fractions and degradation of D1 protein was assayed by immunoblotting after incubation of samples at 4 or 37°C for 6 h. As negative controls, E.coli fractions without expressed DegP2 (control) or fractions with the overexpressed non-proteolytic protein class 1a metallothionein (MT1a) were reconstituted with thylakoid membranes and analysed in parallel. (C) Kinetics of D1 protein degradation in heat-treated thylakoid membranes in the absence (– GTP) or presence (+ GTP) of 2 mM GTP. Thylakoid membranes were reconstituted either with purified recombinant DegP2 (+ DegP2) or with E.coli fractions containing an overexpressed MT1a (– DegP2). (D) Isolated thylakoid membranes were washed with 0.5 M NaCl to remove endogenous DegP2 activity and the D1 protein was photodamaged by illumination at 5000 µmol/m2/s for 90 min at 0°C. The thylakoid membranes were then used for reconstitution with purified recombinant DegP2 and the degradation of D1 protein was assayed by immunoblotting (upper and middle panels). In control assays, degradation of CP43 and D2 proteins was assayed by immunoblotting after 120 min of incubation of high-light-pre-treated samples in the presence (+) or absence (--) of purified recombinant DegP2 (lower panel).
The DegP/HtrA protease in E.coli was reported to have a molecular chaperone function at low temperatures while having a proteolytic activity at elevated temperatures (Spiess et al., 1999). It was demonstrated that below 22°C, bacterial DegP/HtrA was proteolytically inactive. To test whether DegP2 in higher plants shows a similar behaviour, the reconstitution assays were performed at 4 and 37°C in the presence or absence of recombinant DegP2. The results (Figure 5B) revealed that in the presence of purified DegP2, the D1 protein was degraded at both temperatures but the 23 kDa proteolytic fragment was detected only in assays performed at 4°C. This fragment was missing in assays carried out at 37°C, suggesting that either overexpressed DegP2 was involved in a secondary cleavage or endogenous thylakoid proteases retained partial activity. In control assays reconstituted with E.coli extract without recombinant DegP2 or with E.coli extract expressing class 1a metallothionein, only small amounts of the 23 kDa degradation fragment were detected at 37°C. This speaks in favour of the possibility that with incubation time, denatured endogenous DegP2 recovered a residual proteolytic activity. These results show that DegP2 can indeed catalyse the primary cleavage of the D1 protein that has been denatured by heat treatment, and this process is not dependent on temperature. The low temperature, however, significantly decreased the kinetics of the degradation of the 23 kDa proteolytic fragment (Figure 5B), probably by partially inhibiting the FtsH protease.
It was found recently that GTP promoted the primary cleavage within the D–E loop of the D1 protein mediated by an endogenous protease (Spetea et al., 1999, 2000). To test whether the degradation of the D1 protein by recombinant DegP2 is stimulated by the presence of GTP, the kinetics of D1 degradation were measured in the presence and absence of added GTP. The results revealed (Figure 5C, left panel) that the degradation of the D1 protein was stimulated up to 2- to 4-fold by addition of GTP. In the absence of recombinant DegP2, the addition of GTP did not influence the level of D1 protein significantly in heat-inactivated thylakoid membranes (Figure 5C, right panel).
We designed an experiment to investigate whether DegP2 can degrade the D1 protein in thylakoids photoinhibited by strong illumination. Such a treatment is proposed first to induce irreversible oxidative damage, which in turn triggers a conformational change in the D1 protein, thereby exposing sites that are normally shielded from proteolysis (Prasil et al., 1992; Aro et al., 1993). Using dark- or high-light-pre-treated thylakoid membranes, where endogenous proteolytic activity directed against the D1 protein was partially removed by NaCl washes, we show that added recombinant DegP2 can recognize and degrade light-damaged D1 protein (Figure 5D, upper and middle panels). In the absence of recombinant DegP2, only traces of the 23 kDa proteolytic fragment could be detected and were probably related to the activity of remaining endogenous DegP2. In contrast, the D1 protein in the dark control sample was not prone to proteolytic degradation by the overexpressed DegP2 protease. Turnover of the reaction centre counterpart of D1, the D2 protein, and a slow degradation of CP43 have been reported under strong photoinhibitory conditions (Prasil et al., 1992; Zer and Ohad, 1995). We tested whether the proteolytic activity of recombinant DegP2 was also directed at both these proteins. Immunoblot analysis showed (Figure 5D, lower panel) that neither the D2 protein nor CP43 was degraded by DegP2 protease under the conditions tested.
Discussion
Since its discovery 20 years ago (Mattoo et al., 1981), the rapid turnover of the D1 protein has been the subject of intense research. It was shown that the D1 protein in its natural conformation is not prone to proteolytic degradation, and a light-dependent production of oxidative species in PSII upon inactivation of electron transport was proposed to modify the D1 protein irreversibly, trigger it for degradation (Prasil et al., 1992; Aro et al., 1993) and allow for repair through insertion of a new protein copy (Kyle et al., 1984; van Wijk et al., 1996; Melis, 1999). It was demonstrated that the primary cleavage of the damaged D1 protein occurs on the stromal loop connecting the membrane-spanning helices D and E (Greenberg et al., 1987). Subsequently, a proteolytic activity performing this process in isolated thylakoid membranes was characterized in biochemical terms, such as a serine-type catalytic mechanism (Aro et al., 1993), pH optimum (Rintamäki et al., 1996) and GTP dependence (Spetea et al., 1999). Based on fractionation of thylakoid membranes and characteristics of proteolytic cleavage, it was proposed that one of the reaction centre polypeptides might possess a proteolytic activity, and the possibility of autocatalytic cleavage of the D1 protein was discussed (Shipton and Barber, 1991). The lack of an identified protease has even led to models where the D1 protein was suggested to undergo chemical photocleavage of peptide bonds rather than proteolysis by enzymatic activities (Miyao, 1994).
Here we describe the identification and characterization of Arabidopsis DegP2 protease, which can perform the primary cleavage of the photodamaged D1 protein in isolated thylakoid membranes.
DegP2 is a member of a large family of related Deg/Htr serine proteases, which are found in most organisms, including bacteria (Gottesman, 1996), humans (Spiess et al., 1999) and plants (Itzhaki et al., 1998; this work). Database searches revealed that at least 13 different Deg/Htr protease homologues, with various predicted subcellular locations, are present in A.thaliana. The chloroplast DegP2 is derived from a 19 exon gene located on chromosome 2 of Arabidopsis and is constitutively expressed in green plant tissues under ambient environmental conditions (not shown).
The biochemical characteristics of DegP2 protease are in agreement with those reported in the past for an unknown protease directed against D1 protein (for a review see Aro et al., 1993). The DegP2 protease shows a serine-type ATP-independent proteolytic activity (Figure 2B), with an optimum at an alkaline pH (not shown). Furthermore, the presence of GTP accelerates the kinetics of D1 protein degradation by recombinant DegP2 protease (Figure 5C).
The D1 protein degradation measured in isolated thylakoid membranes in the presence of inhibitors of cytoplasmic and chloroplast protein synthesis suggested that the D1 protease is relatively stable and permanently present (Aro et al., 1993). We measured the kinetics of autodegradation of recombinant DegP2 and calculated that at 37°C the half-life of this protein is ∼40 min (not shown). A human homologue of DegP, the HtrA2 protease (Gray et al., 2000), and an E.coli DegP/HtrA protease (Kim et al., 1999) were also reported to be unstable and to undergo rapid autoproteolysis.
The DegP/HtrA protease in E.coli is a stress-induced protein with an increased level upon heat shock and in the presence of misfolded proteins (Lipinska et al., 1989). It was shown that this protease was essential for alleviation of the deleterious effect of some oxidants, and that this protection was directed mainly towards membrane proteins (Skórko-Glonek et al., 1999). The level of human L56/HtrA1 mRNA was also increased ∼7-fold in osteoarthritic tissues, and this indicates the possibility that this protein has a role under the conditions of cellular stress present in disease situations (Hu et al., 1998). Our data (Figure 4) demonstrated that neither the level of the DegP2 transcript nor that of the protein increased under heat shock conditions. In contrast, exposure of leaves to elevated temperature led to a reduction in DegP2 expression. In this respect, the expression pattern of chloroplast DegP2 differs from that of DegP/HtrA and resembles more those of two other E.coli paralogues, DegQ/HhoA and DegS/HhoB (Pallen and Wren, 1997).
A common feature of the Deg/Htr protease family is the presence of PDZ domains. DegP2 contains one PDZ domain located at its C-terminus. PDZ domains have been found to be involved in a variety of protein–protein interactions, including the assembly of multimeric complexes (Fanning and Anderson, 1996). It has been demonstrated for the protease subunit ClpX/Hsp100 that the PDZ domains alone were capable of substrate recognition and binding (Levchenko et al., 1997). Furthermore, deletion of one or two PDZ domains in bacterial DegP/HtrA led to a complete loss of protease activity, indicating the importance of this domain for the enzymatic process (Spiess et al., 1999). Thus, it is likely that the PDZ domain in DegP2 plays a role in recognition and binding of the D–E loop of damaged D1 protein and/or participates in the assembly of oligomeric structures. Biochemical and biophysical experiments have indicated that the purified DegP/HtrA from E.coli exists as a dodecamer consisting of two stacks of hexameric rings (Kim et al., 1999). It was suggested that the PDZ domains contain the information necessary for proper assembly of the functional hexameric structure of this protease (Sassoon et al., 1999). The generation of DegP2 deletion constructs partially or completely depleted in the PDZ domain is in progress, and these constructs will allow the investigation of the significance of this domain for DegP2 structure and proteolytic function.
We demonstrated that DegP2 is associated with the stromal side of the non-appressed region of the thylakoid membranes. The release of DegP2 with high salt washes in the absence of detergent indicates that it is not a transmembrane protein, despite the presence of a predicted hydrophobic C-terminal domain. In contrast, the DegP1 protease from pea and Arabidopsis is peripherally bound to the lumenal side of the thylakoid membrane, and could only be removed by a combination of high salt and non-ionic detergent (Itzhaki et al., 1998). The distribution of two closely related DegP proteases on both the stromal and lumenal sides of the thylakoid membrane might be required for efficient degradation of the polytopic D1 protein. Although the physiological substrates of DegP1 are unknown as yet, it was suggested (Itzhaki et al., 1998) that this protease might be involved in two ATP-independent cleavage events at the lumenal loops of the D1 protein, which occur under conditions that preferably induce donor-side photoinhibition of PSII (Andersson and Barber, 1996).
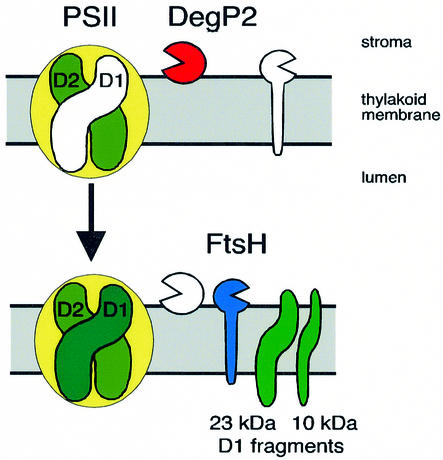
Bacterial DegP/HtrA protease has been implicated in tolerance to various stresses such as oxidation, salt, pH and heat (Spiess et al., 1999). One of the physiological roles of DegP/HtrA in E.coli is to degrade denatured proteins formed in the cellular envelope during stress conditions (Skórko-Glonek et al., 1999). We demonstrated that one physiological role of DegP2 in Arabidopsis is to perform the primary cleavage of the photodamaged D1 protein of PSII prior to its removal by secondary proteolysis. A working model for the proteolytic events occurring during D1 protein degradation is shown in Figure 6. DegP2 catalyses the primary GTP-stimulated cleavage of the damaged D1 protein at the stromal loop connecting the membrane-spanning helices D and E. The N-terminal 23 kDa and C-terminal 10 kDa fragments are then degraded further by the thylakoid membrane FtsH protease (Lindahl et al., 2000), allowing a new copy of the D1 protein to be integrated into the reaction centre and thus allowing restoration of photosynthetic function. It remains to be established how the damaged PSII complexes located in the appressed grana thylakoid regions can interact with the DegP2 protease located in the non-appressed stroma-exposed regions. Possibly, photodamaged PSII centres migrate to the stroma-exposed thylakoid regions, consistent with the finding that the two primary degradation fragments (Barbato et al., 1991) and the FtsH protease (Lindahl et al., 1996) are located in these regions where the newly synthesized D1 protein precursor is also integrated (Aro et al., 1993; van Wijk et al., 1996).
Fig 6. Model for D1 protein degradation. The DegP2 protease peripherally attached to the stromal side of the thylakoid membranes performs the primary cleavage of the photodamaged D1 protein of the PSII reaction centre at the stromal loop between helices D and E, generating 23 and 10 kDa proteolytic fragments. Simultaneously, a newly synthesized functional copy replaces the degraded D1 protein. The 23 kDa fragment is degraded further by an integral membrane metalloendopeptidase, FtsH (Lindahl et al., 2000).
It was reported that E.coli DegP/HtrA protein has both general molecular chaperone and proteolytic activities (Spiess et al., 1999). The chaperone function dominated at low temperatures, while the proteolytic activity was present at elevated temperatures >22°C. Our data show that DegP2 was able to cleave the D1 protein independently of temperature between 4 and 37°C. Whether DegP2 can act as a molecular chaperone still remains to be elucidated. It is very tempting to speculate that having a chaperone activity, DegP2 could be involved not only in the proteolytic phase but also in the assembly phase during the PSII repair cycle.
In conclusion, we propose that the DegP2 protease is the crucial missing link in our understanding of the chain of events leading to damage and repair of oxygen-evolving PSII in photoinhibited plants.
Materials and methods
Growth of plants and stress conditions
Arabidopsis thaliana L. cv. Columbia were grown in a growth chamber on soil at 25°C at a light intensity of 100 µmol/m2/s under short-day conditions.
Light stress treatment was performed on mature leaves, detached from 4- to 5-week-old plants, floating on water and exposed to a light intensity of 2500 µmol/m2/s for 2 h. For cold or heat stress, detached leaves floating on water were transferred for 2 h to incubators set at 4 or 42°C, respectively. Desiccation stress was performed on leaves dehydrated on Whatman 3MM paper at room temperature at a light intensity of 10 µmol/m2/s. After 2 h of incubation, the relative water content of leaves was reduced to 50%. Leaves subjected to a high salt or an oxidative stress were submerged for 2 h in 400 mM NaCl or in 2% H2O2 solutions, respectively. UV-A treatment was performed by illumination of detached leaves with a UV lamp (366 nm) at a light intensity of 25 µmol/m2/s for 2 h. Plant material was collected and either used immediately for extractions or frozen in liquid nitrogen and stored at –70°C for further preparations.
Cloning of the DegP2 gene
Based on the ORF F17A22.33 localized on chromosome 2 predicted to encode a putative DegP2 protease in Arabidopsis, a pair of PCR primers (5′-AGTGCCGCCTCCGTAGCA-3′ and 5′-TTATGCCCACACCAGTCCATC-3′) were designed and used for amplification of the DegP2 ORF from the λ-ZAP (Stratagene) cDNA library (1 × 103 p.f.u.) prepared from Arabidopsis seedlings. The PCR cycling profile (30 cycles) was denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 2 min. The amplified 1821 bp cDNA fragment was purified (Qiaex; Qiagen) and cloned into the pCR2.1 cloning vector (Invitrogen) prior to sequencing of both DNA strands (CyberGene, Stockholm, Sweden). A search of the expressed sequence tag (EST) database with the obtained DegP2 sequence identified the Arabidopsis EST clone 133N20T7 (2280 bp) identical to DegP2 collected into tentative consensus TC54010.
DNA analysis
Genomic DNA was extracted from leaves as described by Doyle and Doyle (1990). DNA (10 µg per reaction) was digested by restriction enzymes, and DNA fragments were separated in 0.8% agarose gel and transferred to Hybond-N+ membrane (Amersham Pharmacia) according to the manufacturer’s protocol. The cDNA probe was labelled with [α-32P]dCTP using a megaprime DNA labelling kit (Amersham Pharmacia). The hybridization and washes were performed under low and high stringency conditions according to Sambrook et al. (1989).
RNA analysis
Total RNA was extracted from control or stress-treated leaves using an RNeasy mini kit (Qiagen) according to the manufacturer’s protocol. After separation of 5 µg of RNA in 1.2% agarose gel, RNA was transferred to a Hybond-N+ membrane prior to the hybridization according to Sambrook et al. (1989).
Overexpression of DegP2 in E.coli and production of polyclonal antibodies
The DegP2 cDNA was amplified using a pair of PCR primers (5′-GCCGCCTCCGTAGCAAAC-3′, 5′-TGCCCACACCAGTCCATCA-3′) and the cDNA plasmid of EST clone 133N20T7 as template. The DegP2 protein was expressed in E.coli as a fusion protein with the N-terminal-attached thioredoxin and C-terminal-attached His tag (His6) using a kit (pBAD/Topo® ThioFusion™ Expression System, Invitrogen), according to the manufacturer’s protocol. Recombinant DegP2 protein was purified under denaturing conditions by affinity chromatography on Ni-NTA–agarose (Qiaexpress; Qiagen) and eluted from the column with 200 mM imidazole. The DegP2-containing fractions were separated by SDS–PAGE and transferred to a nitrocellulose membrane prior to raising polyclonal antibodies in rabbits (BioGenes, Berlin, Germany).
Protein analysis
Isolated thylakoid membrane and stroma proteins were separated by SDS–PAGE (Laemmli, 1970) generally using 10% polyacrylamide mini-gels (Hoefer mini gel system). The gels were loaded on an equal protein basis. Immunoblotting was carried out according to Towbin et al. (1979) using enhanced chemiluminescence (ECL; Amersham International) as the detection system.
Activity assays and inhibitor studies
For activity assays, 1 µg of His-tagged purified DegP2 protease was separated at 4°C on 10% SDS–polyacrylamide gels containing 0.2% gelatine (Type B bloom 75; Sigma) as a non-specific protease substrate. The gels were then incubated in 50 mM Tris pH 8.0, 5 mM MgCl2, 1% Triton X-100 for 30 min at 25°C to replace SDS and to allow the renaturation of the DegP2 protease. The buffer was changed to one that was detergent free and the gel was incubated for an additional 16 h at 37°C to allow degradation of gelatine. After staining the gel with Coomassie Blue solution (40% ethanol, 10% acetic acid, 0.1% Coomassie Blue R-250) and destaining with 40% ethanol and 10% acetic acid solution, a white zone on a blue background was visible, corresponding to the position of the active DegP2 protease.
To classify the DegP2 protease, activity gels were incubated in the absence (control) or presence of protease inhibitors: 100 µg/ml PMSF, 10 µg/ml DCI, 1 µg/ml aprotinin, 1 µg/ml E64 [3-carboxy-trans-2,3-epoxypropyl-leucylamido(4-guanidino)butane], 1 µg/ml pepstatin and 500 µg/ml EDTA according to the manufacturers’ protocols (Boehringer Mannheim and Sigma).
Localization of DegP2 in chloroplast subfractions and its topology in the thylakoid membranes
Intact Arabidopsis chloroplasts were isolated and purified on a Percoll gradient according to Cline (1986). For further fractionation, chloroplasts were ruptured by osmotic shock in 50 mM Tris pH 8.0 and 5 mM MgCl2 at a chlorophyll concentration of 1 mg/ml for 10 min at 4°C, and thylakoid and stromal fractions were separated by centrifugation at 10 000 g for 10 min at 4°C. For preparation of thylakoid lumen, isolated thylakoids were resuspended in the same buffer as above, sonicated 10 times for 30 s at 4°C (power output 8.0; Misonix Inc., Farmingdale, NY) and the thylakoid membranes separated from the soluble lumen fraction by centrifugation at 220 000 g for 1 h at 4°C. Proteins in each fraction were separated by SDS–PAGE and the DegP2 location assayed by immunoblotting.
For isolation of appressed and non-appresed membrane regions, thylakoids were mechanically disrupted by Yeda press, and membrane fragments separated by differential centrifugation and aqueous polymer two-phase partitioning as described by Andersson and Anderson (1980).
For isolation of the peripheral thylakoid proteins, the thylakoid membranes were resuspended in 25 mM MES pH 6.5 at a chlorophyll concentration of 0.1 mg/ml and the extrinsic membrane proteins were extracted with 0.5 M NaCl or 0.5 M CaCl2 for 30 min at 4°C in darkness under gentle stirring. The suspension was centrifuged at 220 000 g for 1 h at 4°C and the pellet (integral membrane proteins) and supernatant (peripheral membrane proteins) were analysed by SDS–PAGE and immunoblotting.
The topology of DegP2 protease was tested by protease protection assay. Isolated thylakoid membranes were resuspended in 50 mM HEPES pH 8.0 and 330 mM sorbitol at a chlorophyll concentration of 1 mg/ml and incubated for 30 min at 4°C with 50 µg/ml trypsin. Protection of DegP2 against degradation by trypsin was tested by immunoblotting.
Reconstitution of recombinant DegP2 with thylakoid membranes
For reconstitution studies, thylakoid membranes were isolated and aliquots containing 9 µg of chlorophyll were denatured for 10 min at 90°C to arrest endogenous proteolytic activities. After addition of His-tagged purified recombinant DegP2 (0.1 µg protein), assays were incubated in 50 mM Tris pH 9.5, 5 mM MgCl2, 2 mM GTP for 2 h at 4 or 37°C. Degradation of D1 protein was assayed by SDS–PAGE and immunoblotting.
For light-triggered D1 protein degradation, isolated thylakoid membranes were washed with 0.5 M NaCl to reduce endogenous DegP2 activity and then incubated either in darkness or in high light (5000 µmol/m2/s) for 90 min at 0°C as described by Spetea et al. (1999). Aliquots of thylakoid membranes were used for reconstitution studies, and the degradation of D1 protein was followed by immunoblotting in the presence or absence of recombinant DegP2 for 120 min at 37°C and a light intensity of 10 µmol/m2/s.
DDBJ/EMBL/GenBank accession number
The cDNA sequence of the DegP2 gene is available in GenBank under accession No. AF245171.
Acknowledgments
Acknowledgements
We thank P.Dessi for critical reading of the manuscript, the Arabidopsis Biological Resource Center at Ohio State University for providing the DegP2 EST cDNA clone (133N20T7), and R.Oelmüeller for providing the Arabidopsis seedling cDNA library and antibodies against the α-subunit of the F1 ATPase. E.-M.Aro and K.J.van Wijk kindly provided the antibodies against D1 protein, and N.H.Chua and A.Trebst provided antibodies against CP43 and D2 proteins, respectively. M.Dunaeva provided recombinant E.coli expressing Arabidopsis class 1a metallothionein as a fusion protein with the N-terminal-attached thioredoxin and C-terminal-attached His tag. This work was supported by research grants from the Carl Tryggers Foundation, the Swedish Natural Science Research Council and the Swedish Strategic Foundation to I.A. and B.A., and the Ekströms fellowship to K.H.
REFERENCES
- Andersson B. and Anderson,J.M. (1980) Lateral heterogeneity in the distribution of chlorophyll–protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta, 593, 427–440. [DOI] [PubMed] [Google Scholar]
- Andersson B. and Barber,J. (1996) Mechanism of photodamage and protein degradation during photoinhibition of photosystem II. In Baker,N.R. (ed.), Advances in Photosynthesis. Photosynthesis and Environment. Vol. 5. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 101–121.
- Andersson B., Larsson,C., Jansson,C., Ljungberg,U. and Åkerlund,H.-E. (1984) Immunological studies on the organization of proteins in photosynthetic oxygen evolution. Biochim. Biophys. Acta, 766, 21–28. [Google Scholar]
- Aro E.-M., Virgin,I. and Andersson,B. (1993) Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta, 1143, 113–134. [DOI] [PubMed] [Google Scholar]
- Barbato R., Friso,G., Giardi,M.T., Rigoni,F. and Giacometti,G.M. (1991) Breakdown of the photosystem II reaction center D1 protein under photoinhibitory conditions: identification and localization of the C-terminal degradation products. Biochemistry, 30, 10220–10226. [DOI] [PubMed] [Google Scholar]
- Barber J. and Andersson,B. (1992) Too much of a good thing: light can be bad for photosynthesis. Trends Biochem. Sci., 17, 61–66. [DOI] [PubMed] [Google Scholar]
- Barber J. and Kühlbrandt,W. (1999) Photosystem II. Curr. Opin. Struct. Biol., 9, 469–475. [DOI] [PubMed] [Google Scholar]
- Canovas P.M. and Barber,J. (1993) Detection of a 10 kDa breakdown product containing the C-terminus of the D1-protein in photoinhibited wheat leaves suggests an acceptor side mechanism. FEBS Lett., 324, 341–344. [DOI] [PubMed] [Google Scholar]
- Cline K. (1986) Import of proteins into chloroplasts. Membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J. Biol. Chem., 261, 14804–14810. [PubMed] [Google Scholar]
- Cserzo M., Wallin,E., Simon,I., von Heijne,G. and Elofsson,A. (1997) Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng., 10, 673–676. [DOI] [PubMed] [Google Scholar]
- Depka B., Jahns,P. and Trebst,A. (1998) β-carotene to zeaxanthin conversion in the rapid turnover of the D1 protein of photosystem II. FEBS Lett., 424, 267–270. [DOI] [PubMed] [Google Scholar]
- Doyle J.J. and Doyle,J.L. (1990) Isolation of plant DNA from fresh tissue. Focus, 12, 13–15. [Google Scholar]
- Emanuelsson O., Nielsen,H. and von Heijne,G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci., 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fanning A.S. and Anderson,J.M. (1996) Protein–protein interactions: PDZ domains networks. Curr. Biol., 6, 1385–1388. [DOI] [PubMed] [Google Scholar]
- Frishman D. and Argos,P. (1996) Incorporation of long-distance interactions into a secondary structure prediction algorithm. Protein Eng., 9, 133–142. [DOI] [PubMed] [Google Scholar]
- Gottesman S. (1996) Proteases and their targets in Escherichia coli. Annu. Rev. Genet., 30, 465–506. [DOI] [PubMed] [Google Scholar]
- Gray C.W. et al. (2000) Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur. J. Biochem., 267, 5699–5710. [DOI] [PubMed] [Google Scholar]
- Greenberg B.M., Gaba,V., Mattoo,A.K. and Edelman,M. (1987) Identification of a primary in vivo degradation product of the rapidly turning-over 32 kD protein of photosystem II. EMBO J., 6, 2865–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S.I., Carozza,M., Klein,M., Nantermet,P., Luk,D. and Crowl,R.M. (1998) Human HtrA, an evolutionary conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J. Biol. Chem., 273, 34406–34412. [DOI] [PubMed] [Google Scholar]
- Itzhaki H., Naveh,L., Lindahl,M., Cook,M. and Adam,Z. (1998) Identification and characterization of DegP, a serine protease associated with the lumenal side of the thylakoid membrane. J. Biol. Chem., 273, 7094–7098. [DOI] [PubMed] [Google Scholar]
- Kim K.I., Park,S.C., Kang,S.H., Cheong,G.W. and Chung,C.H. (1999) Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J. Mol. Biol., 294, 1363–1374. [DOI] [PubMed] [Google Scholar]
- Kyle D.J., Ohad,I. and Arntzen,C.J. (1984) Membrane protein damage and repair: selective loss of a quinone-protein function in chloroplast membranes. Proc. Natl Acad. Sci. USA, 81, 4070–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Levchenko I., Smith,C., Walsh,N., Sauer,R. and Baker,T. (1997) PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell, 91, 939–947. [DOI] [PubMed] [Google Scholar]
- Lindahl M., Tabak,S., Cseke,L., Pichersky,E., Andersson,B. and Adam,Z. (1996) Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem., 271, 29329–29334. [DOI] [PubMed] [Google Scholar]
- Lindahl M., Spetea,C., Hundal,T., Oppenheim,A., Adam,Z. and Andersson,B. (2000) The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell, 12, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska B., Fayet,O., Baird,L. and Georgopoulos,C. (1989) Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol., 171, 1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.P., Humphries,S. and Falkowski,P.G. (1994) Photoinhibition of photosynthesis in nature. Annu. Rev. Plant Physiol. Plant Mol. Biol., 45, 633–662. [Google Scholar]
- Mattoo A.K., Pick,U., Hoffman-Falk,H. and Edelman,M. (1981) The rapidly metabolized 32,000-dalton polypeptide of the chloroplast is the ‘proteinaceous shield’ regulating photosystem II electron transport and mediating diuron herbicide sensitivity. Proc. Natl Acad. Sci. USA, 78, 1572–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A. (1999) Photosystem-II damage and repair cycle in chloroplasts: what modulates the rate of photodamage? Trends Plant Sci., 4, 130–135. [DOI] [PubMed] [Google Scholar]
- Michel H. and Deisenhofer,J. (1986) X-ray diffraction studies on a crystalline bacterial photosynthetic reaction center. In Staehelin,L.A. and Arntzen,C.J. (eds), Encyclopedia of Plant Physiology, New Series, Vol. 19, Photosynthesis III, Photosynthetic Membranes and Light Harvesting Systems. Springer Verlag, Berlin, Germany, pp. 371–381.
- Miyao M. (1994) Involvement of active oxygen species in degradation of the D1 protein under strong illumination in isolated subcomplexes of photosystem II. Biochemistry, 33, 9722–9730. [DOI] [PubMed] [Google Scholar]
- Murata N. and Miyao,M. (1985) Extrinsic membrane proteins in the photosynthetic oxygen-evolving complex. Trends Biochem. Sci., 10, 122–124. [Google Scholar]
- Morrissey P.J., McCauley,S.W. and Melis,A. (1986) Differential detergent-solubilization of integral thylakoid membrane complexes in spinach chloroplasts. Localization of photosystem II, cytochrome b6–f complex and photosystem I. Eur. J. Biochem., 160, 389–393. [DOI] [PubMed] [Google Scholar]
- Nakai K. and Kanehisa,M. (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics, 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen M.J. and Wren,B.W. (1997) The HtrA family of serine proteases. Mol. Microbiol., 26, 209–221. [DOI] [PubMed] [Google Scholar]
- Prasil O., Adir,N. and Ohad,I. (1992) Dynamics of photosystem II: mechanism of photoinhibition and recovery processes. In Barber,J. (ed.), Topics in Photosynthesis. Vol. 11. Elsevier, Amsterdam, The Netherlands, pp. 295–348.
- Rintamäki E., Kettunen,R. and Aro,E.-M. (1996) Differential D1 dephosphorylation in functional and photodamaged photosystem II centers. Dephosphorylation is a prerequisite for degradation of damaged D1. J. Biol. Chem., 271, 14870–14875. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sassoon N., Arie,J.P. and Betton,J.M. (1999) PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol. Microbiol., 33, 583–589. [DOI] [PubMed] [Google Scholar]
- Shipton C.A. and Barber,J. (1991) Photoinduced degradation of the D1 polypeptide in isolated reaction centers of photosystem II. Evidence of an autoproteolytic process triggered by the oxidizing side of the photosystem. Proc. Natl Acad. Sci. USA, 88, 6691–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skórko-Glonek J., Zurawa,D., Kuczwara,E., Woznak,M., Wypych,Z. and Lipinska,B. (1999) The Escherichia coli heat shock protease HtrA participates in defense against oxidative stress. Mol. Gen. Genet., 262, 342–350. [DOI] [PubMed] [Google Scholar]
- Spetea C., Hundal,T., Lohmann,F. and Andersson,B. (1999) GTP bound to chloroplast thylakoid membranes is required for light-induced, multienzyme degradation of the photosystem II D1 protein. Proc. Natl Acad. Sci. USA, 96, 6547–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetea C., Keren,N., Hundal,T., Doan,J.M., Ohad,I. and Andersson,B. (2000) GTP enhances the degradation of the photosystem II D1 protein irrespective of its conformational heterogeneity at the QB site. J. Biol. Chem., 275, 7205–7211. [DOI] [PubMed] [Google Scholar]
- Spiess C., Beil,A. and Ehrmann,N. (1999) A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell, 97, 339–347. [DOI] [PubMed] [Google Scholar]
- Telfer A., Bishop,S.M., Phillips,D. and Barber,J. (1994) Isolated photosynthetic reaction center of photosystem II as a sensitizer for the formation of singlet oxygen. Detection and quantum yield determination using a chemical trapping technique. J. Biol. Chem., 269, 13244–13253. [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 11, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin,T. and Gordon,J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjus S.E. and Andersson,B. (1991) Extrinsic polypeptides of spinach photosystem I. Photosynth. Res., 27, 209–219. [DOI] [PubMed] [Google Scholar]
- Trebst A. (1986) The topology of the plastoquinone and herbicide binding peptides of photosystem II in the thylakoid membranes. Z. Naturforsch., 41c, 240–245. [Google Scholar]
- Van Wijk K.J., Andersson,B. and Aro,E.-M. (1996) Kinetic resolution of the incorporation of the D1 protein into photosystem II and localization of assembly intermediates in thylakoid membranes of spinach chloroplasts. J. Biol. Chem., 271, 9627–9636. [DOI] [PubMed] [Google Scholar]
- Vass I., Styring,S., Hundal,T., Koivuniemi,A. and Aro,E.-M. (1992) Reversible and irreversible intermediates during photoinhibition of photosystem II: stable reduced QA species promote chlorophyll triplet formation. Proc. Natl Acad. Sci. USA, 89, 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin I., Ghanotakis,D.F. and Andersson,B. (1990) Light-induced D1-protein degradation in isolated photosystem II core complexes. FEBS Lett., 269, 45–48. [DOI] [PubMed] [Google Scholar]
- Zer H. and Ohad,I. (1995) Photoinactivation of photosystem II induces changes in the photochemical reaction center II abolishing the regulatory role of the QB site in the D1 protein degradation. Eur. J. Biochem., 231, 448–453. [DOI] [PubMed] [Google Scholar]