Abstract
The minichromosome maintenance (Mcm) proteins 2–7 are required for both the initiation and elongation steps of chromosomal DNA replication. Previous studies have shown that the Mcm complex consisting of the Mcm 4, 6, and 7 proteins contains 3′ to 5′ DNA helicase activity with limited processivity (displacing duplex DNA regions up to 30 nt). In this report, we show that the presence of both 5′ and 3′ single-stranded tails in DNA helicase substrates is essential for the processive helicase activity of the Mcm complex. The presence of both 5′ and 3′ tails facilitated the formation of double heterohexameric complexes of Mcm4/6/7 on substrate DNA, which appeared to be essential for the processive helicase activity. The double heterohexameric complex of Mcm4/6/7, in the presence of a single-strand DNA binding protein, is capable of unwinding duplex DNA region of about 600 bp in length. These results support the hypothesis that the Mcm4/6/7 complex can function as a replication helicase.
The six minichromosome maintenance (Mcm) proteins, Mcm2, Mcm3, Mcm4, Mcm5, Mcm6, and Mcm7, are structurally related proteins that are highly conserved in all eukaryotes, and all six proteins are essential for DNA replication (1–3). The recruitment of these proteins onto replication origins during the G1 phase of the cell cycle is essential for the formation of a prereplicative complex and initiation of DNA replication (4–6). The Mcm proteins interact to form various complexes, including Mcm2/3/4/5/6/7, Mcm4/6/7, Mcm2/4/6/7, or Mcm3/5 (7–11). Biochemical studies with these complexes have shown that only the dimeric complex of the Mcm4/6/7 heterotrimer contained DNA helicase, single-stranded (ss) DNA binding, and DNA-dependent ATPase activities (12–14). It also has been shown that the interaction of Mcm2 or Mcm3/5 with the Mcm4/6/7 complex inhibited the helicase activity of Mcm4/6/7 complex (14, 15). In vivo crosslinking and chromatin immunoprecipitation experiments performed in Saccharomyces cerevisiae showed that the localization of the Mcm proteins (Mcm4 and Mcm7) shifted from origin regions to interorigin regions during S phase (4). Furthermore, studies using mcm degron mutants in S. cerevisiae suggest that Mcm proteins also are required for the progression of the replication fork (16). These observations, taken together with the biochemical properties of Mcm4/6/7 complex, suggest that Mcm proteins may play a role as a replicative helicase in eukaryotes, similar to that of the bacterial DnaB or the large T antigen of simian virus 40 (SV40) (17–19). Consistent with this proposal, the dodecameric complex of the single Mcm protein, isolated from the archaeon Methanobacterium thermoautotrophicum, possesses helicase activity that unwinds duplex DNA regions of 500 bp in length (20–22). In contrast, the eukaryotic Mcm4/6/7 complex showed limited helicase activity, only unwinding very short duplex DNA (12, 14), raising doubts as to whether this complex can act as a replicative helicase. Because of its limited processivity, it was postulated that additional factors or modifications are required for the processive helicase activity of this complex or that the Mcm4/6/7 complex is only the catalytic core of the more fully active and processive helicase complex.
In this study, we examined the helicase activity of the Schizosaccharomyces pombe Mcm4/6/7 complex with several different types of substrates, including forked DNA structures. Analysis of the helicase activity and the interactions of the Mcm4/6/7 complex with these substrates showed that the presence of ss tails in forked DNA substrates stimulates the processive helicase activity of Mcm4/6/7 complex presumably by facilitating the formation of double heterohexameric complex of Mcm4/6/7.
Materials and Methods
Reagents.
Labeled and unlabeled dNTPs and rNTPs were obtained from Amersham Pharmacia. M13mp18 ssDNA was from New England Biolabs. Anti-FLAG M2 Ab-agarose and FLAG peptide were from Sigma. Crosslinking reagent Bis(sulfosuccinimidyl)suberate (BS3) was from Pierce. Escherichia coli ssDNA binding protein (SSB) was from Amersham Pharmacia. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Expression and Purification of Mcm4/6/7 Complex in Insect Cells.
The Mcm4/6/7 complex was expressed and purified from Sf9 insect cells by using Ni-agarose affinity chromatography followed by anti-FLAG M2 Ab-agarose affinity chromatography and glycerol gradient sedimentation as described (14).
DNA Helicase Assay.
For the preparation of substrates used to measure DNA helicase activity, oligonucleotides were synthesized that contained a 37-nt region complementary to the M13mp18(+) strand (nucleotides 6289–6326) and different lengths of oligo(dT) tails (0, 10, 20, 30, or 40 nt) at the 5′ end [5′-(dT)0–40GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT-3′]. After annealing to M13mp18 ssDNA, the 3′ end of the annealed DNA was labeled with [α-32P]dGTP and the Klenow fragment. These labeled M13mp18 DNA substrates were purified by Sepharose CL4B column chromatography. To determine the maximal length of duplex DNA displaced by the Mcm4/6/7 helicase, substrates containing longer duplex regions were prepared by elongating singly primed M13mp18 ssDNA by using Sequenase (United States Biochemical). For this purpose, 5′-(dT)40GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT-3′ was annealed to M13mp18 ssDNA, and the 3′ end of this oligomer was labeled with [α-32P]dGTP and [α-32P]dCTP in the presence of Sequenase, and then elongated in the presence of ddATP and all four dNTPs, according to the manufacturer's protocol. The DNA product was purified by Sepharose CL4B column chromatography. The resulting labeled substrate (15,000 cpm/fmol) contained duplex regions that varied in length between 40 and 600 bp. For the preparation of forked DNA substrates containing various combinations of 5′ and 3′ ss tails, 5′-tailed oligomers containing 0–50 nt of 5′ tails [5′-(dT)0–50GGTTGGCCGATCAAGTGCCCAGTCACGACGTTGTAAAACGAGCCCGAGTG-3′] and 3′-tailed oligomers with 0–60 nt of 3′-tails [5′-CACTCGGGCTCGTTTTACAACGTCGTGACTGGGCACTTGATCGGCCAACC(dT)0–60-3′] were synthesized. Oligomers containing 5′ tails were radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase, annealed to 3′-tailed oligomers, and the annealed products were gel-purified as described (23).
DNA helicase activity was measured in reaction mixtures (15 μl) containing 25 mM Hepes-NaOH (pH 7.5), 25 mM sodium acetate, 10 mM magnesium acetate, 4 mM ATP, 1 mM DTT, 0.1 mg/ml BSA, 5–10 fmol of 32P-labeled substrate (4,000 cpm/fmol), and enzyme fraction. After incubation at 32°C for 1 h, 4 μl of 5× loading buffer (100 mM EDTA/0.5% SDS/0.1% xylene cyanol/0.1% bromophenol blue/25% glycerol) was added, and 7-μl aliquots were loaded onto a 12% polyacrylamide gel in 1× TBE (90 mM Tris/90 mM boric acid/1 mM EDTA) and electrophoresed for 1.5 h at 150 V.
Gel Mobility Shift Assay.
DNA substrates containing a 60-nt 3′ tail alone, a 30-nt 5′ tail alone, or both 3′ and 5′ tails (30 and 60 nt, respectively) were prepared as described above. These substrates were used in the gel mobility shift assays. Enzyme fractions were incubated at 25°C for 30 min in reaction mixtures (15 μl) containing 25 mM Hepes-NaOH (pH 7.5), 50 mM sodium acetate, 10 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml BSA, 0.5 mM adenosine 5′-O-thiotriphosphate (ATP-γ-S), and 20 fmol of 32P-labeled substrate (4000 cpm/fmol). After addition of 2 μl of 50% glycerol, aliquots of reaction mixtures were electrophoresed for 4 h at 120 V through a 4% polyacrylamide gel containing 6 mM magnesium acetate and 5% glycerol in 0.5× TBE at 4°C.
Glycerol Gradient Centrifugation of Mcm/DNA Complexes.
The Mcm4/6/7 complex was incubated in reaction mixtures (150 μl) containing 25 mM Hepes-NaOH (pH 7.5), 50 mM sodium acetate, 10 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml BSA, 0.5 mM ATP-γ-S, and 0.5 pmol of 32P-labeled substrate (4,000 cpm/fmol). After incubation at 25°C for 30 min, reaction mixtures were applied onto 5-ml 15–35% glycerol gradients in buffer A (20 mM Hepes-NaOH, pH 7.5/25 mM sodium acetate/5 mM magnesium acetate/1 mM DTT) containing 0.1 mg/ml BSA. After centrifugation at 48,000 rpm for 6 h in a Beckman SW 50.1 rotor at 4°C, fractions (330 μl) were collected from the bottom of the tube. The distribution of Mcm/DNA complexes or DNA was determined by liquid scintillation counting.
Crosslinking of Mcm4/6/7 Complex on Forked DNA Substrate.
DNA substrates containing biotin at the 3′ end of the 5′-tailed oligomer were used for the isolation of the Mcm4/6/7 complex bound to DNA. The Mcm4/6/7 complex (2 μg) was incubated with 0.2 pmol of the biotinylated DNA substrate at 25°C for 30 min in the same reaction mixture (50 μl) used in the gel-mobility assay. After addition of 5 μl of streptavidin-conjugated magnetic beads (Promega), reaction mixtures were incubated at 4°C for an additional 10 min. Magnetic beads were washed two times with 0.2 ml of buffer A containing 0.1 mM ATP-γ-S and 0.1 mg/ml BSA. Beads then were suspended in 15 μl of buffer A containing 0.1 mM ATP-γ-S. To crosslink Mcm proteins, BS3 was added to 0.05, 0.1, or 0.2 mM, and the mixtures were incubated on ice for 10 min. The crosslinking reaction was stopped by the addition of 1 μl of 1 M Tris⋅HCl, pH 7.5, and proteins were analyzed by SDS/4.5% PAGE followed by staining with silver.
Results
Stimulation of the Helicase Activity of the Mcm4/6/7 Complex by a 5′ ss Tail on an M13 Partial Duplex DNA Substrate.
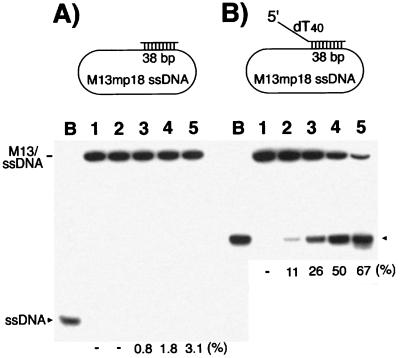
The helicase activity of the S. pombe Mcm4/6/7 complex was examined by using the M13 DNA partial duplex substrate containing a 38-bp duplex region. The Mcm4/6/7 complex hardly displaced the duplex DNA as shown in Fig. 1A. This observation was consistent with the low processivity of the Mcm4/6/7 helicase reported previously (14). However, when the same duplex M13 substrate containing an ss tail on the 5′ end of oligomer DNA was used (Fig. 1B Upper), the presence of a 40-nt oligo(dT) tail markedly stimulated the helicase activity of Mcm4/6/7 complex (Fig. 1B). When M13 substrates containing the same 38-bp duplex region with a 5′ oligo(dT) tail of 0, 10, 20, 30, or 40 nt were used, 200 ng of the Mcm4/6/7 complex displaced 3%, 12%, 38%, 63%, or 67% of the substrate, respectively (data not shown). Thus, the stimulation of helicase activity by the presence of a 5′ tail was proportional to the length of the tail. We also observed stimulation of the Mcm4/6/7 helicase activity by the addition of an oligo(dT) tail at the 5′ end of oligomer DNAs with M13 substrates containing a short 18-bp duplex region (data not shown). However, the stimulatory effect was relatively low (<5-fold) compared with the effects observed with M13 substrates containing a 38-bp duplex region (>20-fold). This difference in the level of stimulation suggests that the presence of a 5′ tail on helicase substrates not only affects the rate of displacement but also increases the processivity of the Mcm4/6/7 helicase.
Figure 1.
Influence of the presence of a 5′ tail on the helicase activity of Mcm4/6/7 complex. DNA helicase activity assays were carried out with increasing amounts of Mcm4/6/7 protein and 5 fmol of the indicated substrates with (B) or without (A) a 40-nt oligo(dT) tail at its 5′ end, as described in Materials and Methods. Lane B, boiled substrate; lane 1, no Mcm4/6/7 protein was added; lanes 2–5 contained 25, 50, 100, or 200 ng of the Mcm4/6/7 complex, respectively.
The Processive Helicase Activity of the Mcm4/6/7 Complex Requires a Fork Structure.
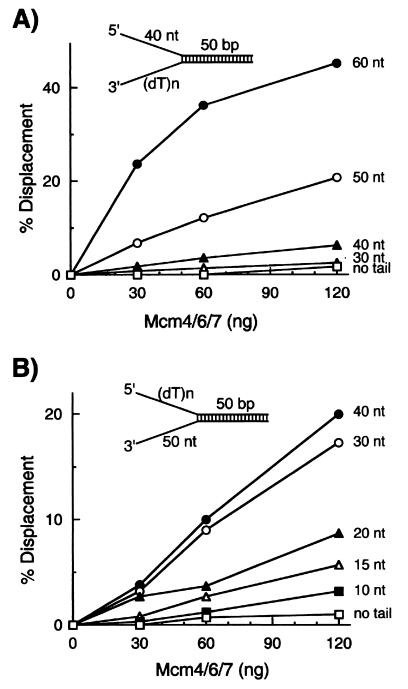
The requirements of 3′ and/or 5′ tails for processive helicase activity of the Mcm4/6/7 complex were determined. For this purpose, small forked DNA helicase substrates that contained a 50-bp duplex region with varying lengths of oligo(dT) tails on its 3′ or 5′ end were used. First, the effects of a 3′ tail were investigated by using forked DNA substrates with a 40-nt oligo(dT) tail on its 5′ end and varying lengths of oligo(dT) residues at the 3′ end. Substrates lacking a 3′ tail or substrates containing a 3′ tail shorter than 30 nt were poor substrates for this Mcm4/6/7 helicase (Fig. 2A), demonstrating that the presence of an oligo(dT) tail at the 3′ end is essential for the helicase activity of the Mcm4/6/7 complex. Under the conditions used, the efficiency of the displacement reaction increased as the 3′-tail length increased up to 60 nt. The influence of a 5′ tail also was investigated with forked DNA substrates containing a 50-nt oligo(dT) tail at the 3′ end and oligo(dT) tails of varying lengths at the 5′ end. In the absence of a 5′ tail, the Mcm4/6/7 complex hardly displaced any of the substrates examined. However, the efficiency of unwinding gradually was increased by the addition of longer 5′ tails of 10–40 nt in length. This stimulatory effect reached a plateau with substrates possessing a 40-nt 5′ tail (Fig. 2B). These results indicate that both 3′ and 5′ ss tails bordering a duplex region are essential for the processive helicase activity of the Mcm4/6/7 complex. The requirement for a relatively long ss tail at the 3′ end of substrates for helicase activity suggested that the loading and stable binding of the Mcm4/6/7 helicase complex, which possesses a 3′ to 5′ polarity, required an ssDNA region longer than 40 nt. However, the length of the 5′ tail required for processive helicase activity differs from the length of the 3′ tail, and this difference suggests that the ss tail at the 5′ end might play a role other than the loading of the Mcm4/6/7 complex onto the substrate DNA.
Figure 2.
DNA substrates containing both 5′ and 3′ ss tails are essential for processive helicase activity of the Mcm4/6/7 complex. (A) Increasing levels of the Mcm4/6/7 complex were incubated with 10 fmol of helicase substrates containing a 40-nt 5′ oligo(dT) ss region and varying lengths of 3′ ss tail as indicated. □, Reactions carried out with a substrate containing no 3′ tail; ◊, substrate with a 30-nt 3′ tail; ▴, substrate containing a 40-nt 3′ tail; ○, substrate with a 50-nt 3′ tail; ●, substrate with a 60-nt 3′ tail. (B) Helicase assays were performed with 10 fmol of DNA containing a 3′ oligo(dT) ss region of 50 nt in length and varying lengths of oligo(dT) at the 5′ end as indicated. □, No 5′ tail; ■, 10-nt 5′ tail; ◊, 15-nt 5′ tail; ▴, 20-nt 5′ tail; ○, 30-nt 5′ tail; and ●, 40-nt 5′ tail.
Formation of the Double Heterohexameric Complex of Mcm4/6/7 on Forked DNA Substrates.
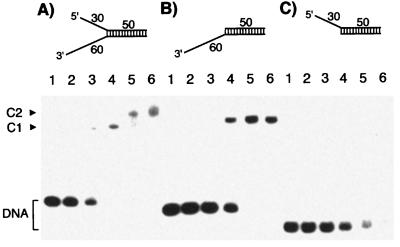
To examine the role played by the ss tails in stimulating the helicase activity of the Mcm4/6/7 complex, the interaction of the complex with different helicase DNA substrates was analyzed by using the gel mobility shift assay. When a substrate DNA containing 3′ tail alone (Fig. 3B Upper) was incubated with increasing levels of the Mcm4/6/7 complex, the majority of the Mcm/DNA complex formed migrated as a single band, reflecting the stable binding of a Mcm4/6/7 dimeric complex with the ss tail (Fig. 3B). As described above, the Mcm4/6/7 complex did not displace the duplex region within this substrate under the same reaction conditions used (Fig. 2B, no tail). These results suggest that the binding of the Mcm4/6/7 dimeric complex to the DNA substrate is not sufficient for processive helicase activity. On the other hand, a substrate containing the 30-nt 5′ tail alone (Fig. 3C Upper) did not appear to yield a stable complex with the Mcm proteins. Even though a substantial portion of the DNA substrate disappeared under these reaction conditions, the Mcm/DNA complex appeared to dissociate during electrophoresis (Fig. 3C, lanes 4–6). In support of this, a weak smear of shifted radioactivity was detected upon longer exposure of the gel during autoradiography (data not shown). When the DNA substrate containing both 3′ and 5′ tails (as shown in Fig. 3A) was used, a faster migrating band (C1) was observed (Fig. 3A, lanes 2–4) at low concentrations of the Mcm complex. Another slower migrating band (C2) was detected only at higher concentrations of protein (Fig. 3A, lanes 5 and 6). Based on these properties, the faster migrating band appeared to be an Mcm/DNA complex containing one Mcm4/6/7 dimeric complex, whereas the slower migrating complex may contain two molecules of the Mcm4/6/7 dimeric complex. Because the Mcm4/6/7 dimeric complex binds only to ssDNA and the forked DNA substrate contains short ssDNA tails that cannot be occupied by two molecules of Mcm4/6/7 dimeric complex, it was reasonable to assume that each dimeric complex interacted with each ss tailed region in this substrate. The weak and unstable interaction of the Mcm4/6/7 dimeric complex with the short 5′ tail, as shown in Fig. 3C, might be stabilized by its interaction with another dimeric complex bound to the 3′-tail region (C2 in Fig. 3A).
Figure 3.
Binding of the Mcm complex to various DNA helicase substrate. Gel mobility shift assays were performed with three different DNA substrates (each at 10 fmol) containing both a 60-nt 3′ tail and a 30-nt 5′ tail (A), a 60-nt 3′ tail alone (B), or a 30-nt 5′ tail alone (C). Lane 1, no Mcm4/6/7 protein was added; lanes 2–6, reactions were carried out with 2.2, 6.7, 20, 60, and 180 ng of the Mcm4/6/7 complex, respectively. C1 and C2 represent DNA–Mcm protein complexes.
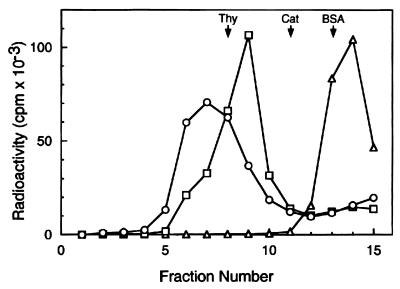
The properties of these Mcm/DNA complexes were analyzed by glycerol gradient sedimentation. For this purpose, 0.5 pmol of the forked DNA substrate containing both 3′ and 5′ tails (same as Fig. 3A Upper) was incubated with 7.5 μg of the Mcm4/6/7 complex (13 pmol of Mcm4/6/7 dimer) to form the C2 complex or with 0.5 μg of the Mcm complex (0.5 pmol) to form the C1 complex by using the identical conditions described for the gel mobility shift assay. After incubation, the formation of the C1 or C2 complex was confirmed by the gel mobility shift assay (data not shown), and these reaction mixtures then were sedimented through a 15–35% glycerol gradient. As shown in Fig. 4, the sedimentation properties of the C1 complex in the glycerol gradient (peaking at fraction 9, 16.4 S) were in keeping with the sedimentation of the substrate DNA alone (peaking at fraction 14, 1.9 S) and the Mcm4/6/7 dimeric complex alone (17.4 S, ref. 14). These findings suggest that the C1 complex most likely contained one molecule of Mcm4/6/7 dimeric complex bound to the DNA substrate. On the other hand, the C2 complex peaked at fraction 7 (21.4 S), suggesting that this complex most likely contained two molecules of Mcm4/6/7 dimeric complex bound to the forked DNA substrate. Gel mobility shift analysis of the complexes detected in the glycerol gradient fractions also was carried out. The Mcm-DNA complex that peaked at 16.4 S was shown to be the complex C1. The Mcm-DNA complex that peaked at 21.4 S was found to consist of a mixture of C1 and C2 complexes (data not shown). These findings, which are in keeping with the results presented in Fig. 3, suggest that the C2 complex is relatively unstable under the conditions used whereas the C1 complex is stable.
Figure 4.
Glycerol gradient sedimentation analysis of Mcm/DNA complexes. Mcm4/6/7 complex was incubated with 0.5 pmol of a 32P-labeled DNA substrate containing a 60-nt 3′ tail and a 30-nt 5′ tail with a 50-bp duplex region (the same substrate described in Fig. 3A) in a reaction mixture (150 μl) containing 25 mM Hepes-NaOH, pH 7.5, 50 mM sodium acetate, 10 mM magnesium acetate, 1 mM DTT, and 0.1 mg/ml BSA. After incubation at 25°C for 30 min, the reaction mixture was loaded onto a 5-ml 15–35% glycerol gradient in a buffer A containing 0.1 mg/ml BSA. After centrifugation at 48,000 rpm for 6 h in a Beckman SW 50.1 rotor at 4°C, fractions (330 μl) were collected from the bottom of the tube. The distribution of Mcm/DNA complexes or DNA were determined by liquid scintillation counting. The marker proteins used were thyroglobulin (Thy, 19S), catalase (Cat, 11.3S), and BSA (BSA, 4.3S). ○, The binding reaction was carried out with 7.5 μg of the Mcm4/6/7 complex (13 pmol Mcm4/6/7 dimer) to assemble the C2 complex; □, 0.3 μg of the Mcm4/6/7 complex was used (about 0.5 pmol Mcm4/6/7 dimer) for the formation of the C1 complex; ◊, DNA substrate alone.
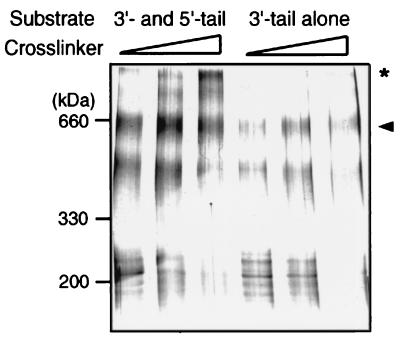
To examine whether the two dimeric complexes of Mcm4/6/7 present in complex C2 are in direct contact with each other, crosslinking experiments were performed. In these experiments, the forked DNA substrate containing both 3′- and 5′-tailed regions (same as described in Fig. 3A Upper) was biotinylated. In addition, a biotinylated DNA substrate containing only the 3′-tailed region (Fig. 3B Upper) also was prepared. Both substrates were incubated with the Mcm4/6/7 complex, using the same conditions used in the gel mobility shift assay. The Mcm/DNA complexes formed were separated from excess free Mcm proteins by using streptavidin-coated magnetic beads. The Mcm complex bound to the DNA substrate was crosslinked by the addition of the crosslinking reagent BS3, and the mixture then was subjected to SDS/PAGE analysis. As shown in Fig. 5, a protein band of about 600 kDa, representing the dimeric complex of Mcm4/6/7, was detected after crosslinking. As expected, this protein band was formed in the presence of both DNA substrates (Fig. 5, arrowhead). However, the overall intensities of the protein bands detected in lanes 1–3 were approximately 2- to 3-fold higher than those observed in lanes 4–6. This suggested that two molecules of the Mcm 4/6/7 dimeric complex were bound to the forked DNA substrate, whereas the 3′-tailed substrate contained only one Mcm4/6/7 dimeric complex. Furthermore, Mcm protein complexes larger than 600 kDa were detected only in reactions that contained the substrate possessing both 3′- and 5′-ss tails (Fig. 5, asterisk). Based on its migration properties, the slowest migrating top band appeared to be a double heterohexameric complex of about 1,100 kDa. This finding, together with the binding assay and sedimentation analysis results, suggest that two dimeric complexes of Mcm4/6/7 loaded onto the forked DNA template interact with each other to form a double heterohexameric complex of Mcm4/6/7.
Figure 5.
Crosslinking of the Mcm4/6/7 complex bound to helicase DNA substrates. The Mcm4/6/7 complex (2 μg) was incubated with 0.2 pmol of the biotinylated DNA containing both a 60-nt 3′ tail and a 30-nt 5′ tail or a substrate containing a 3′ tail alone (the same substrates as described in Fig. 3 A and B) under identical conditions used in the gel mobility shift assay. After binding of the Mcm/DNA complex to streptavidin-coated magnetic beads, the beads were washed twice with buffer A. Crosslinking was carried out at three different concentrations of BS3 (0.05, 0.1, or 0.2 mM, respectively) as described in Materials and Methods. Crosslinked proteins were electrophoresed through SDS/4.5% PAGE and then stained with silver. The arrowhead indicates the position of the dimeric complex of Mcm4/6/7, and the asterisk indicates the position of the double hexameric Mcm4/6/7 complex.
Helicase Processivity of the Double Hexameric Complex of Mcm4/6/7.
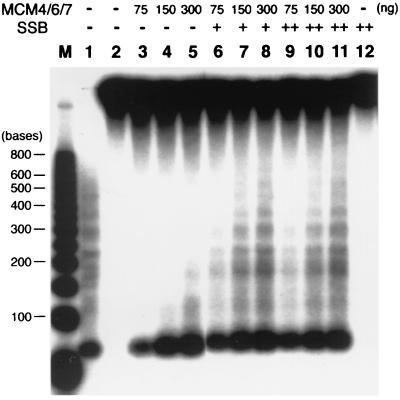
The length of the duplex DNA region displaced by the double hexameric complex of Mcm4/6/7 was determined by using a 5′-tailed DNA/M13 ssDNA substrate that contained duplex DNA regions that varied between 40 and 600 bp in length. Displacement of strands up to 200 nt in length was observed in the presence of 300 ng of the Mcm4/6/7 complex (Fig. 6, lane 5). In the presence of E. coli SSB, the Mcm4/6/7 complex efficiently unwound duplex DNA regions up to 600 bp (Fig. 6, lanes 8 and 11). The stimulatory effects of SSB were observed only when the SSB was added after preincubation of the Mcm complex with the substrate DNA. If DNA was added to reaction mixtures containing both SSB and the Mcm complex, no helicase activity was observed (data not presented). Presumably, the binding of E. coli SSB to ss regions prevents the subsequent binding of the Mcm complex to the ss DNA. The molar excess amounts of SSB protein added to the reactions described above (Fig. 6, lanes 6–11) would be expected to prevent the reassociation of the Mcm complex with ss regions. These results suggest that the double heterohexameric Mcm4/6/7 complex acted as a processive helicase.
Figure 6.
Processivity of the Mcm4/6/7 helicase. DNA helicase activity was assayed by using a partial duplex M13 substrate (5 fmol) containing duplex regions varying in length between 40 and 600 bp and a 40-nt oligo(dT) tail on its 5′ end. Indicated amounts of the Mcm4/6/7 complex were preincubated with DNA substrates in the helicase reaction buffer at 25°C for 10 min, and E. coli SSB was added as indicated. After incubation at 32°C for 1 h, reactions were stopped by the addition of 4 μl 5× reaction stop buffer (100 mM EDTA/0.5% SDS) and proteinase K (2 μg), and reaction mixtures were incubated for an additional 30 min at 37°C. Aliquots then were electrophoresed through a 2.5% low melting agarose gel in 1× TBE at 120 V for 3 h. Lane M, 32P-labeled 50-bp ladder DNA marker (denatured); lane 1, boiled substrate; lanes 2 and 12, reactions without Mcm4/6/7 protein; + and + + denote the addition of 200 and 400 ng of E. coli SSB, respectively.
Discussion
The initiation of chromosomal DNA replication in eukaryotes is a multistep process. It involves the binding of origin recognition complex to replication origins, the recruitment of Cdc6/Cdc18, the Mcm complex, and Cdc45 to form the prereplicative complex, and the activation of the prereplicative complex by Cdc7 and Cdc28 protein kinases. Among the components involved in the formation of the prereplicative complex, the Mcm complex is the most likely candidate to act as the replicative helicase. It was shown that the Mcm proteins appear to move through DNA during S phase, presumably as part of the replication fork (4), Furthermore, functional Mcm proteins are not only essential for the initiation of replication but are also required for the progression of the replication fork (16). In vitro studies also showed that the dimeric complex of Mcm4/6/7 contained DNA helicase activity. However, the helicase activity of the dimeric Mcm4/6/7 complex showed limited processivity (12, 14), a property compromising its putative role as a replicative helicase. In this study, we demonstrated that the double heterohexameric complex of Mcm4/6/7 that assembled on a forked DNA substrate is a processive helicase, thus providing evidence that the Mcm complex may function as a replicative helicase.
The experiments described above suggest that formation of a double hexameric complex may be important for the processive helicase activity of the Mcm4/6/7 complex. The formation of a double hexamer appears to be conserved in evolution because the single Mcm protein of the archaeon M. thermoautotrophicum also forms a double hexameric complex, which contains helicase activity capable of unwinding long regions of duplex DNA (20–22). This biochemical property is also similar to that of the SV40 T antigen. The double hexameric complex of SV40 T antigen also can be assembled on forked DNA substrates and possesses much higher helicase activity than the single hexameric SV40 T antigen complex (24). Electron microscopic analysis of the SV40 origin-dependent DNA unwinding reaction showed that the two replication forks of the plasmid DNA substrate were joined through SV40 T antigen complexes, suggesting that the double hexameric complex of T antigen remained intact during replication (25). Thus, the double hexameric form of T antigen appears responsible for strand separation during fork movement.
Studies from several organisms, including yeast to human, indicate that the Mcm proteins interact to form a stable heterohexameric complex containing all six subunits, which appears to be the predominant form of the Mcm complex found in vivo (7, 10). Mcm proteins also appeared to be recruited to chromatin as a complex containing all six subunits (9, 26). Because this complex lacks detectable enzymatic activities (including helicase activity), these results suggest that alteration of the Mcm complex leading to the activation of helicase activity would be required for the initiation of replication. Genetic and biochemical studies suggest that modifications of the Mcm complex by the two S-phase promoting kinases, Cdc7-Dbf4 and the S-phase cyclin-dependent kinase, may be required for the conversion of the inactive complex to the active helicase complex (3, 27). In S. cerevisiae, a specific mutant allele, Mcm5-bob1, suppressed all mutations in CDC7 or DBF4, suggesting that an alteration of the Mcm5 protein satisfies the essential functions of the Cdc7-Dbf4 complex (28). A number of the Mcm proteins appeared to be modified by these two kinases (29–31). In vitro studies, with the hexameric Mcm complex as a substrate, showed that Mcm2 was directly phosphorylated by the Cdc7-Dbf4 kinase (32, 33). Although these findings suggest that the Mcm complex is the main target of these kinases, it is presently unclear whether their modification of the Mcm complex is sufficient to activate the helicase activity.
In vitro analysis showed that only the Mcm4/6/7 complex contained DNA helicase activity and that the addition of Mcm2 or Mcm3/5 complex inhibited its helicase activity (12, 14, 15). These results suggest that the Mcm4/6/7 complex may be the catalytic core of Mcm complex and Mcm2 or Mcm3/5 may regulate the activity of this complex. Thus, the activation of the six-subunit Mcm complex may require the dissociation of Mcm2 and Mcm3/5. However, recent studies using mcm degron mutants in S. cerevisiae showed that the degradation of any one of the Mcm proteins after hydroxyurea treatment inhibited fork elongation after removal of hydroxyurea (16). These results imply that all six Mcm proteins may be required for the elongation of replication fork, which appears to contradict the proposal that the Mcm4/6/7 complex is the active helicase at the replication fork. One possible explanation is that the elongation complex, including the Mcm helicase complex, dissociates after hydroxyurea addition and exposure of cells to the nonpermissive temperature and the reassembly of the elongation complex onto a stalled replication fork may require the complete six-subunit Mcm complex. In E. coli, conditions that block replication can result in the dissociation of the components at the replication fork. The restart of stalled replication forks requires the primosome assembly factors such as PriA to load the DnaB helicase onto the stalled fork (34). At present, however, we cannot rule out the possibility that the six-subunit Mcm complex may be an active helicase complex at the replication fork. The biochemical activities of the Mcm4/6/7 complex described here may only represent the properties of the catalytic core of the complete replicative helicase complex.
Acknowledgments
We thank Dr. Z.-Q. Pan for critical review of this paper. These studies were supported by National Institutes of Health Grant GM 38559 (to J.H.). J.H. is a Professor of the American Cancer Society.
Abbreviations
- Mcm
minichromosome maintenance
- ss
single-stranded
- ATP-γ-S
adenosine 5′-O-thiotriphosphate
- BS3
bis(sulfosuccinimidyl)suberate
- SV40
simian virus 40
- SSB
ssDNA binding protein
References
- 1.Koonin E V. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kearsey S E, Labib K. Biochim Biophys Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 3.Tye B K. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 6.Zou L, Stillman B. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 7.Thommes P, Kubota Y, Takisawa H, Blow J J. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. J Biol Chem. 1996;271:24115–24122. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 9.Fujita M, Kiyono T, Hayashi Y, Ishibashi M. J Biol Chem. 1997;272:10928–10935. doi: 10.1074/jbc.272.16.10928. [DOI] [PubMed] [Google Scholar]
- 10.Adachi Y, Usukura J, Yanagida M. Genes Cells. 1997;2:467–479. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- 11.Sherman D A, Forsburg S L. Nucleic Acids Res. 1998;26:3955–3960. doi: 10.1093/nar/26.17.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishimi Y. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 13.You Z, Komamura Y, Ishimi Y. Mol Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J K, Hurwitz J. J Biol Chem. 2000;275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- 15.Ishimi Y, Komamura Y, You Z, Kimura H. J Biol Chem. 1998;273:8369–8375. doi: 10.1074/jbc.273.14.8369. [DOI] [PubMed] [Google Scholar]
- 16.Labib K, Tercero J A, Diffley J F. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 17.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 18.Leatherwood J. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- 19.Borowiec J A, Dean F B, Bullock P A, Hurwitz J. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 20.Kelman Z, Lee J K, Hurwitz J. Proc Natl Acad Sci USA. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shechter D F, Ying C Y, Gautier J. J Biol Chem. 2000;275:15049–15059. doi: 10.1074/jbc.M000398200. [DOI] [PubMed] [Google Scholar]
- 22.Chong J P, Hayashi M K, Simon M N, Xu R M, Stillman B. Proc Natl Acad Sci USA. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. . (First Published February 4, 2000; 10.1073/pnas.030539597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Smelkova N V, Borowiec J A. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wessel R, Schweizer J, Stahl H. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter A, Baack M, Holthoff H P, Ritzi M, Knippers R. Biol Chem. 1998;379:1181–1187. doi: 10.1515/bchm.1998.379.8-9.1181. [DOI] [PubMed] [Google Scholar]
- 27.Dutta A, Bell S P. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 28.Hardy C F, Dryga O, Seematter S, Pahl P M, Sclafani R A. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendrickson M, Madine M, Dalton S, Gautier J. Proc Natl Acad Sci USA. 1996;93:12223–12228. doi: 10.1073/pnas.93.22.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato N, Arai K, Masai H. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown G W, Kelly T J. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- 33.Jiang W, McDonald D, Hope T J, Hunter T. EMBO J. 1999;18:5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandler S J, Marians K J. J Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]