Abstract
Rab7 is a small GTPase that controls transport to endocytic degradative compartments. Here we report the identification of a novel 45 kDa protein that specifically binds Rab7GTP at its C-terminus. This protein contains a domain comprising two coiled-coil regions typical of myosin-like proteins and is found mainly in the cytosol. We named it RILP (Rab-interacting lysosomal protein) since it can be recruited efficiently on late endosomal and lysosomal membranes by Rab7GTP. RILP-C33 (a truncated form of the protein lacking the N-terminal half) strongly inhibits epidermal growth factor and low-density lipoprotein degradation, and causes dispersion of lysosomes similarly to Rab7 dominant-negative mutants. More importantly, expression of RILP reverses/prevents the effects of Rab7 dominant-negative mutants. All these data are consistent with a model in which RILP represents a downstream effector for Rab7 and both proteins act together in the regulation of late endocytic traffic.
Keywords: endocytosis/late endosomes/lysosomes/Rab7
Introduction
In the past few years, studies of various membrane trafficking steps have indicated that the machinery responsible for membrane fusion in each compartment is complex and very carefully regulated (Pfeffer, 1999). The specificity of the reaction seems to be ensured by a number of factors including Rabs, SNAREs and tethering proteins. Tethering proteins appear to act prior to the interaction between t- and v-SNAREs to bring the membranes together. SNARE complexes are responsible later on for the stable attachment of the vesicle to the target membrane. Proteins that could be responsible for tethering have been identified in different steps of transport and, surprisingly, they are not related. The Rab family of Ras-related GTPases appears to be essential for the regulation of intracellular membrane traffic in mammalian cells. Rab proteins are anchored to the cytoplasmic surface of specific intracellular membrane compartments via a geranyl-geranyl group that is added to the C-terminal cysteines post-translationally and is important for their function (Casey, 1995; Rando, 1996). Each Rab protein regulates one (or more) specific step of intracellular membrane traffic in eukaryotic cells, probably by assembling the general tethering/docking/fusion machinery (Olkkonen and Stenmark, 1997; Chavrier and Goud, 1999; Pfeffer, 1999; Waters and Pfeffer, 1999). Moreover, several lines of evidence suggest an involvement of Rab proteins in actin- and microtubule-based processes (Peranen et al., 1996; Echard et al., 1998; Nielsen et al., 1999).
To understand the molecular mechanism underlying a step of membrane transport, it is essential to identify all the molecules involved. The early steps of endocytosis (budding of endocytic vesicles from the plasma membrane and delivery to early endosomes) have been studied extensively and are quite well understood (Mukherjee et al., 1997). Indeed, several important factors that function at the level of early endosomes have been isolated (Stenmark et al., 1995; Gournier et al., 1998; Simonsen et al., 1998; Vitale et al., 1998; Christoforidis et al., 1999a,b; McBride et al., 1999; Michaely et al., 1999; Valetti et al., 1999; Nagelkerken et al., 2000). In contrast, relatively little is known about proteins involved in the steps of endocytosis leading to lysosomes. Rab7 is the Rab protein involved in the control of the late steps of endocytosis (Feng et al., 1995; Méresse et al., 1995, 1997; Papini et al., 1997; Vitelli et al., 1997; Press et al., 1998; Bucci et al., 2000). Moreover, it is essential for cellular vacuolation induced by the Helicobacter pylori cytotoxin VacA (Papini et al., 1997) and for the maturation of Salmonella typhimurium-containing vacuoles (Méresse et al., 1999). However, the exact role of Rab7 and its mechanism of action in late endocytic traffic are still not known. Therefore, it is important for understanding the late steps of endocytosis to isolate Rab7-interacting components. The two-hybrid system has been used successfully to identify Rab-interacting proteins. Indeed GDI, Rabaptin4, Rabaptin5, Rabin3, Rabphilin, Rabkinesin6 and PRA1 have been identified using this approach (Brondyk et al., 1995; Janoueix-Lerosey et al., 1995; Stenmark et al., 1995; Echard et al., 1998; Gournier et al., 1998; Bucci et al., 1999; Nagelkerken et al., 2000). We have used a GTPase-deficient mutant (Rab7Q67L) to screen for Rab7-interacting proteins. Here we describe the identification of a novel 45 kDa protein that specifically interacts with the GTP-bound form of Rab7, and demonstrate that this protein, together with Rab7, controls late endocytic transport.
Results
Identification of a Rab7-binding protein
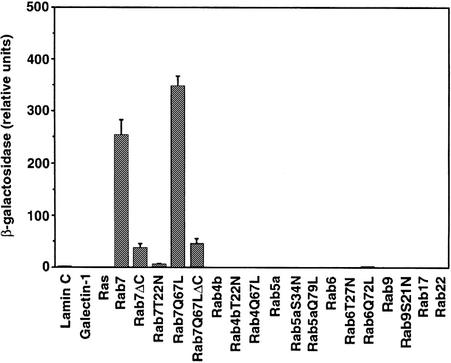
A yeast two-hybrid screen was used to isolate proteins that interact with the Rab7 GTPase. A LexA–Rab7Q67L fusion construct was made in pBTM116 (Bartel et al., 1993) and used in the screen to isolate putative effector proteins. The final three amino acids of Rab7 (Cys-Ser-Cys) were deleted in this construct to prevent C-terminal post-translational prenylation (Farnsworth et al., 1991) since such modification might cause high background in the assay. Expression of the fusion protein was confirmed by western blot analysis of total yeast extracts with an anti-Rab7 antiserum (data not shown). The construct was used to screen a HeLa cell line cDNA library encoding proteins as C-terminal fusions with the transcriptional activation domain of Gal4 (Bartel et al., 1993). From 107 primary transformants, 198 survived the initial selection on medium lacking leucine, tryptophan and histidine, and 120 were also positive for β-galactosidase activity by nitrocellulose filter assay. Of these initial positives, only 12 transformants encoded a true positive that did not activate transcription in the presence of a non-specific test bait (Ras, galectin 1 or lamin C). Ten of these transformants contained a clone coding for the human prenylated Rab acceptor (hPRA1) (Bucci et al., 1999). Two transformants contained a clone encoding part of a novel protein. This clone was 900 bp long and contained only the 3′ portion of a coding sequence, the 3′-untranslated region and the poly(A) tail. We then tested the interaction of this clone (called C33) with different wild-type (wt) and mutant Rab proteins, looking at the growth of the yeast transformants in medium lacking His, and at the development of the blue color due to β-galactosidase activity (data not shown). C33 did not interact with Rab4, Rab5, Rab6, Rab9, Rab17 and Rab22, as shown by the lack of growth on medium lacking His and by the lack of blue color in the yeast colonies. C33 interacted with the Rab7 wt and with the two Rab7 mutant proteins Rab7I41M and Rab7Q67L. These two mutants are active since they have reduced or no GTPase activity. The deletion of the last three amino acids in the Rab7 wt and Rab7Q67L mutant did not abolish the interaction. In contrast, there was no interaction of C33 with the dominant-negative mutant Rab7T22N locked in the GDP-bound form. To quantify the interaction, we used a liquid β-galactosidase assay (Figure 1). No interaction was detected with Ras or with other Rab proteins. A very strong interaction was instead observed with Rab7 wt and Rab7Q67L. The reaction was weaker (three to five times less) if the last three amino acids were removed from the Rab7 wt or Rab7Q67L protein, demonstrating that prenylation is important for the interaction. A very weak interaction was detected with the Rab7T22N mutant (∼40 times less compared with Rab7 wt).
Fig. 1. Specific interaction between Rab7 and C33. The β-galactosidase activity of double transformants was measured using o-nitrophenyl-β-d-galactoside as substrate. Activities are measured as arbitrary relative units and represent mean values ± SEM of four independent transformants.
Altogether, these results indicate that clone C33 encodes part of a protein that, at its C-terminus, specifically binds the GTP-bound and geranyl-geranylated form of the Rab7 protein.
The C33 cDNA encodes a novel protein
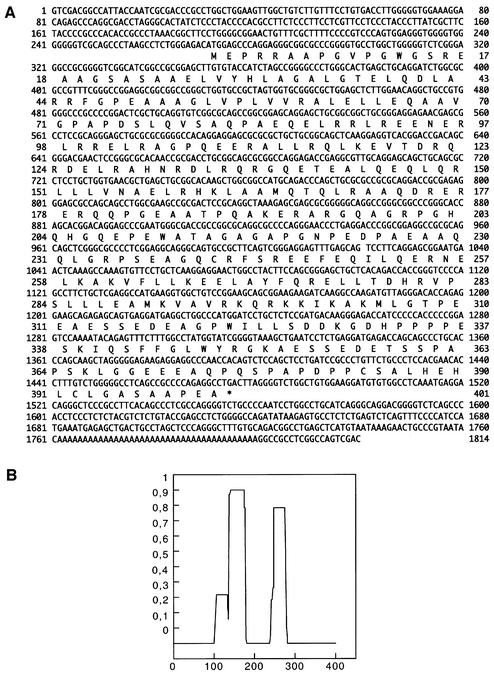
A search of the EMBL nucleotide database revealed no significant homology with any known human protein, but identified several expressed sequence tag (EST) sequences virtually identical to parts of the C33 cDNA. None of these sequences extended further at the 5′ end compared with clone C33. We then screened a HeLa cDNA library and isolated a full-length clone. The complete nucleotide sequence of the clone with the deduced amino acid sequence is shown in Figure 2A. The cDNA is 1814 bp long and contains a coding sequence of 401 amino acids with a predicted mol. wt of 45 kDa. A histogram of the probability of the formation of an α-helical coiled-coil structure in the protein was determined using the Lupas algorithm (Lupas et al., 1991) with a window size of 28 residues. This histogram (Figure 2B) shows that there are two regions in the protein that are likely to be in the coiled-coil conformation: the first between amino acids 140 and 180, and the second between amino acids 245 and 280. We called this protein RILP (Rab-interacting lysosomal protein).
Fig. 2. Primary structure of RILP. (A) Nucleotide and deduced amino acid sequence of RILP. The predicted amino acid sequence is shown below the nucleotide sequence in single-letter code. (B) Histogram indicating the probability of forming an α-helical coiled coil in RILP as determined by the Lupas algorithm. The x-axis indicates the amino acid number while the y-axis shows the probability (0–1).
When the amino acid sequence of RILP was challenged against a protein domain database (ProDom), homology was found only with two hypothetical gene products from Drosophila melanogaster (EG:132E8.4; accession No. AE003420) and Caenorhabditis elegans (C32A3.3; accession No. Z81449). The two products have similar sizes (443 and 433 amino acids, respectively), very close to that of RILP (401 amino acids). The homology between the three proteins was more significant at the level of the N-terminal region, where a protein domain of ∼40 amino acids, cataloged as PD154241 in the database, was mapped. The results of the ProDom database and research tools analysis confirmed that RILP, EG:132E8.4 and C32A3.3 had a similar architecture; in fact, the region encompassing the α-helical coiled-coil structure(s) in the three proteins appeared to belong to a single protein domain, consisting of ∼200 amino acids, cataloged as PD000002.
The presence of RILP in the mouse was documented by extensive homology of our cDNA full-length clone with regions of mouse chromosome 11. Significant homology was also found with a hypothetical murine protein (AB041584) of similar length (406 amino acids) belonging to the ezrin–radixin–moesin (ERM) family. However, homology was restricted to three definite regions encompassing amino acids 28–66 (identities 43.6%, positives 64.1%), 109–134 (identities 61.5%, positives 88.5%) and 308–327 (identities 80%, positives 100%). No extensive homology was found with any known yeast protein.
In vitro interaction of Rab7 with RILP
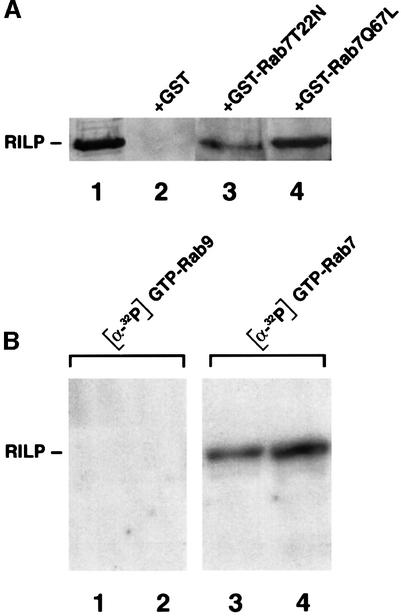
We decided to reconstruct the interaction with RILP in vitro using recombinant Rab7 mutant proteins. For this purpose, we expressed the Rab7T22N and Rab7Q67L proteins in Escherichia coli as glutathione S-transferase (GST) fusions and bound them to glutathione resin. They were loaded with GDP or GTP, and incubated in the presence of HeLa cell lysates as described in Materials and methods. The result of this experiment is shown in Figure 3A. Lane 1 contains extracts of HeLa cells overexpressing RILP as the immunoblotting control. RILP binds efficiently to GST–Rab7Q67L (GTP-bound) (Figure 3A, lane 4) and only weakly to GST–Rab7T22N (GDP-bound) (Figure 3A, lane 3), consistent with the data obtained with the two-hybrid assay. No binding was detected to the GST protein alone (Figure 3A, lane 2).
Fig. 3. Direct interaction between RILP and Rab7 in vitro. (A) GST and GST-tagged Rab7T22N and Rab7Q67L were expressed in BL21 E.coli cells, purified and immobilized on a glutathione resin. They were incubated with 100 µM GDP (Rab7T22N) or GTP (Rab7Q67L) and then with extracts of HeLa cells. Samples were then loaded on an SDS–polyacrylamide gel and subjected to western blot analysis using anti-RILP polyclonal antibodies. (B) One (lanes 1 and 3) or 2 (lanes 2 and 4) µg of GST-tagged RILP expressed in BL21 E.coli cells were loaded onto an SDS–polyacrylamide gel and transferred to nitrocellulose. The blot was then renatured as described in Materials and methods. A 10 µg aliquot of GST–Rab9 (lanes 1 and 2) or GST–Rab7 (lanes 3 and 4) loaded with [α-32P]GTP was applied to the renatured blot.
To demonstrate a direct interaction between Rab7 and RILP, we performed a protein overlay assay. RILP was expressed in E.coli as GST fusions, loaded onto an SDS–polyacrylamide gel and transferred to nitrocellulose. The blot was then renatured as described in Materials and methods. GST–Rab7 and GST–Rab9 were loaded with [α-32P]GTP and then applied to the renatured blot. No binding of [α-32P]GTP-Rab9 to RILP was detected (Figure 3B, lanes 1 and 2). In contrast, [α-32P]GTP-Rab7 bound to RILP in a dose-dependent fashion (Figure 3B, lanes 3 and 4). These experiments demonstrate that RILP interacts specifically and directly with Rab7.
Expression analysis of RILP
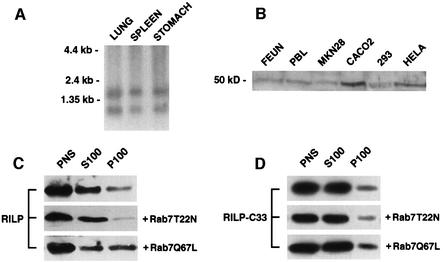
To determine the expression of RILP in different tissues, we used a 1800 bp SalI fragment as a probe in a northern blot of poly(A)+ RNA from three different human tissues (Figure 4A). This analysis showed the presence of two mRNAs of 1.8 and 1.2 kb in the three tissues examined. In order to follow the expression of the protein, we produced a rabbit antiserum against a bacterially produced recombinant RILP protein. This antibody was probed against total extracts of HeLa (human uterus carcinoma), CaCo2 (human colon carcinoma), MKN28 (human gastric tubular adenocarcinoma), FEUN (human skin fibroblast), 293 cells and peripheral blood lymphocytes (Jones et al., 1975; Knowles et al., 1980; Pinto et al., 1983; Romano et al., 1988; Burkhardt et al., 1993). While no bands were detected with the pre-immune serum in any extract (data not shown), a protein of ∼50 kDa was detected in all the extracts tested (Figure 4B). The protein was more abundant in CaCo2 cells. To determine whether the protein was cytosolic or membrane associated, we fractionated the post-nuclear supernatant (PNS) of HeLa cells into a high speed pellet (P100) and a supernatant (S100), and analyzed the distribution of the protein using western blot analysis (Figure 4C). The protein was mainly cytosolic, but a fraction of it (∼5%) was membrane associated.
Fig. 4. Expression analysis of RILP and recruitment on membranes upon expression of Rab7Q67L. (A) Northern blot analysis. Human poly(A)+ RNA (10 µg) from lung, spleen and stomach was electrophoresed, transferred to a nylon membrane, hybridized to a 32P-labeled 1800 bp SalI fragment, washed and autoradiographed as described in Materials and methods. The relative migration of the RNA markers is indicated on the left of the panel. (B) Western blot analysis. Total extracts from FEUN (human embryonic fibroblast), PBL (peripheral blood lymphocytes), MKN28, CaCo2, 293 and HeLa cells were run on an SDS–polyacrylamide gel and analyzed by western blotting using affinity-purified anti-RILP polyclonal antibodies. The molecular weight of the immunoreactive protein is shown. (C) The PNS of HeLa cells, transfected with Rab7T22N or Rab7Q67L or not transfected, was fractionated into a high speed pellet (P100) and supernatant (S100). Proportional amounts of supernatant, pellet and PNS were analyzed by western blotting using affinity-purified anti-RILP polyclonal antibodies. (D) The same experiment as in (C) was performed using HeLa cells transfected with RILP-C33.
Recruitment of RILP on late endosomal/lysosomal membranes by Rab7GTP
To determine whether the expression of Rab7 affected the distribution of the RILP protein, we looked at the distribution of RILP in HeLa cells after transfection with a dominant-negative (Rab7T22N) or a constitutively active mutant (Rab7Q67L) of Rab7 (Figure 4C). Similar to what happens in the case of Rabaptin5, which is recruited on the membrane by active Rab5, the amount of membrane-associated RILP increased when Rab7 wt (not shown) or Rab7Q67L, but not Rab7T22N, was transfected. In a similar manner, the transfected RILP-C33 truncated construct was recruited on membranes by the Rab7 wt (not shown) or Rab7Q67L mutant protein (Figure 4D). These data indicate that active Rab7 is able to recruit RILP as well as its truncated form, RILP-C33, on membranes.
RILP localizes to late endosomal/lysosomal membranes
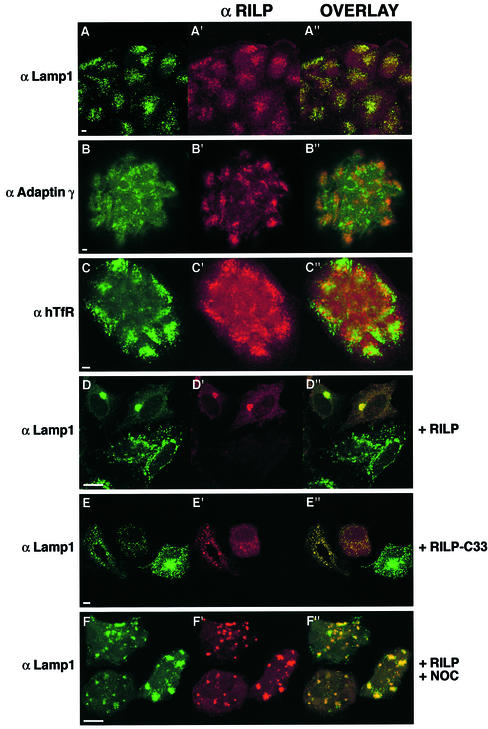
We next examined the intracellular localization of this protein. Antibodies made against this protein and affinity purified gave a diffuse cytosolic staining on HeLa, BHK and CaCo2 cells, confirming that the protein is mainly cytosolic (data not shown). We then performed immunofluorescence, permeabilizing the cells with saponin before fixation to wash out excess cytosolic proteins. Under these conditions, we were able to analyze the membrane-associated fraction of endogenous RILP. We found a high degree of co-localization of RILP and the late endosomal/lysosomal marker Lamp1 in CaCo2 cells where the protein is expressed at higher levels (Figure 5A). Similar results were obtained with Lamp2 and CathD (data not shown), demonstrating that RILP localizes to a Lamp-, CathD-positive compartment. In contrast, no co-localization was detected with adaptin γ (Figure 5B), human transferrin receptor (hTfR; Figure 5C), early endosomal autoantigen 1 (EEA1; data not shown) and protein disulfide isomerase (PDI; data not shown). These data demonstrate that RILP specifically localizes to late endosomal/lysosomal membranes. As expected, overexpressed RILP in HeLa cells (Figure 5D) or RILP-C33 (Figure 5E) co-localized mostly with Lamp1 and not with other cellular markers such as adaptin γ, hTfR, EEA1 and PDI (data not shown). In addition, the late endosomal/lysosomal compartment seems to be strongly affected by the expression of these proteins. Indeed, overexpression of RILP causes a high degree of aggregation of the late endosomal/lysosomal compartment in the perinuclear region, as documented by the Lamp1 staining (Figure 5D), while expression of RILP-C33 seems to cause a dispersion of the compartment (Figure 5E). Indeed, the normal perinuclear accumulation of Lamp-positive vesicles is not observed (Figure 5E). However, in cells transfected with RILP-C33, in some cases individual late endosomal/lysosomal vesicles actually appear enlarged compared with control. The striking perinuclear clustering of the late endosomal/lysosomal compartment caused by overexpression of RILP can be reversed by treatment with nocodazole, demonstrating that this process is microtubule dependent (Figure 5F).
Fig. 5. Confocal immunofluorescence analysis of cells expressing endogenous or high levels of RILP. Low magnification fields of several CaCo2 cells stained with anti-RILP at a dilution of 1:10 (A′, B′ and C′; red) and anti-Lamp1 (A; green), anti-adaptin γ (B; green) or anti-hTfR (C; green). HeLa cells transfected for 5 h with RILP (D and F) or RILP-C33 (E) and stained with anti-RILP at a dilution of 1:1000 (D′, E′ and F′; red) and with anti-Lamp1 (D–F; green). In (F), cells were treated for 30 min with 10 µM nocodazole after transfection. A few cells are shown in each field. The overlays of the two colors are in (A′′–F′′). Coverslips were viewed with a Leica confocal microscope. Bars = 10 µm.
These data together demonstrate that RILP specifically localizes to the late endosomal/lysosomal membranes and that its overexpression affects the normal distribution of these compartments in the cell.
RILP is able to overcome the morphological effects of the Rab7T22N dominant-negative mutant on the late endosomal/lysosomal compartment
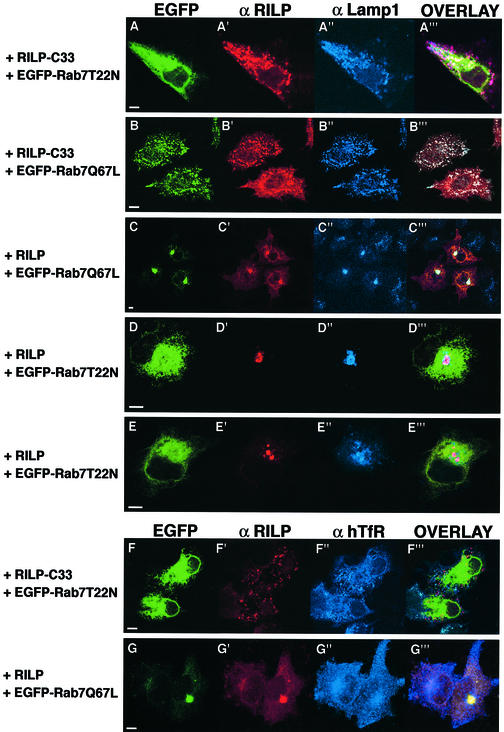
We have demonstrated previously that expression of Rab7 dominant-negative mutants causes selective dispersal of perinuclear lysosomes while expression of Rab7 wt or Rab7Q67L increases the degree of aggregation. In addition, the dispersed lysosomes obtained after expression of Rab7 dominant-negative mutants are no longer acidic and they can not be reached by endocytic markers (Bucci et al., 2000). Interestingly, while overexpressed RILP causes perinuclear aggregation of late endosomes and lysosomes, similar to what occurs in the case of Rab7Q67L expression (Figure 5D), expression of RILP-C33, the truncated form of RILP, causes the dispersal of the perinuclear late endosomal/lysosomal aggregate, similar to the case of cells expressing Rab7 dominant-negative mutants (Figure 5E). To analyze better the effects of RILP and RILP-C33 on the late endosomal/lysosomal compartment, we expressed these proteins in HeLa cells together with the Rab7 dominant-negative or constitutively active mutants (Figure 6). As expected, co-expression of Rab7T22N and RILP-C33 resulted in the dispersion of the Lamp-positive compartment (Figure 6A), which remained well separated from the early endosomal compartment labeled by hTfR (Figure 6F). In contrast, co-expression of Rab7Q67L and RILP resulted in the perinuclear aggregation of these structures, which were even more compact as compared with cells transfected with the active Rab7 active mutant (Figure 6C; Bucci et al., 2000). The perinuclear clustering caused by RILP and Rab7 involves only the late endosomal/lysosomal compartment since the distribution of other compartments was not affected. Indeed, adaptin γ-, PDI-, EEA1- and TfR-labeled vesicles maintained their normal distribution, clearly distinct from the perinuclear cluster of late endosomes and lysosomes (data not shown and Figure 6G). Interestingly, in cells co-expressing Rab7T22N and RILP, lysosomes are not dispersed but clustered in the perinuclear region as documented by the Lamp1 staining, indicating that RILP is able to prevent/reverse the action of the Rab7 dominant-negative mutant (Figure 6D and E). In contrast, dispersed lysosomes caused by expression of RILP-C33 are present even in the presence of the Rab7Q67L constitutively active mutants (Figure 6B).
Fig. 6. Dispersal of lysosomes in Rab7 dominant-negative mutant-expressing cells is prevented/reversed by expression of the RILP protein but not of the RILP-C33 protein. HeLa cells were transfected with RILP-C33 and EGFP–Rab7T22N (A and F), with RILP-C33 and EGFP–Rab7Q67L (B), with RILP and EGFP–Rab7Q67L (C and G), or with RILP and EGFP–Rab7T22N (D and E). Cells were stained with anti-RILP (A′–G′; red) and with anti-Lamp1 (A′′–E′′; blue) or anti-hTfR (F′′–G′′; blue), while EGFP–Rab7 mutants were green. Overlays of the three colors are shown in (A′′′–G′′′). Coverslips were viewed with a Leica confocal microscope. Bars = 10 µm.
Altogether, these data demonstrate that RILP together with Rab7 plays a fundamental role in the organization of the late endosomal/lysosomal compartment, and suggest that RILP could act as a downstream effector of Rab7 since its expression is able to bypass the effects on the lysosomal compartment caused by the Rab7 dominant-negative mutants.
RILP controls transport to lysosomes
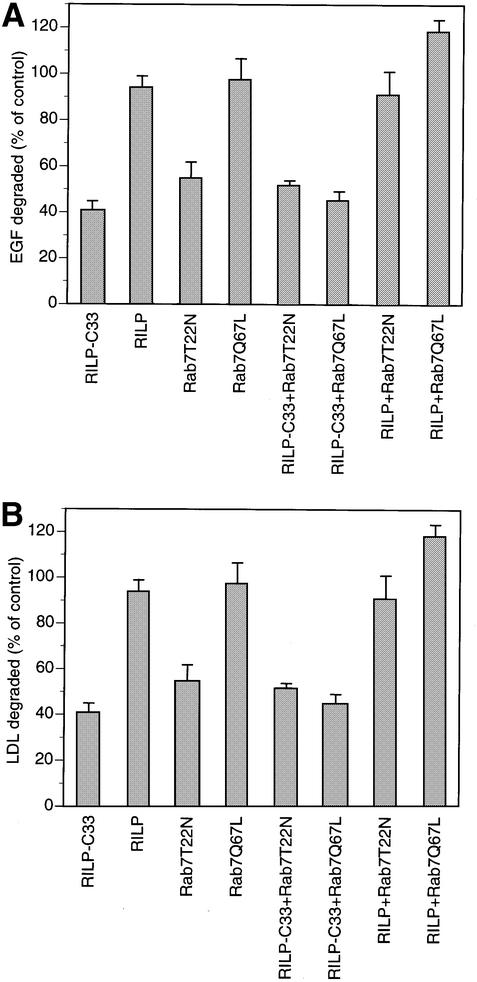
We next investigated the function of RILP in the endocytic pathway by analyzing the effect on uptake and degradation of [125I]low-density lipoprotein (LDL) and [125I]epidermal growth factor (EGF) in HeLa cells expressing RILP and its truncated form RILP-C33. No effect on [125I]LDL or [125I]EGF kinetics of internalization was observed when RILP or RILP-C33 were expressed, indicating that RILP, like Rab7, does not affect the early stages of the endocytic pathway (data not shown). In contrast, a strong inhibition of [125I]LDL and [125I]EGF degradation was detected when RILP-C33, but not RILP, was expressed (Figure 7). This inhibition was comparable with that obtained when expressing the Rab7 dominant-negative mutants and indicated that RILP plays an important role in the regulation of the transport to degradative compartments of the endocytic pathway. We next investigated whether expression of functional RILP was able also to reverse/prevent the inhibitory effect of the dominant-negative mutants of Rab7 on LDL or EGF degradation. We therefore measured the degradation in cells doubly transfected with a Rab7 dominant-negative mutant, Rab7T22N (Figure 7), and the full-length or truncated form (RILP-C33) of RILP. Degradation was normal in cells co-expressing Rab7 dominant-negative mutants and RILP, while it was inhibited if Rab7 dominant-negative mutants were expressed with RILP-C33. In addition, expression of the Rab7-activating mutant was not able to reverse/prevent the inhibitory effect caused by the RILP-C33 protein. Co-expression of RILP and Rab7Q67L did not result in a statistically significant increase in degradation compared with control. All these data together indicate that Rab7 and RILP work sequentially in the regulation of the late endocytic pathway, and suggest that RILP could represent the downstream effector of Rab7.
Fig. 7. Inhibition of EGF and LDL degradation by expression of RILP-C33 and rescue of the inhibition caused by the Rab7 dominant-negative mutants by expression of RILP. (A) Cells transfected with the indicated constructs were allowed to internalize [125I]EGF for 1 h, then were washed extensively at 4°C and reincubated at 37°C for 2 h. The amount of [125I]EGF degraded was quantified. (B) Transfected cells were allowed to internalize [125I]LDL for 5 h and the amount of [125I]LDL degraded was determined. Data are plotted as a percentage of control cells transfected with the empty vector. Error bars represent the range between four independent experiments.
Discussion
Rab7 plays a central role in the transport to degradative compartments and in the maintenance of their organization. Here we describe the isolation and characterization of a novel 45 kDa protein that interacts specifically with Rab7, and is essential for late endosomal/lysosomal organization and for transport to these compartments. We have called this protein RILP since the membrane-bound fraction shows a high degree of co-localizaton with Lamp1, Lamp2 and CathD. We have proved by three different methods that the protein interacts with Rab7: (i) two-hybrid assay; (ii) in vitro direct interaction of the two proteins; and (iii) recruitment of the protein on the late endosomal/lysosomal membranes by the constitutively active mutant of Rab7.
We have also demonstrated that the protein specifically interacts with the GTP-bound form of Rab7, suggesting that we have isolated a Rab7 effector protein. In addition, the two-hybrid interaction data indicate that the geranyl-geranyl group is important in the interaction. Recruitment of cytosolic factors seems to be common to several GTPases and in particular to Rab proteins (Vojtek et al., 1993; Li et al., 1994; Stenmark et al., 1995). Indeed, the current view on Rab proteins postulates that Rabs recruit membrane tethering, docking and fusion factors, therefore regulating the assembly of the complex machinery that will then be responsible for docking and fusion (Pfeffer, 1999; Waters and Pfeffer, 1999; Somsel Rodman and Wandinger-Ness, 2000). In addition, recent data suggest that Rabs may recruit cytoskeletal proteins, establishing a link between vesicle docking and the actin- and microtubule-based cytoskeletons (Peranen et al., 1996; Echard et al., 1998; Nielsen et al., 1999). Our data are consistent with this model since the active (GTP-bound) form of Rab7 recruits RILP on membranes. Nevertheless, no homology has been found between RILP and other Rab-interacting proteins. This is not surprising, since most of the known Rab-interacting proteins are not related to each other and perform distinct functions (Pfeffer, 1999; Waters and Pfeffer, 1999; Somsel Rodman and Wandinger-Ness, 2000).
What is the function of RILP? We have demonstrated that this protein is important in late endocytic traffic. Indeed, expression of its truncated form inhibits degradation and disperses lysosomes similarly to what happens in cells expressing the dominant-negative mutant of Rab7. In contrast, overexpression of the full-length protein increases perinuclear aggregation of the late endosomal/lysosomal compartment similarly to cells expressing the Rab7 wt or the Rab7Q67L constitutively active mutant. We think that RILP could represent a downstream effector of Rab7 since its expression is able to prevent/reverse the dramatic effects on the late endocytic pathway caused by the expression of dominant-negative mutants of Rab7 and, on the contrary, expression of Rab7Q67L is not able not prevent/reverse the effect of RILP-C33. Co-expression of the Rab7 dominant-negative mutant and RILP-C33 does not yield synergy in the inhibition of transport. This is also consistent with the hypothesis that both RILP and Rab7 act sequentially on the same pathway.
RILP appears to be poorly conserved evolutionarily. A search for homologous proteins in yeast was unsuccessful. A specific search for similarity between RILP and proteins known to interact with the Rab7 yeast homolog Ypt7p, including Vam2p and Vam6p, Vps33p and Vam7p (the vacuolar SNAP-23/25 homolog), did not give positive results. Vacuole-associated Vam2p and Vam6p are components of a 65S complex containing SNARE proteins, which, upon priming by Sec18p/NSF and ATP, are released as a 38S subcomplex (which also includes Vps33p and other class C Vps proteins) that binds Ypt7p to initiate docking (Seals et al., 2000; Ungermann et al., 2000). No homology was found with Vam3p or Nyv1p, members of the SNARE complex and required for homotypic vacuole fusion in vitro with Ypt7p, Sec18p/NSF, Sec 17p/αSNAP and LMA1 (Nichols et al., 1997; Sato and Wickner, 1998).
Using protein domain identification research tools, we identified hypothetical gene products in D.melanogaster and C.elegans sharing common structural features with RILP, including the PD154241 and PD000002 domains (encompassing the two α-helical coiled-coil structures). Nothing is known about the function of the PD154241 domain, while the protein domain PD000002 is typical of muscle and non-muscle heavy chain myosin tails, where it is arranged in tandem, and of paramyosins. Interestingly, this domain is present in several proteins that are involved in microtuble-associated motility. These include CLIP-170, an intermediate filament-associated protein that links endocytic vesicles to microtubules (Pierre et al., 1992; Griparic and Keller, 1998) and that has also been shown to be part of hetero-motor complexes interacting with myosin (Goode et al., 2000), EEA1, an early endosome-associated protein (Mu et al., 1995), and, more interestingly, p150Glued, the largest subunit of the dynactin complex. p150Glued plays a critical role in binding membranous organelles to motor complexes (Gill et al., 1991; Karki and Holzbaur, 1995; Vaughan and Vallee, 1995; Echeverri et al., 1996; King, 2000). Interestingly, RILP exhibits similarity to a hypothetical murine protein (AB041584) of similar length (406 amino acids) belonging to the ERM family. ERM proteins are a group of closely related membrane–cytoskeleton linkers, which play a role in the Rho and Rac signaling pathway (Bretscher, 1999; Mangeat et al., 1999). The degree of similarity between RILP and the hypothetical murine ERM protein is very high at the level of three regions not involved in α-helical coiled-coil structures.
Based on these considerations, RILP might be involved in targeting Rab7-containing organelles to the motor complex and/or cytoskeleton. The identification of RILP provides the basis for the elucidation and characterization of the molecular machinery responsible for late endocytic trafficking. Further work will be necessary to address the molecular mechanism of action of this protein.
Materials and methods
Two-hybrid screening
The pLexA-Rab7Q67LΔC construct was used to screen a HeLa cDNA library obtained from Clontech using the Saccharomyces cerevisiae L40 reporter strain (Schiestl and Giest, 1989; Hill et al., 1991; Bartel et al., 1993). The transformants were plated on synthetic medium lacking histidine, leucine and tryptophan. After 5 days of growth at 30°C, His+ colonies were picked and assayed for β-galactosidase activity. A total of 120 transformants were LacZ+His+. Plasmid DNA from these transformants was obtained and used to transform HB101 E.coli cells (Bartel et al., 1993). The recovered library plasmids were then tested for interaction with laminC, Ras and pLexA-Rab7Q67LΔC. Twelve clones transactivated the reporters only in the presence of the ‘bait’ (pLexA-Rab7Q67LΔC). Ten of them encoded hPRA1 (Bucci et al., 1999). Two (called C33) encoded part of a novel protein and were tested further for interaction with other Rab wt and mutant proteins. To quantify the interactions, we performed a β-galactosidase assay using o-nitrophenyl-β-d-galactoside as substrate (Guarente, 1983).
Library screening, cDNA cloning and sequencing analysis
A HeLa cDNA library was screened and 5 × 105 phage plaques were plated, lifted onto nitrocellulose filters and screened using the fragment extracted with EcoRI–XhoI from the pGADGH-C33 clone. Hybridization and subsequent washing were carried out as described (Church and Gilbert, 1984). Sequence analysis of a positive clone showed that it contained the full-length open reading frame in a 1800 bp SalI fragment. This cDNA clone was sequenced by the dideoxynucleotide chain termination method on both strands (Sanger et al., 1977).
Protein sequence analysis
Amino acid sequences were analyzed using ProDom, which consists of an automatic compilation of homologous domains. Current versions of ProDom are built using a novel procedure based on recursive PSI-BLAST searches (Altschul et al., 1997; Gouzy et al., 1999). Large families are processed much better with this new procedure than with the former DOMAINER program (Sonnhammer and Kahn, 1994). The program was NCBI-BLASTP2 with the Graphic Output option, or SMART (simple modular architecture research tool).
Cell culture and transient transfection
Tissue culture reagents were from Gibco-BRL. HeLa or CaCo2 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, 100 U/ml penicillin and 10 µg/ml streptomycin, and grown in a 5% CO2 incubator at 37°C. HeLa cells were infected with the vT7 recombinant vaccinia virus and transfected as described (Fuerst et al., 1986; Bucci et al., 1992). The transfection reagent was Fugene from Boehringer Mannheim. Cells were transfected for 4–6 h and then processed for immunofluorescence, western blotting or biochemical assays.
Antibodies and confocal immunofluorescence microscopy
The mouse monoclonal antibodies anti-Lamp1 (H4A3) and anti-Lamp2 (H4B4) were developed by J.T.August and J.E.K.Hildreth, and were obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa (Department of Biological Sciences, Iowa City, IA 52242) under contract NO1-HD-7-3263 from the NICHD. The mouse monoclonal antibodies against adaptin γ and hTfR were from Transduction Laboratories and Boehringer, respectively. Unless indicated otherwise, the concentration of primary antibodies used was 1:500. The anti-RILP antibody was raised in rabbits using recombinant RILP-C33 protein. Cells grown on 11 mm round glass coverslips were permeabilized and fixed as previously described (Bucci et al., 1992). The secondary fluorescein isothiocyanate (FITC)-, tetramethylrhodamine isothiocyanate (TRITC)- or Cy5-conjugated anti-mouse or anti-rabbit antibodies were from Sigma or Amersham, and were diluted 1:300. Cells were viewed with a Leica confocal microscope.
Production of GST–Rab7 wt and mutant proteins and in vitro interaction with RILP
Rab7 wt, Rab7T22N and Rab7Q67L mutated cDNAs (Vitelli et al., 1997) were cloned into the pGEX-4T3 vector to produce GST fusion proteins. These constructs were transformed into E.coli strain BL21. GST–Rab7 wt and mutants proteins were purified using GST–Sepharose 4B resin according to the instructions of Pharmacia Biotech.
For the in vitro interaction of Rab7 and RILP, confluent plates of HeLa cells were lysed in 20 mM HEPES pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1% NP-40 at 4°C for 30 min. Cells were passed through a 26 gauge needle and centrifuged at 10 000 g for 10 min at 4°C. GST–Rab7T22N and GST–Rab7Q67L resin-bound proteins were loaded with 100 µM GDP or GTP, respectively. The HeLa lysate was then incubated with the recombinant proteins bound to the resin at 4°C for 1 h. The resin was washed several times with 20 mM HEPES pH 7.4, 150 mM NaCl, 5 mM MgCl2 at 4°C, and then the samples were analyzed by SDS–PAGE and immunoblotting using anti-RILP antibodies.
Protein overlay assay
A 1–2 µg aliquot of RILP was loaded on an SDS–polyacrylamide gel and transferred to nitrocellulose. The blot was incubated in renaturation buffer [50 mM HEPES pH 7.2, 5 mM MgAc, 100 mM KAc, 3 mM dithiothreitol (DTT), 10 mg/ml bovine serum albumin (BSA), 0.1% Triton X-100, 0.3% Tween-20] overnight at 4°C with gentle agitation.
A 15 µg aliquot of GST–Rab7 or of GST–Rab9 was loaded with radiolabeled [α-32P]GTP at a working concentration of 0.1 µM in 20 mM HEPES pH 7.2, 4 mM EDTA, 10 mM MgCl2 for 1 h at 37°C. The nucleotide-loaded protein was added to 10 ml of binding buffer (12.5 mM HEPES pH 7.2, 1.5 mM MgAc, 75 mM Kac, 1 mM DTT, 2 mg/ml BSA, 0.05% Triton X-100), which was subsequently applied to the renatured blot and incubated at room temperature for 2 h. The blot was washed six times for 10 min with 20 mM Tris–HCl pH 7.4, 100 mM NaCl, 20 mM MgCl2, 0.005% Triton X-100, and then exposed to X-ray film.
Western blot
Cells were lysed in standard SDS sample buffer and 25 µg of the extracts were electrophoresed on 12% SDS–polyacrylamide gels. Separated proteins were transferred onto nitrocellulose membrane. Primary rabbit polyclonal anti-RILP antibody was added at a 1:100 dilution and incubated for 2 h at room temperature. The filter was washed, incubated with a secondary anti-mouse horseradish peroxidase (HRP)-conjugated antibody (at a dilution of 1:5000) for 1 h at room temperature, and the bands visualized using the enhanced chemiluminescence system (ECL; Amersham).
Estimation of [125I]LDL degradation
Human LDL (1.019 < density < 1.063 g/ml) was obtained by preparative ultracentrifugation from serum of normolipidemic subjects. The LDL was then concentrated and purified further by double ultracentrifugation at a density of 1.063 g/ml. Lipoprotein-deficient serum (LPDS) was prepared from the same serum samples used for LDL preparation by ultracentrifugation at a density of 1.210 g/ml. LDL and LPDS were then dialyzed extensively against 0.15 M NaCl, 0.24 mM disodium EDTA pH 7.4. LDL was labeled with 125I and the specific activity of [125I]LDL ranged between 200 and 400 c.p.m./ng of protein. Radioiodinated LDL was always used within 2 weeks of preparation. Cells were incubated for 24 h in medium supplemented with human LPDS before transfection. Transfected cells were allowed to internalize [125I]LDL that was added to the medium at a concentration of 20 µg/ml for 5 h, and the amount of [125I]LDL that was surface bound, internalized and degraded was estimated as described (Brown and Goldstein, 1975).
Estimation of [125I]EGF recycling and degradation
Human [125I]EGF was purchased from Amersham Biotech. Transfected cells were incubated with [125I]EGF at 37°C for 1 h, washed extensively on ice with cold phosphate-buffered saline (PBS)/0.1% BSA and reincubated at 37°C for 2 h. Briefly, to determine the amount of [125I]EGF degraded, the medium was collected and treated with trichloroacetic acid (TCA). The EGF degraded (acid-soluble) and recycled (acid-insoluble) was detected with a γ-counter.
Northern blot analysis
Poly(A)+ RNAs from three different tissues (Clontech Laboratories, Inc.) were electrophoresed, transferred to a nylon membrane and hybridized with a 32P-labeled 1800 bp SalI fragment, spanning the coding region of RILP. The filter was then washed as described (Church et al., 1984). The filters were autoradiographed using Kodak XAR5 film for 16 h at –70°C.
DDBJ/EMBL/GenBank accession number
The accession number for the RILP sequence is AJ404317.
Acknowledgments
Acknowledgements
We thank Drs Philippe Chavrier, Toshi Kobayashi and Jean Gruenberg for the kind gift of anti-Rab7 antibodies, and Suzanne Pfeffer for the gift of the Rab9 plasmids used in this study. This work was supported in part by grants the from Italian CNR (PF BIOTECNOLOGIE), and from the European Economic Community (grant no. ERBFMRXCT960020).
REFERENCES
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P.L., Chien,C.-T., Sternglanz,R. and Fields,S. (1993) Using the two-hybrid system to detect protein–protein interactions. In Hartley,D.A. (ed.), Cellular Interaction in Development: A Practical Approach. Oxford University Press, Oxford, UK, pp. 153–179.
- Bretscher A. (1999) Regulation of cortical structure by the ezrin–radixin–moesin protein family. Curr. Opin. Cell Biol., 11, 109–116. [DOI] [PubMed] [Google Scholar]
- Brondyk W.H., McKiernan,C.J., Fortner,K.A., Stabila,P., Holz,R.W. and Macara,I.G. (1995) Interaction cloning of Rabin3, a novel protein that associates with the Ras-like GTPase Rab3a. Mol. Cell. Biol., 15, 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.S. and Goldstein,J.L. (1975) Regulation of the activity of the low density lipoprotein receptor in human fibroblasts. Cell, 6, 307–316. [DOI] [PubMed] [Google Scholar]
- Bucci C., Parton,R.G., Mather,I.H., Stunnenberg,H., Simons,K., Hoflack,B. and Zerial,M. (1992) The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell, 70, 715–728. [DOI] [PubMed] [Google Scholar]
- Bucci C., Chiariello,M., Lattero,D., Maiorano,M. and Bruni,C.B. (1999) Interaction cloning and characterization of the cDNA encoding the human prenylated Rab acceptor (PRA1). Biochem. Biophys. Res. Commun., 258, 657–662. [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen,P., Nicoziani,P., McCarthy,J. and van Deurs,B. (2000) Rab7: a key to lysosome biogenesis. Mol. Biol. Cell, 11, 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J., Wiebel,F.A., Hester,S. and Argon,Y. (1993) The giant organelles in beige and Chediak–Higashi fibroblasts are derived from late endosomes and mature lysosomes. J. Exp. Med., 178, 1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey P.J. (1995) Mechanism of protein prenylation and role in G protein function. Biochem. Soc. Trans., 23, 161–166. [DOI] [PubMed] [Google Scholar]
- Chavrier P. and Goud,B. (1999) The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol., 11, 466–475. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., McBride,H.M., Burgoyne,R.D. and Zerial,M. (1999a) The Rab5 effector EEA1 is a core component of endosome docking. Nature, 397, 621–625. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska,M., Ashman,K., Wilm,M., Zhao,L., Yip,S.C., Waterfield,M.D., Backer,J.M. and Zerial,M. (1999b) Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nature Cell Biol., 1, 249–252. [DOI] [PubMed] [Google Scholar]
- Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A., Jollivet,F., Martinez,O., Lacapère,J.-J., Rousselet,A., Janoueix-Lerosey,I. and Goud,B. (1998) Interaction of a Golgi-associated kinesin-like protein with Rab6. Science, 279, 580–585. [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal,B.M., Vaughan,K.T. and Vallee,R.B. (1996) Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol., 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth C.C., Kawata,M., Yoshida,Y., Takai,Y., Gelb,M.H. and Glomset,J.A. (1991) C terminus of the small GTP-binding protein smg p25A contains two geranylgeranylated cysteine residues and a methyl ester. Proc. Natl Acad. Sci. USA, 88, 6196–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Press,B. and Wandinger-Ness,A. (1995) Rab7: an important regulator of late endocytic membrane traffic. J. Cell Biol., 131, 1435–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T.R., Niles,E.G., Studier,F.W. and Moss,B. (1986) Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 83, 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.R., Schroer,T.A., Szilak,I., Steuer,E.R., Sheetz,M.P. and Cleveland,D.W. (1991) Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J. Cell Biol., 115, 1639–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Drubin,D.G. and Barnes,G. (2000) Functional cooperation between the microtubules and actin cytoskeletons. Curr. Opin. Cell Biol., 12, 63–71. [DOI] [PubMed] [Google Scholar]
- Gournier H., Stenmark,H., Rybin,V., Lippe,R. and Zerial,M. (1998) Two distinct effectors of the small GTPase Rab5 cooperate in the endocytic membrane fusion. EMBO J., 17, 1930–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy J., Corpet,F. and Kahn,D. (1999) Whole genome protein domain analysis using a new method for domain clustering. Comput. Chem., 23, 333–340. [DOI] [PubMed] [Google Scholar]
- Griparic L., Volosky,J.M. and Keller,T.C. (1998) Cloning and expression of chicken CLIP-170 and restin isoforms. Gene, 206, 195–208. [DOI] [PubMed] [Google Scholar]
- Guarente L. (1983) Yeast promoters and LacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol., 101, 181–189. [DOI] [PubMed] [Google Scholar]
- Hill J., Donald,K.A. and Griffins,D.E. (1991) DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res., 19, 5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I., Jollivet,F., Camonis,J., Marche,P.N. and Goud,B. (1995) Two-hybrid system screen with the small GTP-binding protein Rab6. J. Biol. Chem., 270, 14801–14808. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Laug,W.E. and Benedict,W.F. (1975) Fibrinolytic activity in a human fibrosarcoma cell line and evidence for the induction of plasminogen activator secretion during tumor formation. Cell, 6, 245–252. [DOI] [PubMed] [Google Scholar]
- Karki S. and Holzbaur,E.L. (1995) Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J. Biol. Chem., 270, 28806–28811. [DOI] [PubMed] [Google Scholar]
- King S.M. (2000) The dynein microtubule motor. Biochim. Biophys. Acta, 1496, 60–75. [DOI] [PubMed] [Google Scholar]
- Knowles B.B., Howen,C.C. and Aden,D.P. (1980) Human hepatocellular carcinoma cell lines secrete the major plasma protein and hepatitis B surface antigen. Science, 209, 497–499. [DOI] [PubMed] [Google Scholar]
- Li C., Takei,K., Geppert,M., Daniell,L., Stenius,K., Chapman,E.R., Jahn,R., De Camilli,P. and Sudhof,T.C. (1994) Synaptic targeting of Rabphilin-3A, a synaptic vesicle Ca2+/phospholipid binding protein, depends on Rab3A/3C. Neuron, 13, 885–898. [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Mangeat P., Roy,C. and Martin,M. (1999) ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol., 9, 187–192. [DOI] [PubMed] [Google Scholar]
- McBride H.M., Rybin,V., Murphy,C., Giner,A., Teasdale,R. and Zerial,M. (1999) Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell, 98, 377–386. [DOI] [PubMed] [Google Scholar]
- Méresse S., Gorvel,J.-P. and Chavrier,P. (1995) The Rab7 GTPase resides on a vesicular compartment connected to lysosomes. J. Cell Sci., 108, 3349–3358. [DOI] [PubMed] [Google Scholar]
- Méresse S., Andre,P., Mishal,Z., Barad,M., Brun,N., Desjardins,M. and Gorvel,J.P. (1997) Flow cytometric sorting and biochemical characterization of the late endosomal rab7-containing compartment. Electrophoresis, 18, 2682–2688. [DOI] [PubMed] [Google Scholar]
- Méresse S., Steele-Mortimer,O., Finlay,B.B. and Gorvel,J.P. (1999) The Rab7 GTP-ase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J., 18, 4394–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaely P., Kamal,A., Anderson,R.G. and Bennett,V. (1999) A requirement for ankyrin binding to clathrin during coated pit budding. J. Biol. Chem., 274, 35908–35913. [DOI] [PubMed] [Google Scholar]
- Mu F.T. et al. (1995) EEA1, an early endosome-associated protein, is a conserved α-helical peripheral membrane protein flanked by cysteine ‘fingers’ and contains a calmodulin-binding IQ motif. J. Biol. Chem., 270, 13503–13511. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Ghosh,R.N. and Maxfield,F.R. (1997) Endocytosis. Physiol. Rev., 77, 759–803. [DOI] [PubMed] [Google Scholar]
- Nagelkerken B., Van Anken,E., Van Raak,M., Gerez,L., Mohrmann,K., Van Uden,N., Holthuizen,J., Perlkmans,L. and van der Sluijs,P. (2000) Rabaptin 4, a novel effector of the small GTPase Rab4a is recruited to perinuclear recycling vesicles. Biochem. J., 346, 593–601. [PMC free article] [PubMed] [Google Scholar]
- Nichols B.J., Ungermann,C., Pelham,H.R.B., Wickner,W.T. and Haas,A. (1997) Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature, 387, 199–202. [DOI] [PubMed] [Google Scholar]
- Nielsen E., Severin,F., Backer,J., Hyman,A. and Zerial,M. (1999) Rab5 regulates motility of early endosomes on microtubules. Nature Cell Biol., 1, 376–382. [DOI] [PubMed] [Google Scholar]
- Olkkonen V. and Stenmark,H. (1997) Role of Rab GTPases in membrane traffic. Int. Rev. Cytol., 176, 1–85. [DOI] [PubMed] [Google Scholar]
- Papini E., Satin,B., Bucci,C., de Bernard,M., Telford,J.L., Manetti,R., Rappuoli,R., Zerial,M. and Montecucco,C. (1997) The small GTP-binding protein Rab7 is essential for cellular vacuolation induced by Helicobacter pylori cytotoxin. EMBO J., 16, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranen J., Auvinen,P., Virta,H., Wepf,R. and Simons K. (1996) Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J. Cell Biol., 135, 153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S.R. (1999) Transport-vesicle targeting: tethers before SNAREs. Nature Cell Biol., 1, E17–E22. [DOI] [PubMed] [Google Scholar]
- Pierre P., Scheel,J., Rickard,J.E. and Kreis,T.E. (1992) CLIP-170 links endocytic vesicles to microtubules. Cell, 70, 887–900. [DOI] [PubMed] [Google Scholar]
- Pinto M. et al. (1983) Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell, 47, 323–330. [Google Scholar]
- Press B., Feng,Y., Hoflack,B. and Wandinger-Ness,A. (1998) Mutant Rab7 causes the accumulationn of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol., 140, 1075–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando R.R. (1996) Chemical biology of protein isoprenylation/methylation. Biochim. Biophys. Acta, 1300, 5–16. [DOI] [PubMed] [Google Scholar]
- Romano M., Razandi,M., Sekhon,S., Krause,W.J. and Ivey,K.J. (1988) Human cell line for study of damage to gastric epithelial cells in vitro.J. Lab. Clin. Med., 111, 430–440. [PubMed] [Google Scholar]
- Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. and Wickner,W. (1998) Functional reconstitution of Ypt7p GTPase and a purified vacuole SNARE complex. Science, 281, 700–702. [DOI] [PubMed] [Google Scholar]
- Schiestl R.H. and Giest,R.D. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet., 16, 339–346. [DOI] [PubMed] [Google Scholar]
- Seals D.F., Eitzen,G.,Margolis,N., Wickner,W.T. and Price,A. (2000) A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl Acad. Sci. USA, 97, 9402–9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A. et al. (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature, 394, 494–498. [DOI] [PubMed] [Google Scholar]
- Somsel Rodman J. and Wandinger-Ness,A. (2000) Rab GTPases coordinate endocytosis. J. Cell Sci., 113, 183–192. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L. and Kahn,D. (1994) Modular arrangement of proteins as inferred from analysis of homology. Protein Sci., 3, 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Vitale,G., Ullrich,O. and Zerial,M. (1995) Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell, 83, 423–432. [DOI] [PubMed] [Google Scholar]
- Ungermann C., Price,A. and Wickner,W. (2000) A new role for a SNARE protein as a regulator of the Ypt7/Rab-dependent stage of docking. Proc. Natl Acad. Sci. USA, 97, 8889–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valetti C., Wetzel,D.M., Schrader,M., Hasbani,M.J., Gill,S.R. and Kreis,T.E. (1999) Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol. Biol. Cell, 10, 4107–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K.T. and Vallee,R.B. (1995) Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol., 131, 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale G., Rybin,V., Christoforidis,S., Thornqvist,P.-O., McCaffrey,M., Stenmark,H. and Zerial,M. (1998) Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J., 17, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli R., Santillo,M., Lattero,D., Chiariello,M., Bifulco,M., Bruni,C.B. and Bucci,C. (1997) Role of the small GTP-ase Rab7 in the late endocytic pathway. J. Biol. Chem., 272, 4391–4397. [DOI] [PubMed] [Google Scholar]
- Vojtek A.B., Hollenberg,S.M. and Cooper,J.A. (1993) Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell, 74, 205–214. [DOI] [PubMed] [Google Scholar]
- Waters M.G. and Pfeffer,S.R. (1999) Membrane tethering in intracellular transport. Curr. Opin. Cell Biol., 11, 453–459. [DOI] [PubMed] [Google Scholar]