Abstract
The RUNX family genes are the mammalian homologs of the Drosophila genes runt and lozenge, and members of this family function as master regulators of definitive hematopoiesis and osteogenesis. The RUNX genes encode the α subunit of the transcription factor PEBP2/CBF. The β subunit consists of the non-RUNX protein PEBP2β. We found that RUNX1/AML1, which is essential for hematopoiesis, is continuously subjected to proteolytic degradation mediated by the ubiquitin–proteasome pathway. When PEBP2β is present, however, the ubiquitylation of RUNX1 is abrogated and this causes a dramatic inhibition of RUNX1 proteolysis. Heterodimerization between PEBP2β and RUNX1 thus appears to be an essential step in the generation of transcriptionally competent RUNX1. Consistent with this notion, RUNX1 was barely detected in PEBP2β–/– mouse. CBF(PEBP2)β– SMMHC, the chimeric protein associated with inv(16) acute myeloid leukemia, was found to protect RUNX1 from proteolytic degradation more efficiently than PEBP2β. These results reveal a hitherto unknown and major role of PEBP2β, namely that it regulates RUNX1 by controlling its turnover. This has allowed us to gain new insights into the mechanism of leukemogenesis by CBFβ–SMMHC.
Keywords: AML1/PEBP2β/proteolytic degradation/Runx1/ubiquitylation
Introduction
The Runt domain transcription factor, PEBP2/CBF, is a heterodimeric transcription factor which is one of the major targets of transforming growth factor-β (TGF-β) and bone morphogenetic protein (BMP) (Hanai et al., 1999; Zhang et al., 2000). It is involved in the regulation of gene expression in a variety of biological activities, most notably hematopoiesis and osteogenesis (Ito, 1997, 1999). Its β subunit, denoted as PEBP2β/CBFβ, is homologous to the Drosophila Runt-binding proteins, Brother and Big brother. It does not bind to DNA, but the α subunit of PEBP2, which is homologous to the Drosophila gene products Runt and Lozenge, does have DNA-binding properties.
The α subunit is encoded by RUNX, the mammalian homolog of the runt gene from Drosophila. RUNX contains an evolutionarily conserved 128 amino acid region termed the Runt domain, which is required both for DNA binding and heterodimerization with the β subunit (Kagoshima et al., 1993). In mammals, three α subunit genes exist, namely RUNX1, RUNX2 and RUNX3 (also referred to, respectively, as PEBP2αB, PEBP2αA and PEBP2αC, or CBFA2, CBFA1 and CBFA3, or AML1, AML3 and AML2) (Ito, 1999). RUNX1, which is frequently altered by the chromosome translocations associated with human leukemia (Look, 1997), is essential for inducing definitive hematopoiesis. It is also critical in regulating hematopoietic cell-specific genes in a variety of blood cells (Ito, 1997, 1999). RUNX2 is essential for the generation and maturation of osteoblasts (Komori et al., 1997; Otto et al., 1997) and it plays pivotal roles in regulating the expression of bone-specific genes such as osteocalcin and osteopontin (Ducy et al., 1996). Haplo-insufficiency of RUNX2 causes cleidocranial dysplasia, a human autosomal-dominant bone disease characterized by malformation of the clavicle and skull (Mundlos et al., 1997). RUNX3 is required for immunoglobulin class switching from IgM to IgA (Shi and Stavnezer, 1998).
When either RUNX1 or PEBP2β are disrupted in the mouse, the resulting phenotypes are nearly identical, indicating that both subunits are involved in a common function and that both must be intact for this function to be maintained (Okuda et al., 1996; Wang et al., 1996a,b; Niki et al., 1997). The localization of the two subunits differs, however. The RUNX proteins (human RUNX and mouse Runx proteins are virtually identical and will both be referred to here as RUNX) bear nuclear localization signals and are considered to be nuclear proteins (Kanno,T. et al., 1998), whereas PEBP2β is found mainly in the cytoplasm, at least when it is overexpressed in rodent fibroblasts (Lu et al., 1995). Even when the two subunits are simultaneously overexpressed in the same cell, they do not colocalize. That the two subunits are distributed in different subcellular compartments strongly suggests that their dimerization is controlled by a specific mechanism.
The N- and C-terminal regions of the RUNX proteins each contain a negative regulatory domain for heterodimerization, denoted NRHn and NRHc, respectively. These domains block the association of PEBP2β with the Runt domain (Kim et al., 1999). Artificial deletion of either NRHn or NRHc allows PEBP2β to interact fully with the Runt domain (Lu et al., 1995). The ability of the Runt domain to bind to DNA is also blocked intramolecularly by additional N- and C-terminal regions. These regions are termed the negative regulatory domains for DNA binding and are denoted as NRDBn and NRDBc. The binding of PEBP2β to the Runt domain unmasks the DNA-binding interface of the Runt domain and thus allows the complex to bind stably to DNA (Kim et al., 1999). Thus, the dimerization of PEBP2β with RUNX is the critical step in the activation of PEBP2.
We observed previously that the RUNX proteins are susceptible to proteolytic degradation (Ogawa et al., 1993b). In this study, we show that the ubiquitin– proteasome system is largely responsible for this degradation. We also show that when PEBP2β dimerizes with RUNX it inhibits the ubiquitylation of RUNX, which is necessary for the protein to be targeted for proteolysis by the proteasome. These observations indicate that PEBP2β plays a major role in PEBP2 function. This in turn highlights the importance of RUNX–PEBP2β heterodimerization in regulating the biological processes mediated by PEBP2.
Results
RUNX1 is protected from proteolytic degradation by proteasome-specific inhibitors
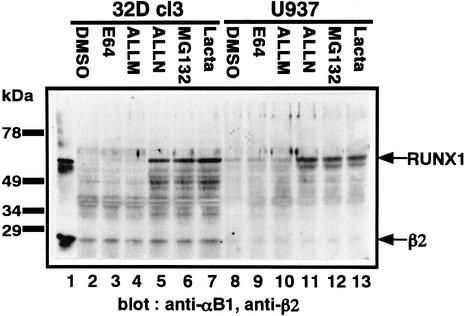
The C-terminal part of RUNX is rich in proline, glutamic acid, serine and threonine (Figure 1), which is characteristic of proteins that are susceptible to proteolysis (Ogawa et al., 1993b). The rapid proteolytic degradation of RUNX proteins explains why they are often difficult to detect as intact protein within cells (Kamachi et al., 1990; Ogawa et al., 1993b). To assess which proteases are responsible for RUNX1 degradation, we tested a variety of protease inhibitors for their ability to stabilize RUNX1. A murine myeloid progenitor cell line, 32D cl3, and a human monocytic leukemia cell line, U937, were treated overnight with five different protease inhibitors, after which whole-cell extracts were run on an SDS gel and probed with specific antibodies against RUNX1 and PEBP2β (Figure 2). Of the five inhibitors, the proteasome-specific inhibitors MG132 and lactacystin, as well as the calpain and proteasome inhibitor ALLN, protected RUNX1 from proteolysis and allowed full-length RUNX1 to be detected (Figure 2), unlike the controls (Figure 2, lanes 2 and 8). Treatment with the calpain inhibitor ALLM or the lysosomal inhibitor E64 did not reveal a distinct RUNX1 band (Figure 2). The inhibitors had equivalent effects regardless of the cell studied, and indeed, stabilization of RUNX1 by the proteasome inhibitor MG132 was also observed to occur in the Kasumi-1, SKNO-1 and HL-60 human myeloid leukemia cell lines (data not shown). Thus, RUNX1 appears to be degraded through the ubiquitin–proteasome system.
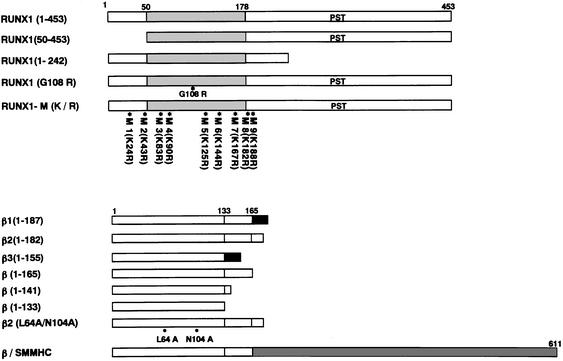
Fig. 1. Diagrammatic representations of RUNX1- and PEBP2β-derived constructs used in this study. The positions of Lys in RUNX1 are indicated by asterisks with numbered markers, M1–9. The positions of the mutation, G108R, in RUNX1 and those of L64A and N104A in PEBP2β are also indicated. PST, the region rich in Pro, Ser and Thr.
Fig. 2. Stabilization of RUNX1 by protease inhibitors. 32D cl3 and U937 were treated with five different protease inhibitors and DMSO (solvent) overnight, after which whole-cell extracts were prepared, run on an SDS gel and probed with anti-αB1 and anti-β2 polyclonal antibodies. The positions of RUNX1 and PEBP2β2 (shown as β2) on the gel are indicated by arrows.
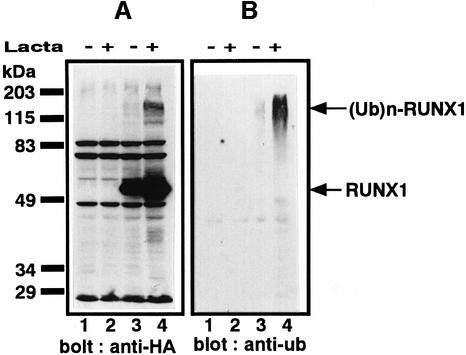
To confirm that RUNX1 is a substrate for the ubiquitin–proteasome system, we assessed whether RUNX1 is ubiquitylated within the cell. RUNX1 tagged with HA at the N-terminus and His6 at the C-terminus was synthesized in P19 embryonal carcinoma cells; in these cells, endogenous RUNX1 is not expressed at significant levels. The resulting RUNX1 protein was purified from the P19 cell extracts with NTA resin and analyzed by western blotting using the anti-HA antibody. When proteolysis was blocked by lactacystin, not only full-length RUNX1 but also multiple size classes of proteins with molecular masses higher than that of the full-length RUNX1 were observed (Figure 3A, lanes 3 and 4). When a similarly prepared blot was probed with the anti-ubiquitin antibody, these higher molecular mass species of RUNX1 were also detected, confirming that RUNX1 is ubiquitylated within the cell (Figure 3B).
Fig. 3. Ubiquitylation of RUNX1. P19 cells were transfected with the plasmids of expression vector alone or plasmid expressing RUNX1 tagged with HA at the N-terminus and His6 at the C-terminus. Twenty-four hours after transfection, the cells were treated with lactacystin and incubated for an additional 12 h. RUNX1 proteins were purified from the cell extract by NTA resin under denaturing conditions. Western blotting was performed with anti-HA (A) or anti-ubiquitin (ub) (B): lane 1, vector alone without lactacystin; lane 2, vector alone with lactacystin; lane 3, HA-tagged RUNX1 without lactacystin; lane 4, HA-tagged RUNX1 with lactacystin. The positions of RUNX1 and ubiquitylated RUNX1 [(Ub)n-RUNX1] are indicated by arrows.
RUNX1 stability is increased by substitution of its lysines with arginines
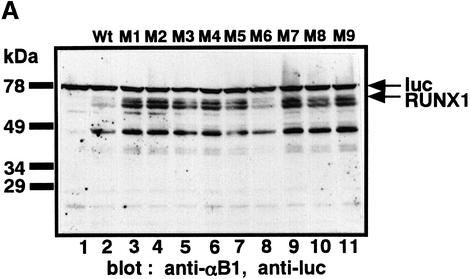
In susceptible proteins, the target of ubiquitylation is lysine (Lys) (Hershko and Ciechanover, 1998). Thus, we examined how the stability of RUNX1 could be affected by mutations of the nine Lys residues (Figure 1) into arginine (Arg), one by one. RUNX1 and the nine mutants were expressed in P19 cells and the proteins were detected by western blotting using a polyclonal antibody against RUNX1 (Figure 4A). All proteins except for K144R (M6) were more stable than wild-type RUNX1 (Figure 4A), while K144R was only as stable as wild-type RUNX1 (Figure 4A, compare lane 2 with lane 8). These results suggest that the Lys residues at positions 24, 43, 83, 90, 125, 167, 183 and 188 are likely targets of ubiquitylation. Notably, the Lys residues in RUNX1 are well conserved among the Runt-domain proteins from species ranging from Caenorhabditis elegans to humans (Figure 4B). In addition, they are centered around the Runt domain.
Fig. 4. Stabilization of RUNX1 by substitution of Lys by Arg. (A) P19 cells were transfected with the plasmids expressing wild-type RUNX1 (Wt, lane 2) or mutants with a single amino acid substitution of Lys to Arg (mutants M1–9, lanes 3–11). Whole-cell extracts were analyzed by western blotting using anti-αB1 and anti-luciferase (anti-luc) as an internal control. The positions of RUNX1 and luc are indicated by arrows. (B) Amino acid sequence comparison of the Runt domains of different animal species. Conserved Lys are indicated by bold letter K. The amino acid sequence accession No. of each protein in the Swiss-Prot and DDBJ/EMBL/GenBank databases is given in parentheses: Runx1 (D13802), residues 50–177; Runx2 (D14636), residues 94–221; RUNX1 (Q01196), residues 50–177; RUNX2 (Q08775), residues 102–219; RUNX3 (Q13761), residues 54–181; xAML1 (O73725), residues 50–177; spRUNT (Q26628), residues 57–184; ceRUNT (O01834), residues 10–137; dLOZENGE (Q24183), residues 278–405; and dRUNT (Q24709), residues 106–233.
However, it is rather surprising that single Lys→Arg substitutions among multiple potential target sites could so significantly decrease the proteolytic susceptibility of the protein. Thus, only a part of these eight Lys residues might be actual targets of ubiquitylation and the remainder could be involved in some other rate-limiting step(s), for instance, the recognition of RUNX1 by its cognate ubiquitylation system. Further intensive studies will be needed to resolve this enigma.
RUNX1 is stabilized by its heterodimerization with the β subunit
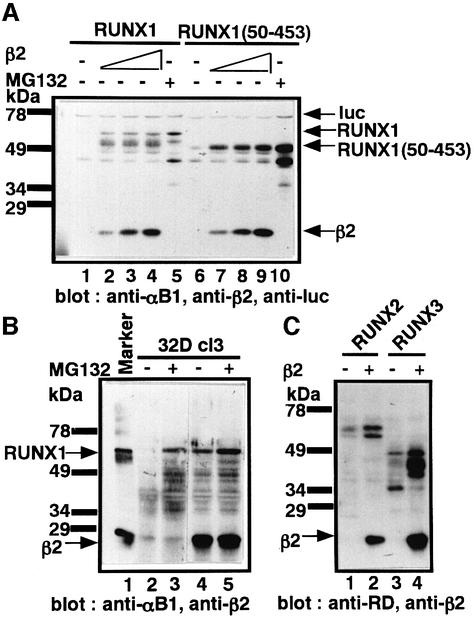
As the Lys residues congregate around the Runt domain, which mediates β subunit binding, it is possible that heterodimerization of RUNX1 with the β subunit impedes its degradation. To test this, we first examined whether RUNX1 in the α/β heterodimer is more stable than free RUNX1. We thus co-expressed RUNX1 and the β2 isoform of PEBP2β in P19 cells and subjected the cellular proteins to western blot analysis with polyclonal antibody against RUNX1 and PEBP2β (Figure 5A). A dramatic stabilization of RUNX1 was observed in a manner dependent on the dosage of PEBP2β, as seen from the appearance of a full-length RUNX1 together with one major and several minor degradation intermediates lacking ∼25, 45, 60 and 100 amino acids (Figure 5A, compare lane 1 with lanes 2–4). Similar PEBP2β-mediated protection was also observed with RUNX1 derivatives partially deleted of N-terminal or C-terminal regions but still retaining the Runt domain [RUNX1(50–453), lanes 6–10 in Figure 5A; RUNX1(1–242), data not shown].
Fig. 5. Stabilization of RUNX1 by heterodimerization with PEBP2β. (A) Transient expression of RUNX1 (lanes 1–5) without (lane 1) or with increasing amounts (lanes 2–4) of PEBP2β. Lane 5 represents the cells treated with MG132. Whole-cell extracts of P19 cells were analyzed using anti-αB1, anti-β2 and anti-luc (an internal control). (B) Effects of exogenously expressed PEBP2β2 and proteasome inhibitor MG132 on the stability of endogenous RUNX1 in 32D cl3 cells. Whole-cell extracts were analyzed by using anti-αB1 and anti-β2. (C) Stabilization of RUNX2 and RUNX3 by PEBP2β. RUNX2 or RUNX3 with or without PEBP2β2 (shown as β2) was exogenously expressed in P19 cells and whole-cell extracts were analyzed by anti-Runt domain, anti-RD(10B7G8) and anti-β2.
We further examined the effect of exogenously expressed PEBP2β on endogenous RUNX1 using 32D cl3 cells as the host cells. As already shown in Figure 2, RUNX1 in 32D cl3 cells could be stabilized by the proteasome inhibitor MG132 (Figure 5B, lanes 2 and 3). When the 32D cl3 cells stably expressed exogenous PEBP2β (Figure 5B, lanes 4 and 5), RUNX1 became detectable even in the absence of MG132. The other RUNX proteins, RUNX2 and RUNX3, could also be stabilized by PEBP2β (Figure 5C).
Mutants of RUNX1 and PEBP2β that cannot heterodimerize do not protect RUNX1
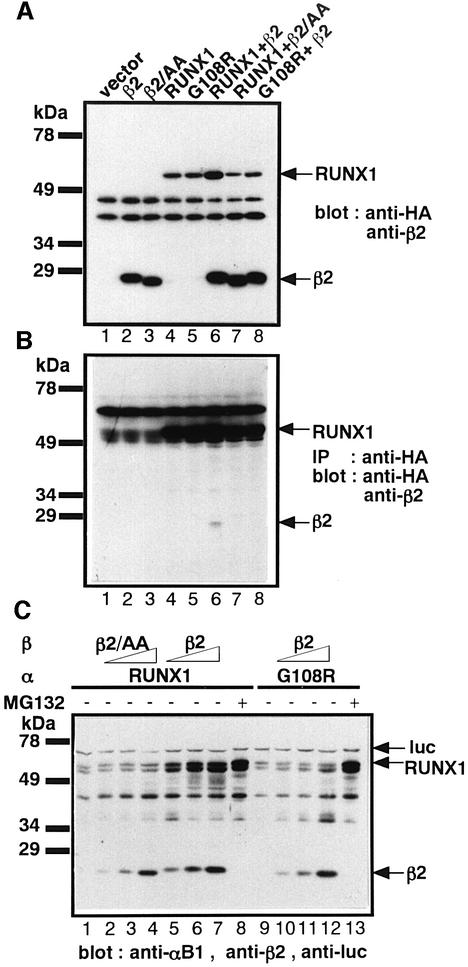
We then examined the effect on RUNX1 proteolysis of mutations that abrogate heterodimerization. Two mutants known to be incapable of heterodimerization, namely, RUNX1(G108R) (Akamatsu et al., 1997b) and PEBP2β(L64A/N104A) (Tang et al., 2000), were studied. That neither mutant could form heterodimers with the wild-type proteins was verified by immunoprecipitation followed by western blotting (Figure 6B, lane 6). When PEBP2β was co-expressed with RUNX1 in P19 cells, RUNX1 was well protected, as expected (Figure 6A, lane 6). In contrast, RUNX1(G108R) could not be stabilized by wild-type PEBP2β (Figure 6A, lane 8), nor could PEBP2β(L64A/N104A) stabilize wild-type RUNX1 (Figure 6A, lane 7). These observations were studied more quantitatively by co-transfecting a constant amount of the RUNX1 expression plasmid with increasing amounts of the PEBP2β expression plasmid (Figure 6C). These experiments again show that RUNX1 is stabilized well by PEBP2β (Figure 6C, compare lane 1 with lanes 5–7), but that RUNX1(G108R) was only weakly protected by PEBP2β (Figure 6C, compare lane 9 with lanes 10–12). PEBP2β(L64A/N104A) does not stabilize RUNX1 at all (Figure 6C, compare lane 1 with lanes 2–4).
Fig. 6. Mutations that abrogate heterodimerization eliminate the ability of PEBP2β to protect RUNX1 from proteolysis. (A) Transient expression of RUNX1, PEBP2β2 and their derivatives alone or in combinations in P19 cells. Whole-cell extracts were prepared from half of the transfected cells and analyzed by western blotting using anti-HA and anti-β2. Notations above the gel pattern indicate: β2/AA, PEBP2β2(L64A, N104A); β2, PEBP2β2; G108R, RUNX1(G108R). (B) The remaining half of the transfected cells in (A) were lysed and immunoprecipitated with polyclonal anti-HA antibody and then analyzed by western blotting using monoclonal anti-HA and polyclonal anti-β2. The strong band present in all lanes in the upper part of the gel is non-specific. (C) RUNX1 or RUNX1(G108R) was transiently expressed in P19 cells with increasing amounts of PEBP2β2 (lanes 2–4) or PEBP2β2(L64A, N104A) (lanes 5–7). Where indicated, proteasome inhibitor MG132 was added to the cell culture medium 24 h after transfection. The notations above the panel are the same as in (A). Whole-cell extracts were prepared after 36 h of incubation and analyzed by western blotting using anti-αB1, anti-β2 and anti-luc (internal control).
PEBP2β inhibits the in vitro ubiquitylation of the Runt domain
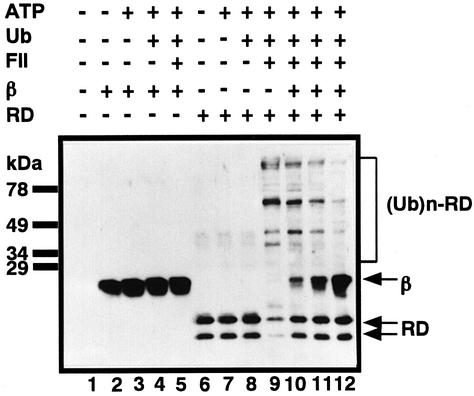
To test directly whether the heterodimerization of RUNX1 with PEBP2β abrogates its ubiquitylation, we assessed the in vitro ubiquitylation of the purified Runt domain with and without PEBP2β. Upon incubation with ubiquitin, ATP and fraction II of HeLa cells, the Runt domain was effectively ubiquitylated as shown by the appearance of species with higher molecular weights (Figure 7, lanes 6–9). When increasing amounts of purified PEBP2β were added to this system, the formation of ubiquitylated Runt-domain molecules was progressively inhibited (Figure 7, lanes 9–12). Purified PEBP2β itself was not ubiquitylated in this system at all (Figure 7, lanes 2–5). These observations strongly suggest a new role for PEBP2β, namely that it inhibits RUNX1 ubiquitylation, and hence proteasome cleavage, by heterodimerizing with it.
Fig. 7. Inhibition of in vitro ubiquitylation of the Runt domain by PEBP2β. Purified His6-tagged PEBP2β (β) or the Runt domain (RD) were ubiquitylated in vitro. In the case of lanes 10–12, the Runt domain was incubated with increasing amounts of PEBP2β (at ratios of 1:1, 1:5, 1:10) 2 h before ubiquitylation. After ubiquitylation was stopped, western blotting was performed with anti-αB1 and anti-β2.
The presence of the β subunit extends the half-life of RUNX1
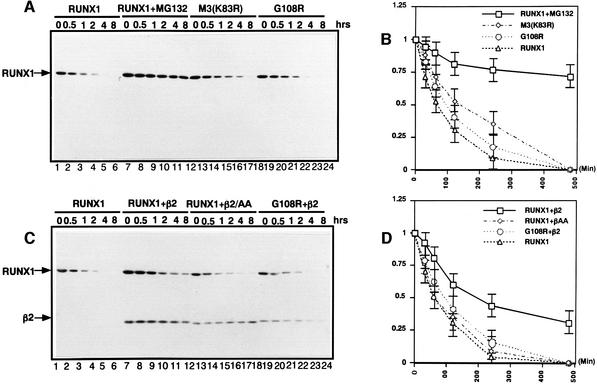
To confirm and extend our observations described above, the effect of PEBP2β on the half-life of RUNX1 was assessed by pulse–chase experiments. The half-life of RUNX1 in its free form is ∼60 min (Figure 8B). The proteasome inhibitor MG132 strongly protected RUNX1 from degradation, as 75% of the protein remained intact after 500 min (Figure 8B). The half-life of RUNX1 (K83R) (M3) was 130 min, more than twice as long as that of RUNX1, consistent with the result of the stabilization (Figure 4A). The heterodimerization-negative mutant, RUNX1(G108R), had a half-life similar to that of wild-type RUNX1 (Figure 8B). In the presence of PEBP2β, however, the half-life of RUNX1 was markedly extended to 200 min, 3-fold longer than that of RUNX1 on its own (Figure 8D). PEBP2β(L64A/N104A) did not alter the half-life of RUNX1 at all, nor did PEBP2β prolong the half-life of RUNX1(G108R) (Figure 8D). All these observations are entirely consistent with the notion that PEBP2β protects RUNX1 from proteolytic degradation.
Fig. 8. The half-life of RUNX1 in the absence or presence of PEBP2β. (A) HA-tagged RUNX1, RUNX1(K83R, M3) or RUNX1(G108R) was transiently expressed in P19 cells. Thirty hours after transfection, cells were pulsed with [35S]methionine for 1 h and chased in the presence of unlabeled methionine. Cell lysates were prepared at the times indicated. Immunoprecipitation was performed with anti-HA antibody. In lanes 7–12, the cells had been treated with MG132. After running on SDS–PAGE, RUNX1 bands were visualized by autoradiography. The intensities of protein bands were quantified by BAS2000 (Fuji). (B) Graphic representation of the data obtained in (A). (C) HA-tagged RUNX1 or RUNX1(G108R) was transiently expressed in P19 cells in the presence or absence of PEBP2β2 or PEBP2β2(L64A, N104A). Pulse–chase experimentation was performed as described for (A). The intensities of protein bands were quantified by BAS2000. (D) Graphical representation of the data obtained in (C). In (B) and (D), mean values from five independent experiments, with statistical deviation, are indicated.
RUNX1 is barely detectable in PEBP2β–/– mouse
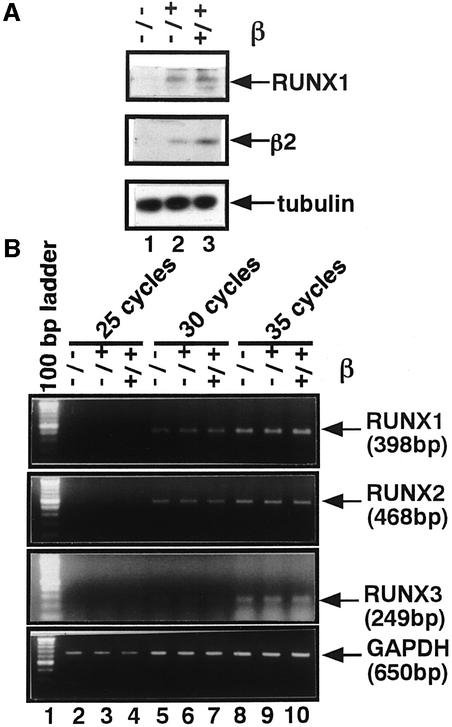
Since we found that PEBP2β protects RUNX1 from proteolysis, we decided to examine the amount of RUNX1 molecule present in the PEBP2β–/– mouse. Whole embryo extracts were prepared from PEBP2β–/– embryos at 11.5 days post-conception (d.p.c.) as well as from PEBP2β+/+ and PEBP2β+/– littermate embryos. RUNX1 was detectable in wild-type and heterozygous mutant embryos but not in the homozygous mutant (Figure 9A).
Fig. 9. Reduced stability of RUNX1 in PEBP2β–/– mouse embryo. (A) Whole-cell extracts were prepared from 11.5 d.p.c. mouse embryos of various zygosities in PEBP2β, and western blotting was performed using anti-αB1, anti-β2 and anti-tubulin (internal control): lane 1, β–/–; lane 2, β+/–; lane 3, β+/+. (B) Transcripts from the three RUNX genes in the embryo extracts as obtained above were quantified by RT–PCR assay. GAPDH was used as a control. cDNAs amplified for the cycles indicated were separated by agar gel electrophoresis and vizualized by staining with ethidium bromide: lanes 2, 5 and 8, β–/–; lanes 3, 6 and 9, β+/–; lanes 4, 7 and 10, β+/+.
In view of the recent proposal that the transcription of RUNX2, and possibly also of the other RUNX genes, is subject to a positive autoregulation (Ducy et al., 1999), we also measured transcripts from the three RUNX genes in the above embryo extracts by an RT–PCR assay. For all the RUNX genes, the levels of transcripts remained virtually constant regardless of whether PEBP2β was mutated or not (Figure 9B). This indicates that the autoregulation of transcription is not operating for either RUNX gene, at least for the embryonic stage as examined.
Taken together, these observations support the view that PEBP2β protects RUNX1 from proteolysis in vivo, thus re-emphasizing the newly postulated physiological role of PEBP2β as a regulator of RUNX1.
β/SMMHC stabilizes RUNX1 more efficiently than PEBP2β
The chromosome rearrangement inversion 16 [inv(16)], which is associated with the FAB M4 Eo subtype of acute myeloid leukemia, produces a chimeric protein consisting of almost the entire PEBP2β molecule (amino acids 1–165) N-terminally fused to the C-terminal coiled-coil region of smooth muscle myosin heavy chain (SMMHC) (Liu et al., 1993). The minimum region of PEBP2β required for heterodimerization with RUNX1 is contained within the first 135 amino acids and is, therefore, present in the chimeric protein (Kagoshima et al., 1996).
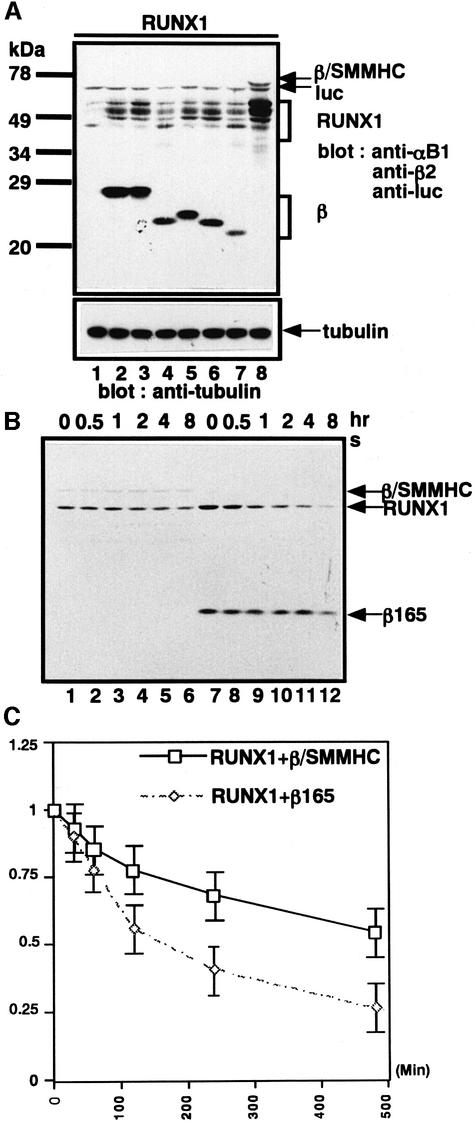
We have suggested previously that PEBP2β–SMMHC (hereafter referred to as β/SMMHC) may dimerize with RUNX1 more efficiently than PEBP2β (Lu et al., 1995). Thus we were prompted to compare β/SMMHC with several naturally occurring variants or artificially generated deletion mutants of PEBP2β for their ability to stabilize RUNX1. Of the three naturally occurring isoforms of PEBP2β, β1 and β2 have been found to perform indistinguishably in dimerization and transactivation assays, whereas β3 functions poorly (Ogawa et al., 1993a). Consistent with these dimerization capacities, β1 and β2 were found to be efficient in stabilizing RUNX1, while β3 was not (Figure 10A, lanes 1–4). We also tested the C-terminally truncated β2 proteins β2(1–165), β2(1–141) and β2(1–133). The first two are known to heterodimerize with RUNX1 as efficiently as β1 and β2, while β2(1–133) is less efficient (Ogawa et al., 1993a). As expected, the first two stabilized RUNX1 as efficiently as β1 and β2 (Figure 9A, lanes 5 and 6), whereas β2(1–133) did so much less efficiently (Figure 10A, lane 7). Compared with all the β subunit-related proteins tested here, β/SMMHC protected RUNX1 most efficiently, resulting in much greater quantities of the full-length RUNX1 being retained in the cells (Figure 10A, lane 8). To confirm this more quantitatively, we examined the half-life of RUNX1 when heterodimerized with either β2(1–165) or β/SMMHC. The half-life of RUNX1 in the former complex was ∼150 min, while its half-life in the latter complex was >500 min (Figure 10B and C).
Fig. 10. β/SMMHC stabilizes co-expressed RUNX1 far more efficiently than any natural and artificially truncated β subunits. (A) RUNX1 was co-expressed with the β derivatives indicated, and whole-cell extracts were analyzed by western blotting with anti-αB1, anti-β2 and anti-luc (internal control). Anti-tubulin was used as a loading control. Lane 1, the backbone vector; lane 2, β1; lane 3, β2; lane 4, β3; lane 5, β165; lane 6, β141; lane 7, β133; lane 8, β/SMMHC. (B) β/SMMHC increases the half-life of RUNX1 much more than its β-derived portion, β165. RUNX1 was co-expressed with β/SMMHC or β165 in P19 cells, and pulse–chase experiments were performed as described in the legend to Figure 9A. (C) Graphical representation of the data obtained from (B). In (B) and (D), mean values from five independent experiments, with statistical deviation, are indicated.
Discussion
Regulation of PEBP2 by the ubiquitin–proteasome system
We have shown here that RUNX1 is proteolytically degraded predominantly by the ubiquitin–proteasome system because inhibitors of this system, but not other protease inhibitors, allowed intracellular detection of this otherwise hardly detectable protein.
The 26S proteasome is an ATP-dependent multicatalytic protease complex that plays a major role in selective protein degradation in eukaryotic cells. It is distributed widely throughout the cell and plays an essential role in rapidly eliminating many short-lived key regulatory proteins such as cell cycle proteins (cyclins, CDKIs), rate-limiting enzymes (ornithine decarboxylase), transcriptional regulators (IκB–NFκB complex, Jun, Fos, p53, MyoD, Myc, E2F, etc.), abnormal proteins and membrane proteins (Ciechanover, 1998; Hershko and Ciechanover, 1998). Proteins targeted for proteolysis are ligated with multiple ubiquitin molecules. The ligation involves a series of enzymic reactions where ubiquitin is first activated with ATP by the ubiquitin-activating enzyme (E1) and then transferred onto a ubiquitin carrier protein (E2). In the third step, the ubiquitin is covalently conjugated by ubiquitin ligase (E3) onto lysine residues of the target protein. Multiply ubiquitylated proteins are recognized by the 19S regulatory subunit of proteasome and subsequently rapidly degraded into short peptides in the 20S catalytic subunit of proteasome.
We found that single mutations at eight of the nine RUNX1 Lys residues to Arg served to protect RUNX1 from degradation. The Lys residues of RUNX1 cluster within or around the highly conserved Runt domain with which the β subunit interacts. Thus, we speculated that the heterodimerization of PEBP2β with RUNX1 might prevent the ubiquitylation of RUNX1 and hence stabilize it. Ample evidence for this notion was obtained, as co-expression of PEBP2β with RUNX1 was found to prevent the degradation of RUNX1 and to extend the intracellular half-life of RUNX1, while a mutant of PEBP2β that could not dimerize with the wild-type RUNX1 could not stabilize it either. Furthermore, PEBP2β inhibited ubiquitylation of the Runt domain of RUNX1 in vitro. In addition, RUNX1 was found to be barely detectable in the PEBP2β–/– embryo, which provides in vivo evidence that supports our hypothesis.
A new function of PEBP2β as the anti-proteolytic regulator for RUNX
The role played by PEBP2β in RUNX1 regulation thus appears to be even more dynamic and profound than previously envisioned. It seems as though RUNX1 is continuously synthesized and degraded and, when the functional form is required, the β subunit, which is metabolically stable and ubiquitously expressed, quickly forms a heterodimer. This role of PEBP2β is not unique to RUNX1, as it appears to be applicable to RUNX2 and RUNX3 as well (Figure 5C). Thus, the protection of the α subunit must be considered to be a major role played by the β subunit of PEBP2. Since haplo-insufficiency of RUNX1 causes familial platelet disorder with predisposition to acute myeloid leukemia (FPD/AML), and that of RUNX2 cleidocranial dysplasia (CCD) (Werner et al., 1999), dosage of functional PEBP2 would have to be tightly regulated. Stabilization of RUNX proteins by heterodimerization with PEBP2β would be very appropriate for this purpose.
As the mechanism of inhibition of ubiquitylation of RUNX1 by PEBP2β, the most simple imaginable possibility may be that PEBP2β physically masks Lys residues in RUNX1 upon heterodimerization. Contrary to this expectation, however, the three-dimensional structure of the Runt domain reveals that most of its Lys residues are located away from the interface that makes contact with PEBP2β (Nagata et al., 1999; Warren et al., 2000). Since the Runt domain by itself is a good substrate for ubiquitylation (Figure 7), a ubiquitin ligase (E3) must also interact with the Runt domain. It is conceivable that PEBP2β may make the Runt domain inaccessible to an E3 molecule either through conformational influence or steric obstruction. Of note here is that PEBP2β itself is completely resistant to ubiquitylation, even though it contains several Lys residues. Thus, PEBP2β appears to be aptly structured for its suggested role in protecting the Runt domain from attack by E3.
Regarding the interaction between the RUNX and PEBP2β in vivo, two critical questions arise as to where their heterodimerization takes place and by what mechanism this process is controlled. On the first point, we found previously that RUNX1 is a nuclear protein whereas PEBP2β is a cytoplasmic protein, at least when these proteins are overexpressed in rodent fibroblasts (Lu et al., 1995). Their differential subcellular localizations do not change regardless of whether they are expressed individually or simultaneously. Similarly, even when RUNX1 was forced to colocalize with PEBP2β in the cytoplasm by C-terminally linking it to the ligand-binding domain of glucocorticoid receptor, no nuclear transport of PEBP2β was elicited by a subsequent translocation of RUNX1 upon the addition of dexamethasone (Kanno,Y. et al., 1998). These observations suggest, though do not prove, that the heterodimerization of the two subunits may take place in the nucleus rather than the cytoplasm. Evidence favoring this notion came from our recent observation that PEBP2β accumulates in the nucleus after treatment of cells with an inhibitor of nuclear export, leptomycin B (our unpublished data). Also, a substantial fraction of PEBP2β is found in the nuclear fraction of leukemic cell lines (Kanto et al., 2000). These observations suggest that PEBP2β by itself can enter into the nucleus and that its preferential cytoplasmic localization is effected by its active export from the nucleus.
On the second point, our previous work suggested that the intact RUNX1 protein naturally takes a functionally cryptic conformation which does not allow it to interact fully with DNA or heterodimerize with PEBP2β (Kim et al., 1999). It is conceivable that, when RUNX1 function is required, some specific signal is induced that promotes its stable heterodimerization with PEBP2β into PEBP2. The nature of this signal is not known. Although MAP kinase and TGF-β/BMP-activated Smads have been shown to stimulate the transactivation activity of RUNX proteins (Tanaka et al., 1996; Hanai et al., 1999), none of them seems to trigger the dimerization (unpublished observations). Interestingly, heterodimerization of RUNX1 with PEBP2β is promoted when RUNX1 interacts with Ets-1 (Kim et al., 1999). However, Ets-1 by itself did not protect RUNX1 from degradation (unpublished data).
The main cause of embryonic lethality of PEBP2β–/– mouse may be degradation of RUNX1
Gene disruption of the RUNX1 and PEBP2β genes results in very similar phenotypes (Okuda et al., 1996; Wang et al., 1996a,b; Niki et al., 1997). Our data indicate that, as PEBP2β is indispensable for the function of RUNX1 because of its ability to protect RUNX1 from degradation, disruption of the PEBP2β gene is essentially equivalent to the disruption of RUNX1. Consistent with this notion is that RUNX1 is only poorly detectable in the PEBP2β–/– mouse, unlike its wild-type and heterozygous littermates (Figure 9A). Leading on from this, it is reasonable to suggest that the effective amount of RUNX proteins present within the cell is dependent on the existing amount of PEBP2β. Thus, it might be more accurate to consider PEBP2β as a regulator of the amount of functional RUNX protein within the cell. It has been reported that the expression of the PEBP2β gene is stimulated when cells are activated by TGF-β or BMP (Li et al., 1998; S.-C.Bae, personal communication). It would be of great interest to determine whether, as we expect, the amount of RUNX protein in the cell rises as PEBP2β accumulates due to TGF-β or BMP stimulation. If this occurs, it would indicate that control of PEBP2β expression is one of the most critical steps in PEBP2 regulation.
β/SMMHC protects RUNX1 from proteolysis more efficiently than PEBP2β
We found in a previous study that the leukemogenic chimeric protein β/SMMHC appears to have dimerized readily with RUNX1 (Lu et al., 1995), indicating that RUNX1’s negative regulatory domains for heterodimerization, NRHn and NRHc, are unable to prevent β/SMMHC from interacting with the Runt domain. The present work further demonstrated that β/SMMHC is more potent in protecting RUNX1 from proteolysis. These observations collectively suggest that β/SMMHC may make an extra contact with the negative regulatory domains of RUNX1 in addition to the Runt domain, thereby gaining an increased ability to interact with RUNX1 than from direct contact with β/SMMHC. We are currently examining the details of RUNX1 and β/SMMHC interaction, as we believe that the molecular mechanism by which β/SMMHC overrides the inhibitory effect of NRHn and NRHc must be the basis of its leukemogenic potential.
Mutational suppression of RUNX1 degradation could contribute to leukemogenesis
The results of our single amino acid substitution experiments targeting Lys residues draw our attention to the special implication of such mutations in leukemogenesis. Indeed, two mutations have been reported to occur in association with human diseases among the eight Lys residues that are potential targets of ubiquitylation (Figure 1): a K83N mutation in RUNX1 in an acute myeloid leukemia patient (Osato et al., 1999) and a K210N mutation in RUNX2 (corresponds to K167N in the numbering for RUNX1) in a familial case of cleidocranial dysplasia (Werner et al., 1999). Both mutant proteins are defective in DNA binding and hence are unable to transactivate. These results suggest that the two mutations are not simple loss-of-function mutations but that they generate abnormal RUNX1 proteins which may act as dominantly interfering molecules. One way they could achieve this is by sequestering putative co-activator proteins that interact with RUNX1. This phenomenon was observed previously for AML1ΔN, which is the product of an alternatively spliced RUNX1 mRNA that lacks the N-terminal region right up to the middle of the Runt domain. By sequestering co-activators, AML1ΔN may inhibit the transactivation function of RUNX1 (Zhang et al., 1997). It is possible that full-length RUNX1 complexed with β/SMMHC and RUNX1(K83N) would also sequester interacting proteins and thus dominantly inhibit the function of RUNX1. This effect would contribute to the leukemogenic potential of these molecules.
Overview of multiple roles of PEBP2β
PEBP2β is known to regulate RUNX in several ways. As proposed previously, PEBP2β increases the affinity of RUNX proteins for DNA by heterodimerizing with RUNX without interacting with the DNA by itself. Molecular analysis of this heterodimerization has allowed us to understand better why the affinity of RUNX for DNA is increased by PEBP2β binding. RUNX1 alone binds very poorly to its cognate DNA site because of masking of the DNA-binding surface of the Runt domain by NRDBn and NRDBc. When the C-terminal 80 amino acids of RUNX1 corresponding to NRHc are cleaved off, leaving NRDBc intact for dimerization with PEBP2β, the resulting dimer binds strongly to DNA. We believe that the dimerization induces structural changes in RUNX1 in such a way that the DNA-binding surface is unmasked. The unmasking of the DNA-binding surface is thus an important role of PEBP2β (Kim et al., 1999). Heterodimerization also increases the affinity of the minimum DNA-binding domain not having NRHs and NRDBs for DNA. This must also be considered another role of PEBP2β (Kagoshima et al., 1996). Finally, as shown by structural analysis, PEBP2β maintains the SH form of the cysteine residue at position 81 in the Runt domain of RUNX1, which may contribute to better DNA binding by the Runt domain (Akamatsu et al., 1997a; Warren et al., 2000).
Here, we propose a novel role for PEBP2β in regulating RUNX proteins, namely that PEBP2β regulates RUNX turnover. In concert with the other biochemical activities of PEBP2β listed above, this role should be of prime biological significance in mobilizing RUNX on demand precisely and dynamically. The discovery of this new role of PEBP2β also significantly alters our understanding of PEBP2, which is an important transcription factor in an ever increasing variety of biological systems. The discovery of this new role of PEBP2β may also reveal new avenues in the development of drugs aiming to control human diseases caused by PEBP2 abnormalities.
Materials and methods
Chemicals and reagents
Lysosomal protease inhibitor E64, calpain inhibitor ALLM, calpain and proteasome inhibitor ALLN, proteasome inhibitors MG132 and lactacystin were purchased from MBL (Nagoya, Japan). To inhibit cell proteolysis, E64 (50 mM), ALLM (25 mM), ALLN (25 mM) and MG132 (25 mM) dissolved in dimethylsulfoxide (DMSO), and lactacystin (10 mM) dissolved in water were diluted 1:1000 into the cell culture medium overnight. Polyclonal rabbit antibodies against αB1 and β2 have been described previously (Zhang et al., 1997). Polyclonal and monoclonal anti-HA were purchased from Santa Cruz Biotech, anti-ubiquitin from MBL, anti-tubulin from Roche Molecular Biochemicals, anti-His6 from Qiagen, protein A and protein G immobilized on beads from Pharmacia, and [35S]methionine from Amersham Phamacia Biotech. Monoclonal antibodies against the Runt domain, anti-RD (10B7G8) and anti-RD (3D9H3) were produced by N.Nozaki, K.Sasaguri and Y.Yamagi. Both of these monoclonal antibodies recognize the epitope located between amino acids 110 and 120, and detect all three RUNX proteins by western blotting. Neither of them were useful for immunoprecipitation.
Plasmids and recombinant protein constructs
The pEF-Runx1 and pEF-β series have been described previously (Lu et al., 1995; Kanno,T. et al., 1998). To create a mammalian expression vector encoding HA-epitope-tagged RUNX1, the sequence encoding residues 2–453 of human RUNX1 was amplified by PCR and inserted into the EcoRI–NotI site of the mammalian expression vector pEF-HA. All deletions and point mutations (Lys substituted with Arg) were generated by PCR-based mutagenesis. The sequences of the resulting constructs were verified by sequencing.
Cell culture
The mouse embryonal carcinoma cell line P19 was cultured in a 1:1 mixture of Dulbecco’s modified Eagle’s medium (Gibco-BRL) and Ham’s F12 medium (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS). Mouse 32D cl3 cells were cultured in RPMI 1640 (Gibco-BRL) supplemented with 10% FBS and 5% WEHI-3B conditioned medium. The human leukemia cell lines U937 were cultured in RPMI 1640 supplemented with 10% FBS.
Gene targeted mice
Mice in which the PEBP2β gene has been targeted have been described previously (Niki et al., 1997).
RT–PCR assay of RUNX transcripts
Total RNA was prepared from 11.5 d.p.c. whole embryos using TRIzol (Gibco-BRL). cDNA was synthesized using Superscript Preamplification System (Gibco-BRL). PCR was carried out under standard conditions. The primers used were: 5′-TCAGTGAATTGGAGCAGCTGCGG-3′ and 5′-CGGCGAGTAGGTGAAGGCGCCT-3′ for Runx1; 5′-AGTGATTTAGGGCGCATTCCTCAT-3′ and 5′-CGGGGTGTAGGTAAAGGTGGCT-3′ for Runx2; 5′-ACCGCTTTGGAGACCTGCGCATG-3′ and 5′-CGCTGTAGGGGAAGGCGGCAGA-3′ for Runx3; and 5′-CGTATTGGGCGCCTGGTCAC-3′ and 5′-CCAGTGAGCTTCCCGTTCAC-3′ for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Transfection and western blotting
P19 cells grown in six-well microplates were transfected with the desired vectors using FuGENE6 (Roche Molecular Biochemicals), and the cells were harvested 30–36 h after transfection. Whole-cell extracts were prepared by sonicating the cells in 2× SDS sample buffer. After boiling for 5 min, whole-cell extracts were electrophoresed on 12 or 15% SDS–polyacrylamide gels. Western blotting was performed according to a standard protocol, and proteins were detected by chemiluminescence according to the manufacturer’s protocol (ECL, Amersham Pharmacia Biotech).
Immunoprecipitation
Cells were lysed in 100 µl of immunoprecipitation (IP) buffer [10 mM PIPES-buffered saline pH 6.8, 300 mM sucrose, 100 mM KCl, 2 mM MgCl2, 4 mM EGTA, 1 mM NaF, 1 mM sodium vanadate, 0.25% (v/v) Triton X-100, 1/1000 vol of the protease inhibitor mixture (Roche Molecular Biochemicals), 10 µg/ml phenylmethylsulfonyl fluoride (PMSF)] by freezing and thawing, followed by centrifugation at 20 000 g. The pellet was re-extracted by sonication in 20 µl IP buffer with 1% SDS and re-centrifuged. The supernatants from the first and second extractions were combined and subjected to immunoprecipitation. Briefly, 20 µl of a 1:1 mixture of protein A and G beads were first added to the extract and allowed to pre-absorb for 1 h, after which 2 µl of antibody and another 20 µl of the protein A and G bead mixture were added. Incubation was continued at 4°C with agitation for 2 h or overnight. After washing four times with 1 ml IP buffer, the beads were suspended in 2× SDS sample buffer, boiled for 5 min and centrifuged. The proteins in the resulting supernatant were separated by SDS–PAGE and subjected to western blotting with the appropriate antibodies (described above).
In vitro ubiquitylation
To facilitate purification using NTA resin, the β165 protein C-terminally tagged with His6 and the Runt domain (50–190) C-terminally tagged with His6 were individually cloned into pQE9 expression vectors. They were expressed in Escherichia coli strain BL21 and purified with Ni2+-chelating resin under denaturing conditions according to the manufacture’s protocol (Invitrogen). For their in vitro ubiquitylation, the proteins were mixed with HeLa cell fraction II (MBL, Nagoya, Japan) and incubated at 37°C for 90 min in ubiquitylation buffer (total volume 40 µl) containing 40 mM Tris–HCl pH 7.6, 2 mM dithiothreitol, 10 mM KCl, 5 mM MgCl2, 1 mM ATP, 1 µg/µl ubiquitin, 0.5 µg of ubiquitin aldehyde, 1 µl lactacystin and 1 µl protease inhibitor mixture. The reaction was terminated by the addition of an equal volume of 2× SDS sample buffer. A 10 µl aliquot of the sample was subjected to SDS–PAGE followed by western blotting using anti-αB1 and anti-β2 antibodies.
Half-life measurements
In order to detect only the full-length RUNX1 protein, RUNX1 was designed to be N-terminally HA-tagged. P19 cells were transfected with the pEF-HA vector encoding either the wild-type or mutated RUNX1 with and without the pEF vector encoding a β derivative essentially as described above. After 24 h of transfection, the cells were pulsed with 200 µCi [35S]methionine for 1 h and then chased with 2 mM cold methionine for the periods indicated. Protein extracts were prepared by vortexing the cells in RIPA buffer (1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.01 M sodium phosphate pH 7.2, 2 mM EDTA, 50 mM NaF, 0.2 mM sodium vanadate, 1/1000 vol of the protease inhibitor mixture, 10 µg/ml PMSF) for 1 min in the presence of proteasome inhibitor MG132. Immunoprecipitation and subsequent analyses were performed as described above.
Acknowledgments
Acknowledgements
We thank P.P.Liu for providing us with CBFB/MYH11(1–611) cDNA; N.Nozaki, K.Sasaguri and Y.Yamagi for producing monoclonal antibodies anti-RD (10B7G8) and anti-RD (3D9H3); Wei-Hui Guo for helping with the RT–PCR analysis; and Zhi-Jian Xiao for constructing expression plasmids for PEBP2β(L64A, N104A). This work was supported by Grant-in-Aid 12213058 to Y.I. for Priority Area in Cancer Research from the Ministry of Education, Science, Sports and Culture, of Japan.
REFERENCES
- Akamatsu Y., Ohno,T., Hirota,K., Kagoshima,H., Yodoi,J. and Shigesada,K. (1997a) Redox regulation of the DNA binding activity in transcription factor PEBP2. The roles of two conserved cysteine residues. J. Biol. Chem., 272, 14497–14500. [DOI] [PubMed] [Google Scholar]
- Akamatsu Y., Tsukumo,S., Kagoshima,H., Tsurushita,N. and Shigesada,K. (1997b) A simple screening for mutant DNA binding proteins: application to murine transcription factor PEBP2α subunit, a founding member of the Runt domain protein family. Gene, 185, 111–117. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. (1998) The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J., 17, 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P., Geoffroy,V. and Karsenty,G. (1996) Study of osteoblast-specific expression of one mouse osteocalcin gene: characterization of the factor binding to OSE2. Connect. Tissue Res., 35, 7–14. [DOI] [PubMed] [Google Scholar]
- Ducy P., Starbuck,M., Priemel,M., Shen,J., Pinero,G., Geoffroy,V., Amling,M. and Karsenty,G. (1999) A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev., 13, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai J. et al. (1999) Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J. Biol. Chem., 274, 31577–31582. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Ito Y. (1997) The runt protein and its companion PEBP2: a close link between this transcription factor and AML. Leukemia, 11, Suppl. 3, 279–280. [PubMed] [Google Scholar]
- Ito Y. (1999) Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells, 4, 685–696. [DOI] [PubMed] [Google Scholar]
- Kagoshima H., Shigesada,K., Satake,M., Ito,Y., Miyoshi,H., Ohki,M., Pepling,M. and Gergen,P. (1993) The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet., 9, 338–341. [DOI] [PubMed] [Google Scholar]
- Kagoshima H., Akamatsu,Y., Ito,Y. and Shigesada,K. (1996) Functional dissection of the α and β subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J. Biol. Chem., 271, 33074–33082. [DOI] [PubMed] [Google Scholar]
- Kamachi Y., Ogawa,E., Asano,M., Ishida,S., Murakami,Y., Satake,M., Ito,Y. and Shigesada,K. (1990) Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J. Virol., 64, 4808–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Kanno,Y., Chen,L.F., Ogawa,E., Kim,W.Y. and Ito,Y. (1998) Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol. Cell. Biol., 18, 2444–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Kanno,T., Sakakura,C., Bae,S.C. and Ito,Y. (1998) Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor α (CBFα) subunit by the leukemia-related PEBP2/CBFβ-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol. Cell. Biol., 18, 4252–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanto S. et al. (2000) The PEBP2β/CBFβ-SMMHC chimeric protein is localized both in the cell membrane and nuclear subfractions of leukemic cells carrying chromosomal inversion 16. Leukemia, 14, 1253–1259. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Sieweke,M., Ogawa,E., Wee,H.J., Englmeier,U., Graf,T. and Ito,Y. (1999) Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J., 18, 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. et al. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell, 89, 755–764. [DOI] [PubMed] [Google Scholar]
- Li J., Tsuji,K., Komori,T., Miyazono,K., Wrana,J.L., Ito,Y., Nifuji,A. and Noda,M. (1998) Smad2 overexpression enhances Smad4 gene expression and suppresses CBFA1 gene expression in osteoblastic osteosarcoma ROS17/2.8 cells and primary rat calvaria cells. J. Biol. Chem., 273, 31009–31015. [DOI] [PubMed] [Google Scholar]
- Liu P., Tarle,S.A., Hajra,A., Claxton,D.F., Marlton,P., Freedman,M., Siciliano,M.J. and Collins,F.S. (1993) Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science, 261, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Look A.T. (1997) Oncogenic transcription factors in the human acute leukemias. Science, 278, 1059–1064. [DOI] [PubMed] [Google Scholar]
- Lu J., Maruyama,M., Satake,M., Bae,S.C., Ogawa,E., Kagoshima,H., Shigesada,K. and Ito,Y. (1995) Subcellular localization of the α and β subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol. Cell. Biol., 15, 1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S. et al. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell, 89, 773–779. [DOI] [PubMed] [Google Scholar]
- Nagata T., Gupta,V., Sorce,D., Kim,W.-Y., Sali,A., Chait,B.T., Shigesada,K., Ito,Y. and Werner,M.H. (1999) Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nature Struct. Biol., 6, 615–619. [DOI] [PubMed] [Google Scholar]
- Niki M. et al. (1997) Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl Acad. Sci. USA, 94, 5697–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa E., Inuzuka,M., Maruyama,M., Satake,M., Naito-Fujimoto,M., Ito,Y. and Shigesada,K. (1993a) Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology, 194, 314–331. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Maruyama,M., Kagoshima,H., Inuzuka,M., Lu,J., Satake,M., Shigesada,K. and Ito,Y. (1993b) PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl Acad. Sci. USA, 90, 6859–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T., van Deursen,J., Hiebert,S.W., Grosveld,G. and Downing,J.R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell, 84, 321–330. [DOI] [PubMed] [Google Scholar]
- Osato M. et al. (1999) Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2αB gene associated with myeloblastic leukemias. Blood, 93, 1817–1824. [PubMed] [Google Scholar]
- Otto F. et al. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell, 89, 765–771. [DOI] [PubMed] [Google Scholar]
- Shi M.J. and Stavnezer,J. (1998) CBF α3 (AML2) is induced by TGF-β1 to bind and activate the mouse germline Ig α promoter. J. Immunol., 161, 6751–6760. [PubMed] [Google Scholar]
- Tanaka T. et al. (1996) The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product and potentially regulates its transactivation ability. Mol. Cell. Biol., 16, 3967–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.Y., Shi,J., Zhang,L., Davis,A., Bravo,J., Warren,A.J., Speck,N.A. and Bushweller,J.H. (2000) Energetic and functional contribution of residues in the core binding factor β (CBFβ) subunit to heterodimerization with CBFα. J. Biol. Chem., 275, 39579–39588. [DOI] [PubMed] [Google Scholar]
- Wang Q., Stacy,T., Binder,M., Marín Padilla,M., Sharpe,A.H. and Speck,N.A. (1996a) Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl Acad. Sci. USA, 93, 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. et al. (1996b) The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell, 87, 697–708. [DOI] [PubMed] [Google Scholar]
- Warren A.J., Bravo,J., Williams,R.L. and Rabbitts,T.H. (2000) Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFβ. EMBO J., 19, 3004–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M.H., Shigesada,K. and Ito,Y. (1999) Runt domains take the lead in hematopoiesis and osteogenesis. Nature Med., 5, 1356–1357. [DOI] [PubMed] [Google Scholar]
- Zhang Y.W., Bae,S.C., Huang,G., Fu,Y.X., Lu,J., Ahn,M.Y., Kanno,Y., Kanno,T. and Ito,Y. (1997) A novel transcript encoding an N-terminally truncated AML1/PEBP2αB protein interferes with transactivation and blocks granulocytic differentiation of 32D cl3 myeloid cells. Mol. Cell. Biol., 17, 4133–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.W. et al. (2000) A RUNX2/PEBP2αA/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl Acad. Sci. USA, 97, 10549–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]