Abstract
Duchenne muscular dystrophy (DMD) is a severe muscle wasting disease arising from defects in the dystrophin gene, typically nonsense or frameshift mutations, that preclude the synthesis of a functional protein. A milder, allelic version of the disease, Becker muscular dystrophy, generally arises from in-frame deletions that allow synthesis of a shorter but still semifunctional protein. Therapies to introduce functional dystrophin into dystrophic tissue through either cell or gene replacement have not been successful to date. We report an alternative approach where 2′-O-methyl antisense oligoribonucleotides have been used to modify processing of the dystrophin pre-mRNA in the mdx mouse model of DMD. By targeting 2′-O-methyl antisense oligoribonucleotides to block motifs involved in normal dystrophin pre-mRNA splicing, we induced excision of exon 23, and the mdx nonsense mutation, without disrupting the reading frame. Exon 23 skipping was first optimized in vitro in transfected H-2Kb-tsA58 mdx myoblasts and then induced in vivo. Immunohistochemical staining demonstrated the synthesis and correct subsarcolemmal localization of dystrophin and γ-sarcoglycan in the mdx mouse after intramuscular delivery of antisense oligoribonucleotide:liposome complexes. This approach should reduce the severity of DMD by allowing a dystrophic gene transcript to be modified, such that it can be translated into a Becker-dystrophin-like protein.
Duchenne muscular dystrophy (DMD), an X-linked recessive disorder, is the most common form of muscular dystrophy, occurring at a frequency of about 1 in every 3,500 live male births (1). Arising from the absence of a functional dystrophin protein, the disease is characterized by severe, progressive muscle wasting and weakness that becomes clinically evident between the ages of 3 and 5 years. Affected boys have difficulty rising from the floor and eventually become restricted to a wheelchair by the age of 12 years, with 90% dying before their 20th birthday because of cardiac and respiratory complications (2). A third of DMD cases are the result of a de novo mutation in the dystrophin gene (3, 4), and consequently this disease can never be eradicated through genetic screening and counseling, placing additional emphasis on the need to develop a treatment for this disorder.
The vast majority of DMD mutations disrupt the dystrophin mRNA reading frame or introduce a stop codon that prematurely ends protein translation (5). In the less severe allelic form of the disease, Becker muscular dystrophy (BMD), dystrophin gene mutations are usually such that the mRNA reading frame is maintained. Thus in BMD patients, some functional gene product, albeit of reduced quantity and/or quality, is synthesized that contributes to the milder phenotype (6).
The mdx mouse (7) is one animal model that has been used to evaluate a variety of therapies for DMD, including myoblast transfer, dystrophin cDNA replacement through viral and plasmid vectors, and up-regulation of a homologous protein such as utrophin (8, 9). The genetic lesion in the mdx dystrophin gene is a nonsense mutation at base 3185 of the mRNA that causes premature termination of translation within exon 23. This nonsense mutation should preclude synthesis of a functional protein, yet rare dystrophin-positive (revertant) fibers have been observed after immunohistochemical staining of mdx dystrophic muscle (10, 11). Revertant fibers have also been observed in many DMD patients (12) and the canine model of DMD (13). Several RNA and protein studies have suggested that a frame-restoring exon-skipping mechanism is the most likely cause of these naturally occurring dystrophin-positive fibers (14, 15). Although the number of revertant fibers increases with age, their frequency is thought by some to be too low to be of any clinical benefit (16). Other studies have shown that some DMD boys with very low levels of dystrophin, as demonstrated by immunostaining, lost mobility some 2 years later than those with no detectable dystrophin (17).
We report a potential therapy for DMD based on the application of 2′-O-methyl antisense oligoribonucleotides (AOs) to induce specific exon skipping of dystrophic pre-mRNA in the mdx mouse. We have induced exon skipping as determined by reverse transcription–PCR (RT-PCR) amplification of RNA extracted from cultured cells after delivering AOs targeted to splice site sequences around the mdx mutation in exon 23. Dystrophin synthesis and correct localization to the sarcolemma of muscle fibers in vivo was demonstrated after intramuscular injections of the same AOs. The removal of dystrophin exon 23 does not disrupt the reading frame, so the induced mRNA can be translated into a Becker-dystrophin-like protein. This slightly shortened product has the potential to minimize the severity of DMD, because some variants of dystrophin in BMD patients correlate with a milder phenotype (17, 18). The consequences of DMD mutations arising from genomic deletions could be reduced by inducing specific removal of one or more adjacent exons to restore the reading frame. Although this type of approach is unlikely to completely cure all cases of DMD, the potential exists for a significant reduction in the severity of symptoms in those patients who do not have mutations involving crucial functional regions of the gene.
Materials and Methods
AOs.
HPLC-purified AOs (Geneworks, Adelaide, Australia) were designed complementary to the sequences available for introns 22 and 23 (GenBank Accession nos. AF062829 and AF062830, respectively). Their locations relative to exon 23 splice sites are indicated in Fig. 1. AO 5′SS-FITC was identical in sequence to AO 5′SS-20 but was also labeled at the 5′ end with fluorescein (IDT, Coralville, IA).
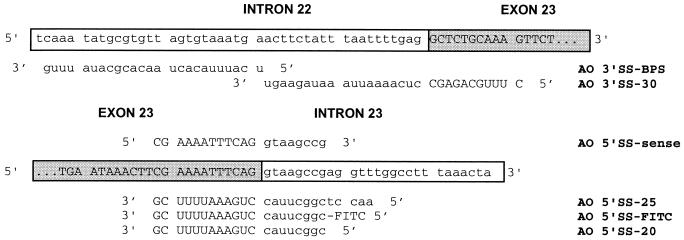
Figure 1.
Sequences and relative binding sites of AOs. The sequence of exon 23 of mouse dystrophin is indicated by capitals and a shaded box, whereas the intronic sequences neighboring the exon are indicated by lowercase and a plain box. The mdx mouse has a nonsense mutation at nucleotide 3185 causing premature termination of translation in this exon. The sequence and orientation of all the AOs used in this study are indicated, except for AO-random (5′-CCAGAUCGGA CGACGUCAGG ACAAC-3′), which was designed not to hybridize to any known sequence as validated by a blast database search. The fluorescein (FITC) group of AO 5′SS-FITC was attached to the 5′ end of the sequence as indicated.
Cell Culture and Transfection.
The H-2Kb-tsA58 mdx myoblasts (19) were proliferated at 33°C under a 10% CO2/90% air atmosphere in high-glucose DMEM supplemented with 20% fetal calf serum and 10% horse serum, 0.5% chicken embryo extract (Life Technologies), and 20 units/ml γ-interferon (Roche Molecular Biochemicals). Cells were then treated with trypsin and plated at 2 × 104 cells per well in 24-well plates coated with 50 μg/ml poly(d-lysine, Sigma) followed by 100 μg/ml Matrigel (Becton Dickinson) for differentiation and fusion at 37°C under 5% CO2 in 5% horse serum DMEM. H-2Kb-tsA58 mdx cells were transfected 48 h after trypsin treatment in a final volume of 0.5 ml of antibiotic- and serum-free Opti-MEM (Life Technologies). Each well was treated with 1 μg (≈300 nM) of appropriate AO complexed with 2 μg of Lipofectin (Life Technologies) according to the supplier's instructions. After 3 h of incubation, the transfection medium was replaced with DMEM supplemented with 5% horse serum.
RNA Extraction and Nested RT-PCR Analysis.
Cells were transfected as duplicate wells with Lipofectin–AO complexes, incubated as specified, and then pooled for total cellular RNA extraction with RNAzol B (Tel-Test, Friendswood, TX) directly on the plate 72 h after transfection. RT-PCR was carried out with 200 ng of total RNA as the starting material for 40 cycles of amplification in the Titan One-Tube RT-PCR system (Roche Molecular Biochemicals). Briefly, forward and reverse primers amplifying from exons 20 and 26, respectively (Ex20Fo, 5′-CAGAATTCTGCCAATTGCTGAG-3′; Ex26Ro, 5′-TTCTTCAGCTTGTGTCATCC-3′), were used at an annealing temperature of 55°C with an extension time of 2 min at 72°C. One microliter of the RT-PCR product was used as template in a 50-μl secondary nested PCR with 0.5 unit of Tth Plus (Biotech, Perth, Australia) and inner primers (Ex20Fi, 5′-CCCAGTCTACCACCCTATCAGAGC-3′; Ex26Ri, 5′-CCTGCCTTTAAGGCTTCCTT-3′). The secondary PCR was carried out for 30 cycles under cycling conditions identical to the primary amplification. Products were examined by electrophoresis in 3% agarose Tris/acetate/EDTA (TAE) gels.
In Vivo Treatments.
Over a 4-week period, 21-day-old C57BL/10ScSn-mdx mice were given weekly intramuscular injections of 1 μg of AO 5′SS-25 complexed with 2 μg of Lipofectin (2:1 weight ratio) prepared in saline. The right quadriceps-femoris complex received the injection on weeks 1, 2, 3, and 4, whereas the left received the injection on weeks 3 and 4 only. A green histological marking dye (Wak Chemie Medical, Hamburg, Germany) was added to the complexes to mark the injection site. One animal underwent a parallel injection regime using only 2 μg of Lipofectin in saline. Mice were killed 1 week after the final injection.
Sectioning and Immunohistochemical Staining.
Muscles were prepared for histological analysis by quenching in liquid nitrogen-cooled isopentane. Five-micrometer sections were cut on a Tissue-Tek II cryostat and fixed on glass slides. Immunostaining was slightly modified from methods described previously (20). Serial sections were sequentially blocked with normal mouse serum, followed by 0.1% avidin solution (Sigma), then 0.01% biotin solution (Sigma), and finally 5 mM levamisole (Sigma). Specific mouse monoclonal antibodies directed against the C terminus of dystrophin, DYS2 (1:30 dilution; NovoCastra, Newcastle, U.K.) and against γ-sarcoglycan (1:30 dilution; NovoCastra) were separately labeled with a biotinylated-anti-mouse Fab fragment (Dako) according to the supplier's instructions. Excess biotinylation agent was blocked with normal mouse serum. Color was detected with alkaline phosphatase-conjugated streptavidin (Dako) followed by Vector red fluorescent substrate (Vector Laboratories) according to the supplier's instructions. Duplicate serial sections from each treated muscle were stained on the same slides as age-matched control sections taken from the respective muscle of an untreated animal. All normal untreated and AO-treated mdx immunostained sections were photographed under the same time and exposure parameters to allow direct comparison. Duplicate slides were also stained in the absence of primary antibody and did not reveal any sarcolemmal fluorescence (data not shown).
Western Blotting.
Muscle samples were homogenized in 19 vol of protein extraction buffer [125 mM Tris⋅HCl, pH 6.8/0.1% SDS/2 M urea/10% (vol/vol) 2-mercaptoethanol/10% (wt/vol) glycerol], and total protein was quantitated with a Bio-Rad DC Protein Assay Kit. Two hundred micrograms of protein from each sample was loaded onto a 4–8% polyacrylamide gel containing 0.2% SDS in Tris–glycine buffer. Samples were electrophoresed overnight at 0.8 mA/cm at 4°C. The fractionated proteins were electroblotted onto a nitrocellulose membrane at 1000 mA for 7 h. Unreacted binding sites on the membrane were blocked with 5% (wt/vol) skim milk solution for 60 min at room temperature. Membranes were probed with DYS2 (1:25) for 60 min followed by three washes in TBST buffer (10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.05% Tween 20) and incubation with a horseradish peroxidase-conjugated secondary antibody (1:1000; Dako) for 60 min. Chemiluminescent detection of proteins was carried out with Lumi-Light Plus substrate (Roche Molecular Biochemicals) according to the supplier's instructions. Membranes were then exposed to Lumi-Film (Roche Molecular Biochemicals).
Results
AO Delivery in Vitro.
We have previously reported the induction of exon 23 skipping in cultured primary mdx myoblasts and C2C12 cells by using AO 5′SS-20 (21). In that series of experiments, an 18-mer directed at the 3′ splice site of intron 22 failed to induce any exon skipping. On the basis of these results, we designed a second generation of longer AOs (Fig. 1) and evaluated their ability to modify exon 23 processing in immortalized H-2Kb-tsA58 mdx cells.
A fluorescein labeled oligoribonucleotide, AO 5′SS-FITC, was used to determine the optimal Lipofectin/AO ratio for nuclear uptake in cultured muscle cells. After titrating the amount of Lipofectin against a constant amount (1 μg) of AO 5′SS-FITC and comparing H-2Kb-tsA58 mdx cell survival versus nuclear fluorescence (data not shown), the optimal ratio was established to be 2:1 (wt/wt) Lipofectin/AO. No nuclear fluorescence was observed before 3 h after transfection, while the complexes were presumably being endocytosed. Neither was nuclear fluorescence observed when H-2Kb-tsA58 mdx cells were transfected in the presence of serum or with equimolar amounts of uncomplexed AO 5′SS-FITC (data not shown). Lipofectin–AO complexes were able to transfect H-2Kb-tsA58 mdx myoblast cells (1 day after plating), or fused myotubes (8 days after plating) and induce exon skipping with no apparent loss of efficiency. Uncomplexed AOs were unable to consistently induce skipping in either cell state (data not shown), suggesting that exon skipping depended on successful delivery rather than state of differentiation. Transfections under optimal conditions were reproducible between preparations of the immortalized H-2Kb-tsA58 mdx cells and routinely yielded transfection efficiencies of ≈100% of the myotubes, many of which exhibited characteristic multiple fluorescent nuclei (Fig. 2).
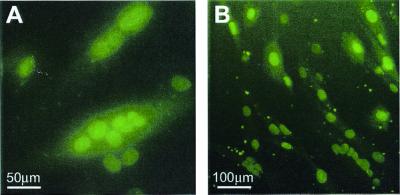
Figure 2.
Efficient nuclear uptake of liposome-complexed fluorescein-labeled AO by H-2Kb-tsA58 mdx cells. Cultured H-2Kb-tsA58 mdx myotubes were assessed for uptake of fluorescence after exposure to complexes of Lipofectin and AO 5′SS-FITC (2:1 ratio). Nuclear fluorescence was observed in ≈100% of the myotubes 3 h after transfection, with some pinpoint foci of fluorescence located in the cytoplasm and at the cell surface (B). Many of the transfected myotubes displayed multiple fluorescent nuclei (A).
AO Design.
The 18-mer directed at the 3′ splice site of intron 22 failed to induce exon 23 skipping (21), suggesting that this AO may have been too short to influence splicing. Consequently, two longer AOs were designed, a 30-mer directed at the splice acceptor site, and a 25-mer directed at the branch point site, AO 3′SS-30 and AO 3′SS-BPS, respectively. Both of these AOs failed to induce any exon skipping, either individually (Fig. 3) or when delivered in combination (data not shown).
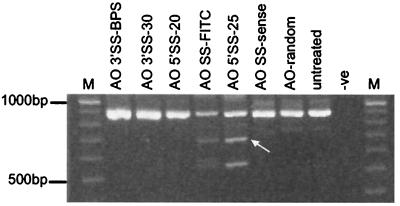
Figure 3.
AO-induced exon skipping in H-2Kb-tsA58 mdx myoblasts. Total RNA was extracted from treated and untreated H-2Kb-tsA58 mdx cells and amplified by nested RT-PCR using primers annealing to exons 20 and 26. Transfections were carried out as described in the text with the indicated AOs. The 901-bp full-length transcript was detected in all samples except the PCR negative (-ve) control. Lane M, size markers. A shorter product of 688 bp (arrow), corresponding to the removal of exon 23, was amplified from cell extracts transfected with AO 5′SS-FITC and AO 5′SS-25.
In marked contrast, a 25-mer, AO 5′SS-25, directed at the 5′ splice site of intron 23, was highly efficient at inducing exon 23 removal during pre-mRNA processing in H-2Kb-tsA58 mdx cells. The 20-mer, AO 5′SS-20, which we previously used to induce exon 23 skipping in primary mdx myoblasts and C2C12 cells, was inconsistent in its ability to cause excision of exon 23 in H-2Kb-tsA58 mdx cells under these transfection conditions (Fig. 3). AO 5′SS-25 annealed to the same exonic region as AO 5′SS-20, but it hybridized to an additional five bases into intron 23 (Fig. 1). This longer AO induced efficient and reproducible exon skipping (Fig. 3) at a concentration of 300 nM with 2 × 104 H-2Kb-tsA58 mdx cells. Under identical transfection conditions, AO 5′SS-FITC was less efficient than AO 5′SS-25, but more efficient than AO 5′SS-20 at causing exon 23 skipping. Although AO 5′SS-FITC has an identical sequence to AO 5′SS-20 (Fig. 1), the fluorescent tag coupled to the 5′ end of the molecule would extend further into intron 23 than the unlabeled AO, suggesting that regions further into the intron may be more sensitive to the influence of these AOs on splicing.
In addition to the removal of exon 23, other shorter RTPCR products were also generated after treatment with AO 5′SS-25. These corresponded to an in-frame transcript arising from the removal of exons 21–23, and a larger out-of-frame transcript skipping exons 22–23. An additional out-of-frame product corresponding to a transcript missing exon 21 was observed intermittently in treated and untreated samples, although it was seen more frequently in treated samples. These transcripts have been observed previously (21). Specificity of the AO action was confirmed after using a sense oligoribonucleotide complementary to AO 5′SS-20/AO 5′SS-FITC and a random sequence oligoribonucleotide, neither of which was observed to induce exon skipping in dystrophin mRNA (Fig. 3).
Dystrophin Expression in Injected mdx Muscle.
Dystrophin immunostaining of mdx muscle treated with AO 5′SS-25 complexed with Lipofectin revealed discontinuous sarcolemmal staining in the vicinity of the green dye in muscle from both the 2- and 4-week injection programs (Fig. 4). There was no discernible difference in staining intensity between the two time points. Mdx muscle treated with uncomplexed AO 5′SS-25 also showed dystrophin expression but at a lower level (data not shown). Regions of treated muscle where no green dye was observed did not stain for dystrophin. Similarly, no staining was observed in untreated muscles (triceps and tibialis anterior) from treated mice, or muscle treated with Lipofectin only.
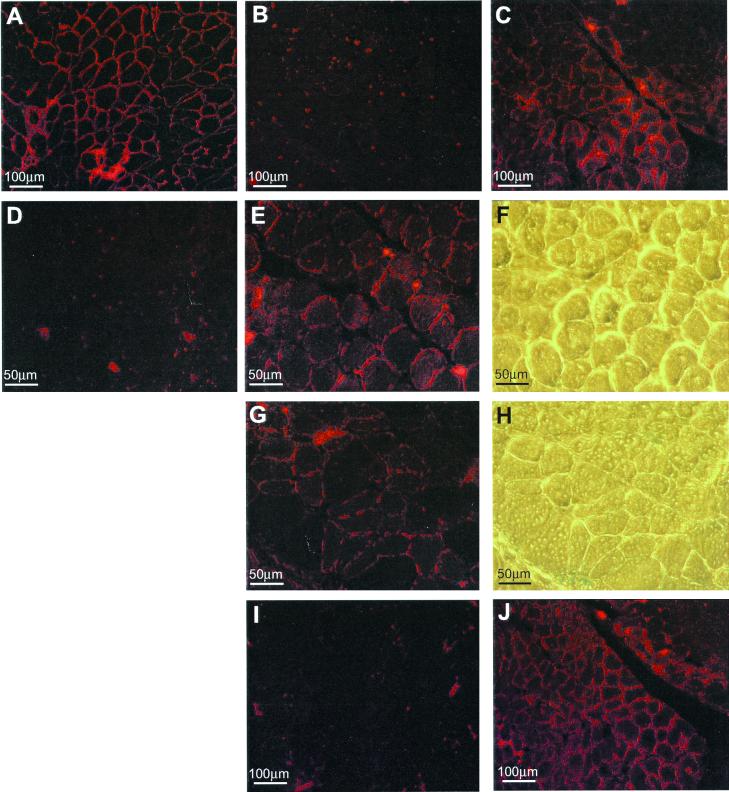
Figure 4.
Demonstration of antisense-induced dystrophin synthesis and restoration of γ-sarcoglycan in C57BL mdx mouse muscle. Immunostaining of normal C57BL mouse muscle (A) and untreated C57BL mdx muscle (B) with antibody DYS2 revealed sarcolemmal staining of dystrophin and a complete absence of the protein, respectively. However, sarcolemmal dystrophin staining was observed in a broad spread of mdx muscle fibers 1 week after injection of Lipofectin–AO 5′SS-25 (2:1 ratio) complexes into the left quadriceps-femoris complex (C). At higher magnification, dystrophin staining was revealed to be discontinuous, reminiscent of BMD staining, in both the left (E) and right (G) legs. Phase-contrast light microscopy of the same region (F and H, respectively) shows the green dye used to locate the site of the injection. No positive dystrophin staining was observed in an age-matched mdx control muscle when stained on the same slide as the treated sections and photographed under identical conditions (D). Serial sections of treated muscle stained with antibodies against γ-sarcoglycan revealed restoration of γ-sarcoglycan to the sarcolemma of muscle fibers that also stained positively for dystrophin (J). No staining was observed in the same muscle of an age-matched mdx control section also stained for γ-sarcoglycan (I).
Similar staining patterns were observed when serial sections were stained with an antibody recognizing γ-sarcoglycan, a component of the dystrophin–glycoprotein complex (DGC) (22). Reduced expression and localization of members of the DGC, including γ-sarcoglycan, are common features of muscular dystrophies (23), so the detection of this protein implies that the induced dystrophin has restored some of the DGC.
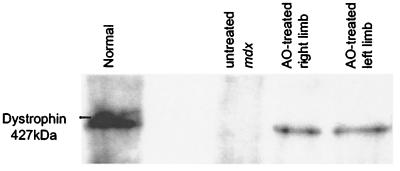
Western blot analysis of total protein demonstrated the presence of a protein of expected size from normal C57BL/10ScSn mouse muscle extracts and both treated mdx muscle samples, but not untreated mdx control muscle (Fig. 5). As with the immunostaining results, there appeared to be little quantitative difference in dystrophin levels between the 2 and 4 week treatments. There was no discernible size difference between normal dystrophin (427 kDa) and AO-induced dystrophin, expected to be 420 kDa, where the absence of exon 23 would remove only 71 amino acids.
Figure 5.
Synthesis of near full-length dystrophin in muscle of mdx mice treated with Lipofectin–AO 5′SS-25 complexes. Western blotting of total protein extracted from the quadriceps-femoris complex of mice treated with Lipofectin–AO 5′SS-25 (2:1 ratio) complexes revealed synthesis of near-full-length dystrophin compared with dystrophin extracted from normal C57BL muscle. No dystrophin protein was detected in muscle samples of untreated mdx mice.
Discussion
Kole and colleagues (24) pioneered the use of AOs to influence splicing by blocking a cryptic splice site in mutated β-globin pre-mRNAs to restore normal processing. We have reinterpreted their approach by blocking normal splice sites in a dystrophic pre-mRNA to induce abnormal splicing to remove an exon carrying a nonsense mutation.
AOs were designed complementary to elements involved in the splicing of dystrophin exon 23 in the mdx mouse. The ability of these AOs to induce efficient exclusion of exon 23 from processed dystrophin mRNA in the immortalized H-2Kb-tsA58 mdx cell line was assayed by RT-PCR. Injection of Lipofectin–AO complexes into the hind limb muscles of mdx mice, followed by RT-PCR and protein analysis, demonstrated specific exon 23 skipping and subsequent synthesis and correct localization of dystrophin and restoration of at least one component of the dystrophin–glycoprotein complex at the muscle fiber periphery. This demonstrates the potential of this alternative genetic therapy for DMD by using antisense chemistry to redirect splicing of mutated dystrophin pre-mRNA, allowing synthesis of a shorter but theoretically semifunctional, or Becker-dystrophin-like protein in dystrophic muscle.
Delivery of the AOs and translocation to the nucleus was shown not to be a limiting factor in vitro, as virtually the whole population of cultured cells displayed fluorescent nuclei after transfection with a fluorescein-labeled AO complexed with Lipofectin. Although delivery of naked plasmid is possible in whole muscle (25), and endocytotic uptake of naked AOs has been demonstrated in a variety of cell types in vitro (26, 27), we failed to observe fluorescence or exon skipping when H-2Kb-tsA58 mdx cells were treated with uncomplexed AOs.
The efficiency of inducing exon skipping in the immortalized H-2Kb-tsA58 mdx cells seemed more dependent on the target site rather than length of the AO, where exon 23 removal was enhanced by using a longer oligoribonucleotide directed at the intron 23 5′ splice site. However, AO length alone will not influence exon skipping if the target site is not sensitive to blocking or is inaccessible because of pre-mRNA secondary or tertiary structure or competition with splicing factors. For example, two AOs directed at splicing motifs at the end of intron 22, AO 3′SS-30 and AO 3′SS-BPS (a 30-mer and a 25-mer, respectively), both failed to induce exon skipping, consistent with our previously reported observations in experiments using an 18-mer AO to this region (21).
There are numerous examples of 3′ and 5′ splice site mutations leading to skipping of only the adjacent exon in the dystrophin gene (28–30). Inducing efficient skipping at the 5′ splice site appeared to be dependent on blocking intronic motifs, rather than only those at the exon–intron junction. The first two bases of the intron are highly conserved and generally considered to be essential for correct splicing (31). Only minimal and sporadic skipping was induced in H-2Kb-tsA58 mdx cells transfected with AO 5′SS-20, whereas AO 5′SS-25 was consistently more efficient at removing exon 23 from dystrophin pre-mRNA transcripts. Furthermore, the fluorescein tag attached to the 5′ end of AO 5′SS-FITC conferred on it the ability to redirect splicing with greater efficiency than AO 5′SS-20, even though the sequences of the two are identical. No splicing elements are known to exist in this intronic region, although consensus sequences crucial to splicing are short and not well conserved (31).
Preliminary in vivo experiments using lower doses of AO injected intramuscularly over 1 to 2 weeks resulted in low but detectable levels of correctly localized dystrophin in some treated mice (data not shown). In the experiments described here a 10-fold higher dose of AO delivered over 2 to 4 weeks resulted in substantial levels of dystrophin. This result was obtained from direct intramuscular delivery 5 weeks after initial treatment without immunosuppression of the animals. More detailed analysis of AO dosage and delivery is needed, along with studies of dystrophin expression and long-term persistence.
Immunostaining for dystrophin demonstrated discontinuous fluorescence around the sarcolemma of numerous successfully treated muscle fibers (Fig. 4C). This pattern of staining, observed in the majority of treated muscles, was reminiscent of that typically observed in sections from BMD patients (32). The discontinuous peripheral staining could be used to distinguish treated fibers from the rare naturally occurring revertants, which usually display continuous sarcolemmal staining. Revertant fibers in the mdx mouse typically occur as single fibers or small clusters undetectable by Western blotting (10). Some cytoplasmic staining of dystrophin was also present in the AO-treated fibers, and this is not typically seen in revertants, a further point of differentiation between AO-treated and natural dystrophin-positive fibers. Cytoplasmic staining has been observed in mdx mouse muscle transfected with dystrophin constructs under the control of Rous sarcoma virus promoters (35). Overexpression of dystrophin has not been associated with abnormalities in muscle fiber architecture or toxicity. The number of fibers visible in the treated section, the discontinuous peripheral and cytoplasmic staining, and the presence of a near-full-length protein in the Western blot confirm that dystrophin has been induced in the mdx mouse after AO therapy. Dystrophin immunostaining was not detected in the mdx muscle that was injected with liposome only, indicating that the induced dystrophin synthesis did not arise from either the delivery agent or repeated physical damage during the multiple injections (data not shown).
Staining of serial sections also demonstrated the partial restoration of γ-sarcoglycan at the periphery of those fibers that also stained for dystrophin. This observation suggested that the induced dystrophin was capable of restoring the complex of membrane glycoproteins, and presumably some function, to the AO-treated muscle.
The Western blot clearly demonstrated a single protein with an electrophoretic mobility approximately that of full-length normal dystrophin in muscle treated for either 2 or 4 weeks. This induced dystrophin isoform could not have arisen from sampling unusually large clusters of revertant fibers. We have shown that revertant fibers result from a spectrum of dystrophin isoforms, the most common of which arise from skipping of 20 or more exons (11). The lack of size difference observed between normal and induced dystrophin on the Western blot was expected, because removal of only exon 23, which codes for 71 amino acids, would not significantly alter the fractionation of such a large (427-kDa) protein through a polyacrylamide gel. Detailed in vivo RT-PCR studies have not yet been undertaken, as the emphasis has been placed on protein studies. Nevertheless, dystrophin transcripts skipping only exon 23 have been detected, and have been confirmed by direct DNA sequencing, 72 h after Lipofectin–AO delivery into mdx muscle (data not shown).
Induction and localization of a Becker-dystrophin-like protein in mdx muscle demonstrates the feasibility of an AO-based approach for treating DMD. As well as representing another therapy for a disease where numerous experimental treatments have met with disappointment (8, 36), this antisense-based approach has several potential advantages over those gene or cell replacement therapies. First, all of the tissue-specific and developmental control elements of dystrophin expression will remain intact (35, 36); liposomes will only have to deliver small payloads, unlike recombinant viral vectors; repeated delivery should be possible because the host should not develop immune responses against transduced cells as happens against virus-based therapies (37); and the costs of AO production should be much lower than those for preparing recombinant viruses. The major disadvantage of this particular AO therapy is that it will not be applicable to all dystrophin mutations, such as very large deletions, or mutations that disrupt or necessitate exclusion of crucial functional domains. Detailed characterization of the dystrophin mutations in BMD patients, especially those with a mild phenotype, will provide a useful guide as to which regions of dystrophin may be amenable to removal from the final product, and to the extent of phenotypic rescue that may be achievable. For example, the patient from whom the dystrophin “minigene” was derived had a mild phenotype despite the loss of 46% of the rod domain (38), whereas another patient with a smaller deletion of exons 3 to 9 was a competitive badminton player until his early 60s and was not diagnosed until his mid-60s (18). By using dystrophin transcripts from such patients as templates, the carefully orchestrated targeting of multiple exons could induce a protein capable of minimizing the severity of DMD. Finally, like all gene therapy protocols, the universal problem of delivery needs to be continually reassessed (8). Systemic delivery rather than intramuscular injection will undoubtedly be required to distribute the AOs if any significant clinical efficacy is to be obtained. Further experiments are necessary to address this problem of delivery, as well as to assess sites more sensitive to AO-targeted exon removal in other regions of the dystrophin gene.
Acknowledgments
We thank Russell Johnson for invaluable assistance with the Western blotting. This work was supported by the Muscular Dystrophy Association of Western Australia, the Neuromuscular Foundation of Western Australia, Inc., and grants from the Muscular Dystrophy Association of the U.S.A. (to S.D.W.).
Abbreviations
- DMD
Duchenne muscular dystrophy
- BMD
Becker muscular dystrophy
- AO
2′-O-methyl antisense oligoribonucleotide
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011408598.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011408598
References
- 1.Koenig M, Monaco A P, Kunkel L M. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E P, Brown R H, Jr, Kunkel L M. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Hodgson S V, Bobrow M. Br Med Bull. 1989;45:719–744. doi: 10.1093/oxfordjournals.bmb.a072354. [DOI] [PubMed] [Google Scholar]
- 4.Zatz M, Lange K, Spence M A. Lancet. 1977;1:759. doi: 10.1016/s0140-6736(77)92211-5. [DOI] [PubMed] [Google Scholar]
- 5.Monaco A P, Bertelson C J, Liechti-Gallati S, Moser H, Kunkel L M. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman E P, Fischbeck K H, Brown R H, Johnson M, Medori R, Loike J D, Harris J B, Waterston R, Brooke M, Specht L, et al. N Engl J Med. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 7.Bulfield G, Siller W G, Wright P A L, Moore K S. Proc Natl Acad Sci USA. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher S, Wilton S D, Howell J M. Curr Opin Neurol. 2000;13:553–560. doi: 10.1097/00019052-200010000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Morgan J E. Hum Gene Ther. 1994;5:165–173. doi: 10.1089/hum.1994.5.2-165. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman E P, Morgan J E, Watkins S C, Partridge T A. J Neurol Sci. 1990;99:9–25. doi: 10.1016/0022-510x(90)90195-s. [DOI] [PubMed] [Google Scholar]
- 11.Lu Q L, Morris G E, Wilton S D, Ly T, Artem'yeva O V, Strong P, Partridge T A. J Cell Biol. 2000;148:985–996. doi: 10.1083/jcb.148.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson L V B, Davison K, Johnson M A, Slater C R, Young C, Bhattacharya S, Gardner-Medwin D, Harris J B. J Neurol Sci. 1989;94:137–146. doi: 10.1016/0022-510x(89)90224-4. [DOI] [PubMed] [Google Scholar]
- 13.Schatzberg S J, Anderson L V, Wilton S D, Kornegay J N, Mann C J, Solomon G G, Sharp N J. Muscle Nerve. 1998;21:991–998. doi: 10.1002/(sici)1097-4598(199808)21:8<991::aid-mus2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 14.Klein C J, Coovert D D, Bulman D E, Ray P N, Mendell J R, Burghes A H M. Am J Hum Genet. 1992;50:950–959. [PMC free article] [PubMed] [Google Scholar]
- 15.Wallgren-Pettersson C, Jasani B, Rosser L G, Lazarou L P, Nicholson L V, Clarke A. J Neurol Sci. 1993;118:56–63. doi: 10.1016/0022-510x(93)90246-u. [DOI] [PubMed] [Google Scholar]
- 16.Fanin M, Danieli G A, Cadaldini M, Miorin M, Vitiello L, Angelini C. Muscle Nerve. 1995;18:1115–1120. doi: 10.1002/mus.880181007. [DOI] [PubMed] [Google Scholar]
- 17.Nicholson L V B, Johnson M A, Bushby K M D, Gardner-Medwin D. Arch Dis Child. 1993;68:632–636. doi: 10.1136/adc.68.5.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heald A, Anderson L V, Bushby K M, Shaw P J. Neurology. 1994;44:2388–2390. doi: 10.1212/wnl.44.12.2388. [DOI] [PubMed] [Google Scholar]
- 19.Morgan J E, Beauchamp J R, Pagel C N, Peckham M, Atalotis P, Jat P S, Nobel M D, Farmer K, Partridge T A. Dev Biol. 1994;162:486–498. doi: 10.1006/dbio.1994.1103. [DOI] [PubMed] [Google Scholar]
- 20.Lu Q L, Partridge T A. J Histochem Cytochem. 1998;46:977–984. doi: 10.1177/002215549804600813. [DOI] [PubMed] [Google Scholar]
- 21.Wilton S D, Lloyd F, Carville K, Fletcher S, Honeyman K, Agrawal S, Kole R. Neuromuscul Disord. 1999;9:330–338. doi: 10.1016/s0960-8966(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 22.Matsumura K, Saito F, Yamada H, Hase A, Sunada Y, Shimizu T. Cell Mol Biol. 1999;45:751–762. [PubMed] [Google Scholar]
- 23.Hack A A, Groh M E, McNally E M. Microsc Res Tech. 2000;48:167–180. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<167::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 24.Dominski Z, Kole R. Proc Natl Acad Sci USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff J A, Malone R W, Williams P, Chong W, Acsadi G, Jani A, Felgner P L. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 26.Yakubov L A, Deeva E A, Zarytova V F, Ivanova E M, Ryte A S, Yurchenko L V, Vlassov V V. Proc Natl Acad Sci USA. 1989;86:6454–6458. doi: 10.1073/pnas.86.17.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loke S L, Stein C A, Zhang X H, Mori K, Nakanishi M, Subasinghe C, Cohen J S, Neckers L M. Proc Natl Acad Sci USA. 1989;86:3474–3478. doi: 10.1073/pnas.86.10.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharp N J H, Kornegay J N, Van Camp S D, Herbstreith M H, Secore S L, Kettle S, Hung W-Y, Constantinou C D, Dykstra M J, Roses A D, Bartlett R J. Genomics. 1992;13:115–121. doi: 10.1016/0888-7543(92)90210-j. [DOI] [PubMed] [Google Scholar]
- 29.Ikezawa M, Minami N, Takahashi M, Goto Y, Miike T, Nonaka I. Brain Dev. 1998;20:165–168. doi: 10.1016/s0387-7604(98)00012-6. [DOI] [PubMed] [Google Scholar]
- 30.Wilton S D, Johnsen R D, Pedretti J R, Laing N G. Am J Med Genet. 1993;46:563–569. doi: 10.1002/ajmg.1320460521. [DOI] [PubMed] [Google Scholar]
- 31.Reed R, Palandjian L. In: Spliceosome Assembly. Krainer A R, editor. Oxford: IRL Press; 1997. pp. 103–129. [Google Scholar]
- 32.Morandi L, Mora M, Confalonieri V, Barresi R, Di Blasi C, Brugnoni R, Bernasconi P, Mantegazza R, Dworzak F, Antozzi C, et al. J Neurol Sci. 1995;132:146–155. doi: 10.1016/0022-510x(95)00147-t. [DOI] [PubMed] [Google Scholar]
- 33.Acsadi G, Dickson G, Love D R, Jani A, Walsh F S, Gurusinghe A, Wolff J A, Davies K E. Nature (London) 1991;352:815–818. doi: 10.1038/352815a0. [DOI] [PubMed] [Google Scholar]
- 34.Morgan J E, Partridge T A. BioEssays. 1992;14:641–645. doi: 10.1002/bies.950140913. [DOI] [PubMed] [Google Scholar]
- 35.Ahn A H, Kunkel L M. Nat Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- 36.Culligan K G, Mackey A J, Finn D M, Maguire P B, Ohlendieck K. Int J Mol Med. 1998;2:639–648. doi: 10.3892/ijmm.2.6.639. [DOI] [PubMed] [Google Scholar]
- 37.Petrof B J, Acsadi G, Jani A, Massie B, Bourdon J, Matusiewicz N, Yang L, Lochmuller H, Karpati G. Am J Respir Cell Mol Biol. 1995;13:508–517. doi: 10.1165/ajrcmb.13.5.7576685. [DOI] [PubMed] [Google Scholar]
- 38.England S B, Nicholson L V B, Johnson M A, Forrest S M, Love D R, Zubrzycka-Gaarn E E, Bulman D E, Harris J B, Davies K E. Nature (London) 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]