Abstract
The yeast UPF1, UPF2 and UPF3 genes encode trans-acting factors of the nonsense-mediated mRNA decay pathway. In addition, the upf1Δ strain demonstrates a nonsense suppression phenotype and Upf1p has been shown to interact with the release factors eRF1 and eRF3. In this report, we show that both upf2Δ and upf3Δ strains demonstrate a nonsense suppression phenotype independent of their effect on mRNA turnover. We also demonstrate that Upf2p and Upf3p interact with eRF3, and that their ability to bind eRF3 correlates with their ability to complement the nonsense suppression phenotype. In vitro experiments demonstrate that Upf2p, Upf3p and eRF1 compete with each other for interacting with eRF3. Con versely, Upf1p binds to a different region of eRF3 and can form a complex with these factors. These results suggest a sequential surveillance complex assembly pathway, which occurs during the premature translation termination process. We propose that the observed nonsense suppression phenotype in the upfΔ strains can be attributed to a defect in the surveillance complex assembly.
Keywords: eRF3/nonsense suppression/surveillance complex/UPF2/UPF3
Introduction
In order to ensure the high fidelity of gene expression, the cell has evolved quality control mechanisms to safeguard against faulty products. A clear example of such a cellular quality control mechanism is the nonsense-mediated mRNA decay (NMD) pathway. NMD is an mRNA surveillance pathway that promotes rapid decay of nonsense-containing mRNAs (reviewed in Czaplinski et al., 1999; Hentze and Kulozik, 1999; Jacobson and Peltz, 2000). A nonsense codon in a given transcript will lead to premature translation termination and usually result in a loss-of-function phenotype. There are >200 genetic disorders in which the disease state can be attributed to nonsense mutations. Understanding the process of NMD and how these nonsense-containing transcripts are translated can lead to rational approaches for the treatment of these disorders.
It has been shown in the yeast Saccharomyces cerevisiae that activation of the NMD pathway requires translation of the nonsense-containing mRNA (reviewed in Czaplinski et al., 1999). In particular, some trans-acting factors that are involved in NMD have been suggested to function in the translation termination process as well (Weng et al., 1996b; Czaplinski et al., 1998; Maderazo et al., 2000). These observations suggest that translation termination is a critical event in determining whether a transcript will be rapidly degraded. The translation termination process consists of a termination codon-dependent hydrolysis of the peptidyl-tRNA bond, resulting in release of the nascent polypeptide chain from the ribosome (reviewed in Nakamura et al., 1996; Welch et al., 2000). In eukaryotes, this process is under the control of two interacting release factors, eRF1 and eRF3 (Stansfield et al., 1995; Zhouravleva et al., 1995). The eRF1 protein has a structure mimicking that of a tRNA molecule. It recognizes the stop codon in the A site of the ribosome and catalyzes the hydrolysis of the peptidyl-tRNA bond (Song et al., 2000). The eRF3 protein is a ribosome- and eRF1-dependent GTPase, and stimulates eRF1 activity in a GTP-dependent manner (Frolova et al., 1996).
A key question in understanding how the NMD pathway functions is to understand what differentiates a premature termination event from a normal one. One hypothesis is that the distance from the termination codon to the poly(A) site is a key determinant as to whether an RNA will be degraded by the NMD pathway (Muhlrad and Parker, 1999a). However, results using reporter genes containing the 5′-untranslated region of the GCN4 transcript, which harbors four upstream open reading frames, showed that increasing the distance from the termination codon to the poly(A) site did not activate the NMD pathway (Ruiz-Echevarria et al., 1996). Rather, these results support the view that activation of NMD requires the presence of a ‘downstream sequence element’ (DSE) located 3′ of the premature termination codon (Peltz et al., 1993; Zhang et al., 1995). The RNA-binding protein Hrp1p has been demonstrated to interact specifically with the DSE, and this recognition is required for the activation of the NMD pathway (Gonzalez et al., 2000).
The yeast UPF1, UPF2 and UPF3 genes have been identified as trans-acting factors of the NMD pathway (Leeds et al., 1991; Cui et al., 1995; Lee and Culbertson, 1995). Mutations in these genes result in the specific stabilization of nonsense-containing mRNAs. Homologs of the UPF1, UPF2 and UPF3 genes have been identified in Schizosaccharomyces pombe, Caenorhabditis elegans and Homo sapiens (Pulak and Anderson, 1993; Perlick et al., 1996; Lykke-Andersen et al., 2000; Mendell et al., 2000; Serin et al., 2001), indicating that the roles of Upf proteins are conserved among higher eukaryotes. Upf1p has been shown to bind RNA and demonstrate an RNA-dependent ATPase/helicase activity (Czaplinski et al., 1995, 1998). Interestingly, Upf1p also interacts with release factors eRF1 and eRF3, and appears to influence the translation termination efficiency (Weng et al., 1996b; Czaplinski et al., 1998). The UPF2 gene encodes a protein rich in acidic residues (Cui et al., 1995). Upf2p has been shown to interact with both Upf1p and Upf3p (He et al., 1997). The UPF3 gene encodes a smaller protein highly rich in basic residues (Lee and Culbertson, 1995). Upf3p has three sequence elements that resemble nuclear localization signals and two that resemble the leucine-rich nuclear export signals (Shirley et al., 1998). Cells harboring single or multiple deletions of the UPF genes demonstrate equivalent stabilization of the nonsense-containing mRNAs, indicating that the Upf proteins either function as a complex or are components of one regulatory pathway (Cui et al., 1995; He et al., 1997).
It has been proposed that the translation termination event triggers the assembly of a surveillance complex, which consists of at least the Upf proteins and release factors (Czaplinski et al., 1998; Gonzalez et al., 2000). Upon assembly, the surveillance complex searches 3′ of the termination codon to look for specific signals that indicate a premature termination event has occurred. If such a signal is encountered, the transcript is rapidly decapped by Dcp1p in a deadenylation-independent manner, and subsequently degraded by the 5→3′ exoribonuclease Xrn1p (reviewed in Czaplinski et al., 1999).
In this report we show that, in addition to upf1Δ, both upf2Δ and upf3Δ strains demonstrate a nonsense suppression phenotype. We further demonstrate that the effect of Upf2p and Upf3p on nonsense suppression is independent of their effect on NMD and the nature of the termination codon. Upf2p and Upf3p are also shown to interact with the release factor eRF3, and their ability to bind eRF3 correlates with their ability to complement the nonsense suppression phenotype. These results provide evidence that Upf2p and Upf3p are also components of the surveillance complex. Furthermore, our results suggest that the surveillance complex undergoes a sequential assembly pathway. We propose that the assembly of the surveillance complex is part of the premature translation termination process and that the observed nonsense suppression phenotype can be attributed to the defect in surveillance complex assembly.
Results
The upf2Δ and upf3Δ strains suppress the nonsense-containing tyr7-1 allele
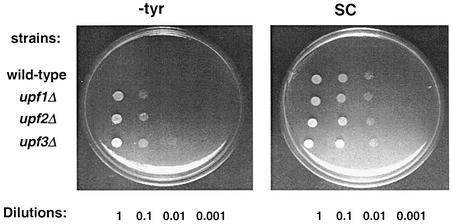
Previous results from our laboratory have demonstrated that Upf1p functions in both nonsense suppression and the NMD pathway (Weng et al., 1996a,b; Czaplinski et al., 1998). Recently, Upf2p and Upf3p have also been shown to prevent suppression of a nonsense allele of the CAN1 gene (can1-100) (Maderazo et al., 2000). We initially examined whether deleting UPF2 or UPF3 would cause suppression of all types of termination codons. The yeast strain KC2 is unable to grow in medium lacking tyrosine (–tyr) because it has a premature UAG codon in its tyr7-1 gene. The UPF2 and UPF3 genes were disrupted individually in this strain, and their growth on –tyr medium was monitored and compared with that of the isogenic wild-type and upf1Δ strains (Figure 1). The upf2Δ and upf3Δ strains both demonstrated a nonsense suppression phenotype as measured by growth on –tyr medium (left panel), similar to that observed with a upf1Δ strain. Deletion of the UPF genes had no apparent effect on cell growth on complete medium (right panel). These results suggest that the Upf proteins have a general effect on the suppression of nonsense alleles.
Fig. 1. The upf2Δ and upf3Δ strains demonstrate a nonsense suppression phenotype. The UPF1, UPF2 and UPF3 genes in a wild-type yeast strain (KC2) were disrupted individually. Ten-fold serial dilutions of mid-log phase cells were plated on synthetic complete (SC) (right panel) and –tyr (left panel) plates, and their growth at 30°C was monitored.
Quantitative analysis of the nonsense suppression activity in the upfΔ strains
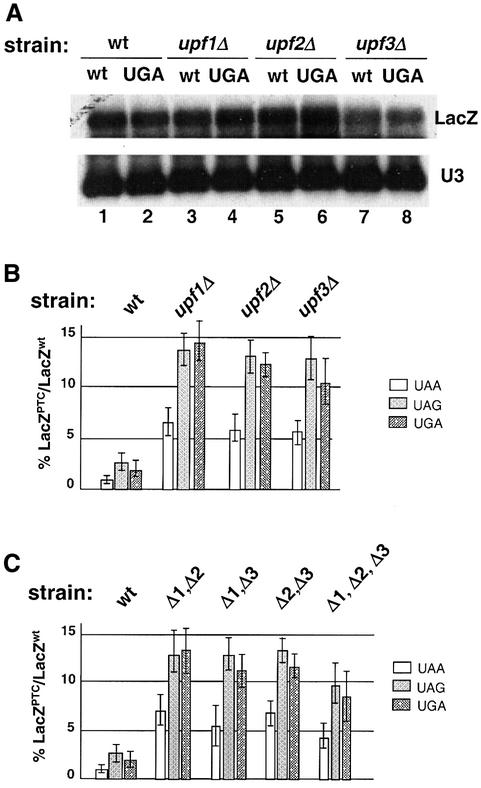
Since the NMD pathway is inactive in all upfΔ strains, the abundance of nonsense-containing tyr7-1 transcripts increased significantly in these strains (data not shown). Previous results have demonstrated that inactivation of NMD is not sufficient to cause nonsense suppression (Weng et al., 1996b; Maderazo et al., 2000). The change in transcript levels, however, will affect the amount of full-length protein produced. Therefore, in order to quantitatively analyze the nonsense suppression activity in upfΔ strains, a β-galactosidase (β-gal) reporter system was utilized. The advantage of using this reporter transcript is that the abundances of wild-type and UGA-containing LacZ mRNAs were nearly identical in UPF+ and upfΔ strains (Figure 2A). The abundances of LacZ transcripts containing UAA and UAG codons were also similar to the abundance of the wild-type LacZ transcript (data not shown), indicating that the nonsense-containing LacZ transcript is not subjected to NMD. Thus, changes in the amount of full-length β-gal synthesized from the nonsense-containing LacZ mRNAs in upfΔ strains are independent of NMD.
Fig. 2. (A) The abundance of wild-type and UGA-containing LacZ transcripts in the wild-type and upfΔ strains. Total RNAs isolated from the specified strains were separated on a 1% agarose gel and probed with 32P-labeled LacZ and U3 probes. (B) The nonsense suppression activity in wild-type and upfΔ strains was assessed quantitatively using a β-gal reporter system. The indicated yeast strains were transformed with either a wild-type LacZ gene or a LacZ gene containing the specified nonsense codon. The assays were performed as described in Materials and methods. (C) Nonsense suppression activity in wild type and strains harboring multiple UPF deletions.
The wild-type, upf1Δ, upf2Δ and upf3Δ strains were transformed with either a wild-type LacZ gene (LacZwt), or a LacZ gene containing the specified premature termination codon (LacZPTC). The nonsense suppression activity was measured as the ratio of β-gal activity in cells harboring LacZPTC and LacZwt (LacZPTC/LacZwt). The results showed that the upf1Δ, upf2Δ and upf3Δ strains demonstrated a significantly higher LacZPTC/LacZwt ratio with all three nonsense codons (Figure 2B), consistent with their ability to suppress the tyr7-1 nonsense allele. The LacZPTC/LacZwt ratio increased ∼6- to 8-fold with all the termination codons, indicating that the effect of UPF genes on nonsense suppression is independent of the nature of termination codon. The high LacZPTC/LacZwt ratio in the absence of UPF genes is most likely a reflection of reduced translation termination efficiency, although it can also be attributed to an increase in the translation efficiency specific to nonsense-containing transcripts (see Discussion).
The nonsense suppression activity of strains harboring various combinations of UPF deletions was also examined. The isogenic upf1Δupf2Δ, upf1Δupf3Δ, upf2Δupf3Δ and upf1Δupf2Δupf3Δ strains were constructed by disrupting the corresponding UPF genes in the wild-type cells. All these strains demonstrated a higher nonsense suppression activity with all three termination codons compared with the wild-type strain (Figure 2C). Importantly, strains harboring single or any combination of multiple deletions of the UPF genes resulted in similar levels of nonsense suppression activity (compare Figure 2B and C), indicating that the effect of Upf proteins on nonsense suppression was not additive.
The Upf2 and Upf3 proteins interact with the release factor eRF3
Previous results have shown that Upf1p interacts with the release factors eRF1 and eRF3 (Czaplinski et al., 1998). Therefore, we reasoned that Upf2p and Upf3p could also function through interactions with the release factors. To test this possibility, we initially performed co-immunoprecipitation analysis. Cytoplasmic extracts of yeast cells expressing FLAG-Upf1p, -Upf2p or -Upf3p were immunoprecipitated with an anti-FLAG antibody. The immunoprecipitated proteins were visualized using an anti-eRF3 antibody. The results showed that eRF3 co-immunoprecipitated with both Upf2p and Upf3p (Figure 3A, lanes 3–4). These interactions were specific since no eRF3 was precipitated when no FLAG-Upf protein was expressed (lane 1). FLAG-Upf1p was used as a positive control since it has previously been shown to interact with both eRF1 and eRF3 (Czaplinski et al., 1998). In contrast, an anti-eRF1 antibody was able to identify eRF1 among the proteins co-immunoprecipitated with Upf1p, but not Upf2p and Upf3p (data not shown).
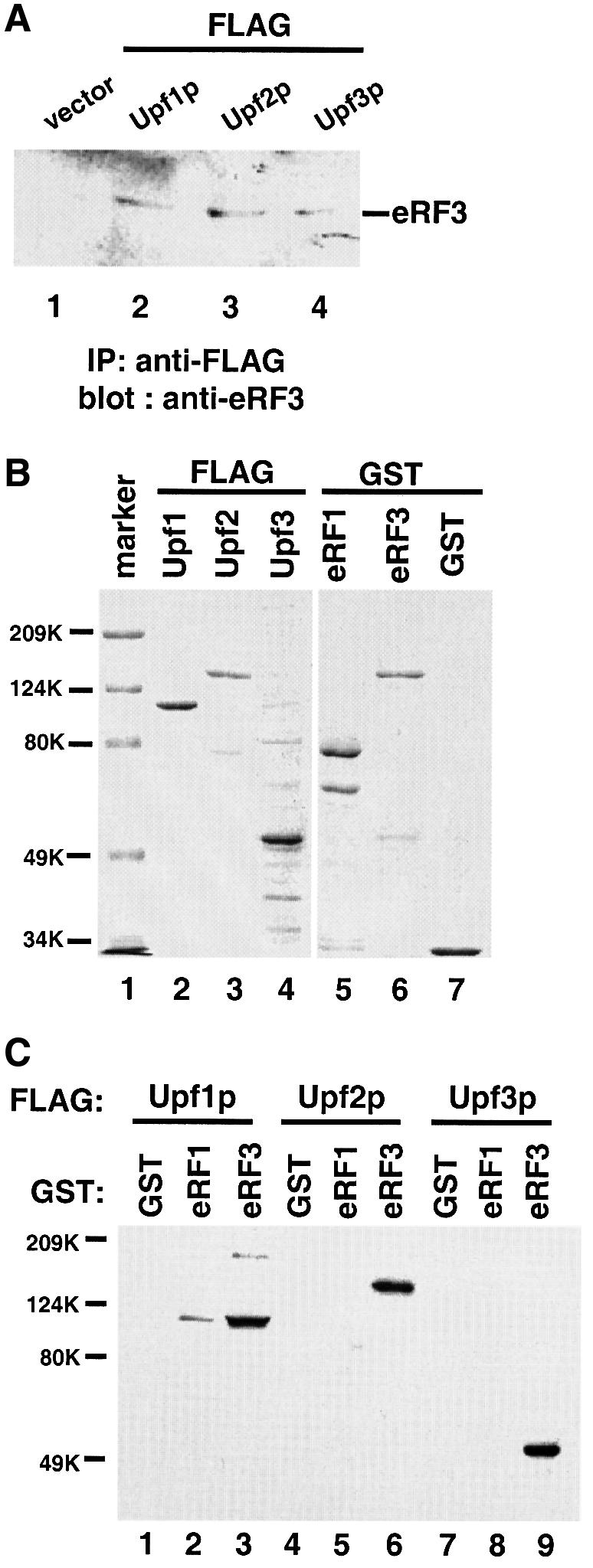
Fig. 3. Upf2p and Upf3p interact with the release factor eRF3. (A) Co-immunoprecipitation. Cells were transformed with either vector alone (lane 1) or vector expressing FLAG-tagged Upf1p, Upf2p or Upf3p (lanes 2–4). Cytoplasmic extracts were prepared and immuno precipitated with an anti-FLAG antibody. The immunoprecipitates were resolved by SDS–PAGE and subjected to immunoblotting with an anti-eRF3 polyclonal antibody. (B) Coomassie Blue staining of the purified fusion proteins. The protein molecular weight markers are as indicated. (C) GST pull-down experiment. Purified GST, GST–eRF1 or GST–eRF3 (1.0 µg each) was combined with glutathione–Sepharose beads and FLAG-Upf1p (1.0 µg), Upf2p (1.0 µg) or Upf3p (0.5 µg). Following incubation and extensive washing, the proteins remaining associated with the beads were analyzed by SDS–PAGE and immunoblotting with an anti-FLAG antibody.
In order to determine whether Upf2p and Upf3p interact with eRF3 in vitro, we carried out a glutathione S-transferase (GST) pull-down experiment with purified GST-tagged release factors and FLAG-tagged Upf proteins. A gel showing the purified proteins used in these experiments is shown in Figure 3B. The GST, GST–eRF1 and GST–eRF3 fusion proteins were immobilized on glutathione–Sepharose beads, and the ability of FLAG-Upf1p, -Upf2p or -Upf3p to complex with the bound GST fusion protein was determined. The proteins associated with the beads were visualized using an anti-FLAG antibody. The results showed that both Upf2p and Upf3p interacted with eRF3 with an affinity similar to that of Upf1p (Figure 3C, lanes 3, 6 and 9). However, unlike Upf1p, neither Upf2p nor Upf3p interacted with eRF1 under these conditions (lanes 2, 5 and 8). These interactions were specific, since no FLAG-tagged Upf protein interacts with the GST alone. Under these conditions, ∼10% of the input Upf proteins can be pulled-down by GST–RF3 (data not shown), indicating that the interactions between the Upf proteins and eRF3 are direct. The in vitro synthesized Upf3p was also shown to bind eRF3 (data not shown), providing further evidence that Upf3p binds to eRF3 directly.
The ability of Upf2p and Upf3p to interact with eRF3 correlates with their ability to complement the nonsense suppression phenotype
We next determined whether the ability of Upf2p and Upf3p to interact with eRF3 correlates with their ability to complement the nonsense suppression phenotype. To test this, several mutants in both the UPF2 and UPF3 genes were constructed.
The C-terminal domain of Upf2p has been identified as the Upf1p-interacting domain (U1I, aa 939–1089) (He et al., 1997). Upf2p also has a highly acidic domain (Ac, aa 886–938) just upstream of its U1I domain (Cui et al., 1995). Both domains are conserved among all known UPF2 homologs (S.pombe, Arabidopsis thaliana, Drosophila melanogaster, Mus musculus and H.sapiens; W.Wang and S.W.Peltz, unpublished result), suggesting that they are important for the function of Upf2p. Two upf2 mutants that lack either the acidic (upf2-ΔAc) or U1I domain (upf2-ΔU1I) were constructed. These mutant proteins were shown to be stable and expressed at the wild-type level in yeast cells (data not shown). Their ability to interact with eRF3 was measured by immunoprecipitation and GST pull-down experiments. The results of both experiments demonstrated that upf2-ΔU1I interacted with eRF3 with an affinity similar to that of the wild-type Upf2p, whereas upf2-ΔAc failed to interact with eRF3 (Figure 4A and B). These results suggest that the acidic, but not the U1I, domain is necessary for Upf2p to interact with eRF3. Interestingly, upf2-ΔAc still interacted with both Upf1p and Upf3p, albeit with a reduced affinity, indicating that this domain is not absolutely required for interactions with Upf1p or Upf3p (data not shown).
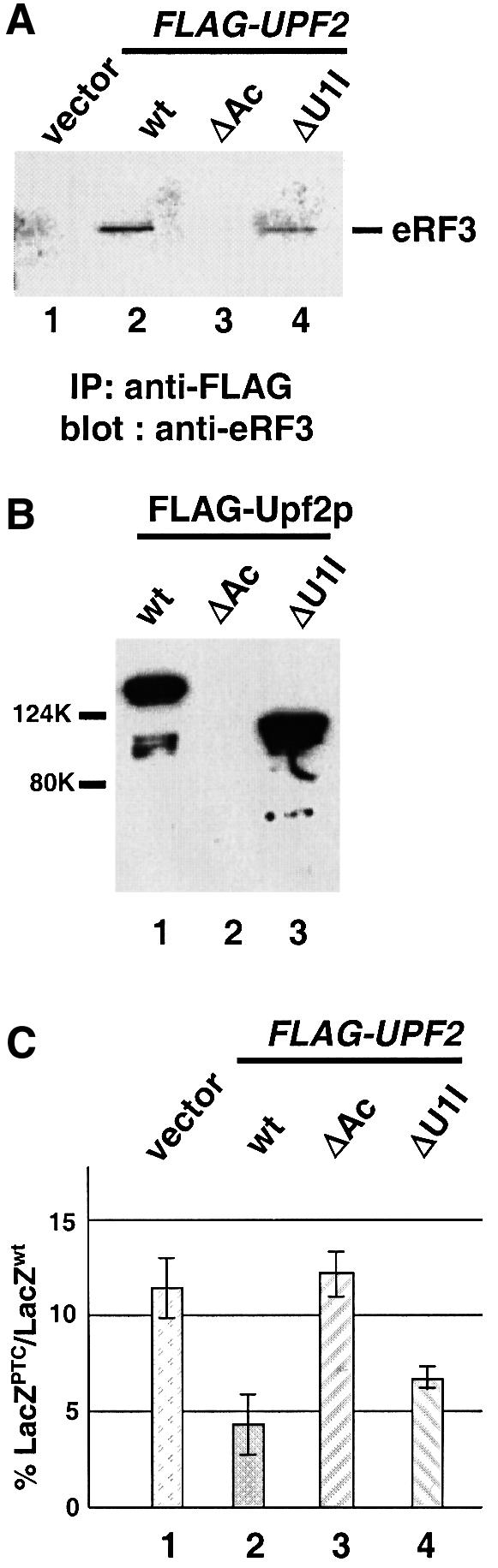
Fig. 4. The ability of Upf2p to complement the nonsense suppression phenotype in a upf2Δ strain correlates with its ability to interact with eRF3. (A) Co-immunoprecipitation. Cells were transformed with either vector alone or vector expressing the specified FLAG-Upf2p. Cytoplasmic extracts were prepared and immunoprecipitated with an anti-FLAG antibody. The immunoprecipitates were separated by SDS–PAGE and immunoblotted using the anti-eRF3 antibody. (B) GST pull-down experiment. Purified wild-type and mutant FLAG-Upf2 proteins (1.0 µg each) were combined with purified GST–eRF3 (1.0 µg) and glutathione–Sepharose beads. Following incubation and extensive washing, the proteins remaining associated with the beads were separated by SDS–PAGE and detected by the anti-FLAG antibody. (C) The ability of upf2 mutants to complement the nonsense suppression phenotype in a upf2Δ strain. Cells harboring either wild type or a UGA-containing LacZ gene were transformed with either vector alone or the vector expressing the specified upf2 gene. The assays were performed as described in Materials and methods.
We next examined whether the upf2-ΔAc and upf2-ΔU1I alleles were able to complement the nonsense suppression phenotype of a upf2Δ strain using the β-gal reporter system. The results showed that the upf2-ΔAc allele was unable to complement the nonsense suppression phenotype of a upf2Δ strain; conversely, the upf2-ΔU1I allele significantly reduced the LacZUGA/LacZwt ratio (Figure 4C). Similar results were obtained with LacZ transcripts containing UAA and UAG codons (data not shown). Taken together, these results suggest that the ability of Upf2p to bind eRF3 is essential for it to complement the nonsense suppression phenotype in a upf2Δ strain. The observation that the upf2-ΔU1I allele is >70% active in complementing the nonsense suppression phenotype in a upf2Δ strain (compare with the wild-type UPF2 allele) suggests that the Upf1p–Upf2p interaction is not critical for this function.
We also determined whether the ability of Upf3p to interact with eRF3 correlated with its ability to complement the nonsense suppression phenotype of a upf3Δ strain. Both the N-and C-terminal domains of Upf3p are rich in basic amino acids. Three upf3 mutants that lack the N-terminus (upf3ΔN, Δaa 1–80), C-terminus (upf3ΔC, Δaa 269–387), or both N- and C-terminus (upf3ΔN,C) were constructed. The polypeptides they encoded were shown to be stable and expressed at a wild-type level in yeast cells (data not shown). The ability of these mutants to interact with eRF3 was measured by immunoprecipitation and GST pull-down experiments. The results showed that upf3ΔN and upf3ΔC interacted with eRF3 with an affinity similar to that of wild-type Upf3p, but upf3ΔN,C completely lost its ability to interact with eRF3 (Figure 5A and B). These results indicate that Upf3p needs at least one of its terminal domains to interact with eRF3.
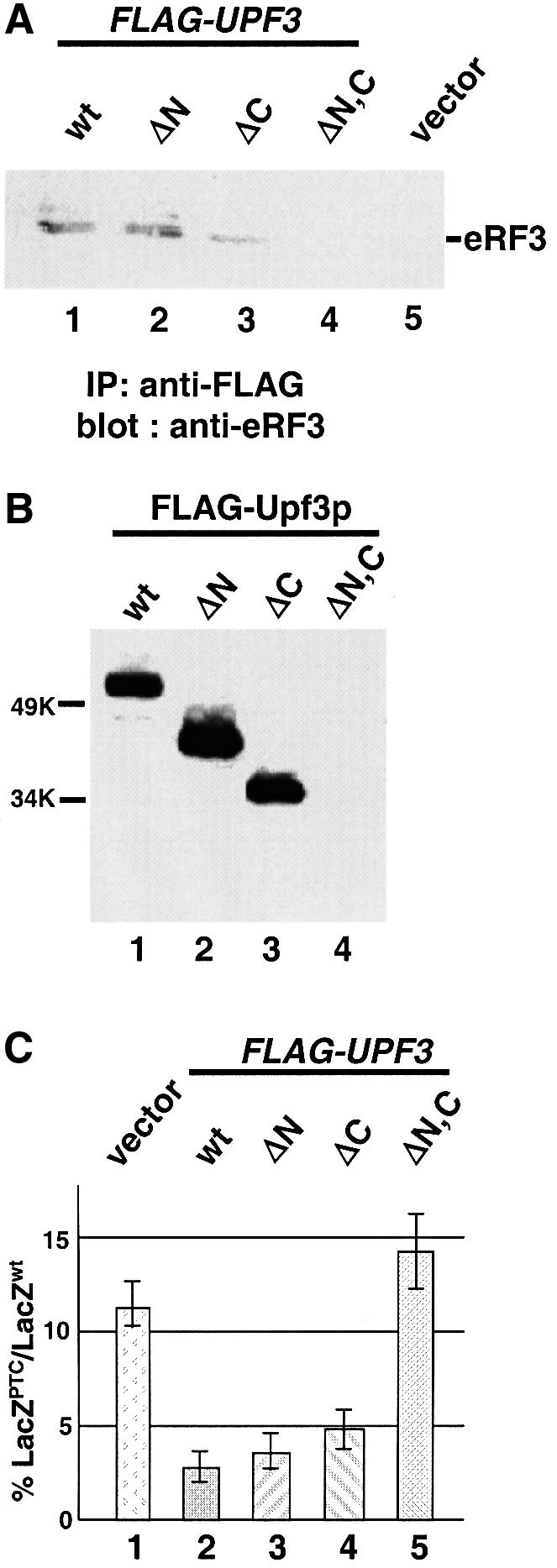
Fig. 5. The ability of Upf3p to complement the nonsense suppression phenotype in a upf3Δ strain correlates with its ability to interact with eRF3. (A) Co-immunoprecipitation. Cells were transformed with either vector alone (lane 5) or a vector expressing the specified FLAG-Upf3p. Cytoplasmic extracts were prepared and immunoprecipitated with an anti-FLAG antibody, and the immunoprecipitates were subjected to SDS–PAGE and immunoblotted by the anti-eRF3 antibody. (B) GST pull-down experiment. Purified wild-type and mutant FLAG-Upf3 proteins (0.5 µg each) were combined with purified GST–eRF3 (1.0 µg) and glutathione–Sepharose beads. Following incubation and extensive washing, the proteins remaining associated with the beads were separated by SDS–PAGE and detected by the anti-FLAG antibody. (C) The ability of upf3 mutants to complement the nonsense suppression phenotype in a upf3Δ strain. Cells harboring either wild-type or a UGA-containing LacZ gene were transformed with either the vector expressing the specified upf3 gene (lanes 1–4) or vector alone (lane 5). The assays were performed as described in Materials and methods.
The ability of these upf3 mutants to complement the nonsense suppression phenotype of a upf3Δ strain was also examined. The results showed that both upf3ΔN and upf3ΔC fully complemented the nonsense suppression phenotype of a upf3Δ strain (Figure 5C, columns 3 and 4). In contrast, upf3ΔN,C, which lost its ability to interact with eRF3, was unable to complement the nonsense suppression phenotype of a upf3Δ strain (column 5). Similar results were also obtained with LacZ transcripts containing UAA and UAG codons (data not shown). Taken together, these results suggest that the ability of Upf3p to interact with eRF3 also correlates well with its ability to complement the nonsense suppression phenotype.
The Upf proteins and eRF1 interact with the essential GTPase domain of eRF3
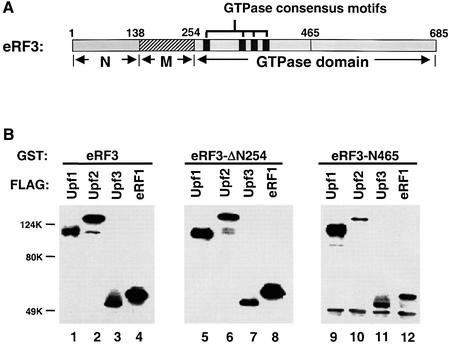
The yeast eRF3 protein (Sup35p) consists of three domains: an N-terminal glutamine-rich domain (aa 1–138; N), a middle hyper-charged domain (aa 139–254; M) and a C-terminal GTPase domain (aa 255–685) (Figure 6A). The C-terminal GTPase domain of eRF3 is conserved among all eukaryotes and is sufficient to support viability in yeast cells (Ter-Avanesyan et al., 1993). The GTPase domain of eRF3 can be further separated into two halves, and the N-terminal half harbors the four GTPase consensus motifs. In order to determine the binding sites for the Upf proteins and eRF1, an eRF3 allele with its N and M domains deleted (eRF3-ΔN254, Δaa 1–254) and another allele with the C-terminal half of the GTPase domain deleted (eRF3-N465, aa 1–465) were constructed. Both proteins were purified in the GST fusion form from Escherichia coli, and their abilities to interact with FLAG-Upf1p, -Upf2p, -Upf3p and -eRF1 were determined by GST pull-down experiments. The results showed that eRF3-ΔN254 interacted with the Upf proteins and eRF1 with similar affinity to the full-length eRF3 protein (Figure 6B). In contrast, although eRF3-N465 bound to Upf1p with an affinity comparable to (or even higher than) that of the full-length eRF3, its affinities for Upf2p, Upf3p and eRF1 were all significantly reduced (Figure 6B). These results showed that the Upf proteins and eRF1 all interact with the essential GTPase domain of eRF3; the N-terminal half of the GTPase domain is sufficient to mediate the interaction with the Upf1p, but not the interactions with Upf2p, Upf3p and eRF1.
Fig. 6. The Upf proteins and eRF1 interact with the essential GTPase domain of eRF3. (A) Schematic diagram of the domain structure of the yeast release factor eRF3 (Sup35p). (B) GST pull-down experiment. Purified GST–eRF3 (1.0 µg, lanes 1–4), eRF3-N254Δ (1.0 µg, lanes 5–8) or eRF3-N465 (1.0 µg, lanes 9–12) was combined with FLAG-Upf1p (1.0 µg, lanes 1, 5 and 9), -Upf2p (1.0 µg, lanes 2, 6 and 10), -Upf3p (0.5 µg, lanes 3, 7 and 11) or -eRF1 (0.5 µg, lanes 4, 8 and 12). Following incubation and extensive washing, the proteins remaining associated with the beads were resolved on 12% SDS–PAGE and detected by the anti-FLAG antibody.
Upf2p, Upf3p and eRF1 compete with each other, but not with Upf1p, for interaction with eRF3
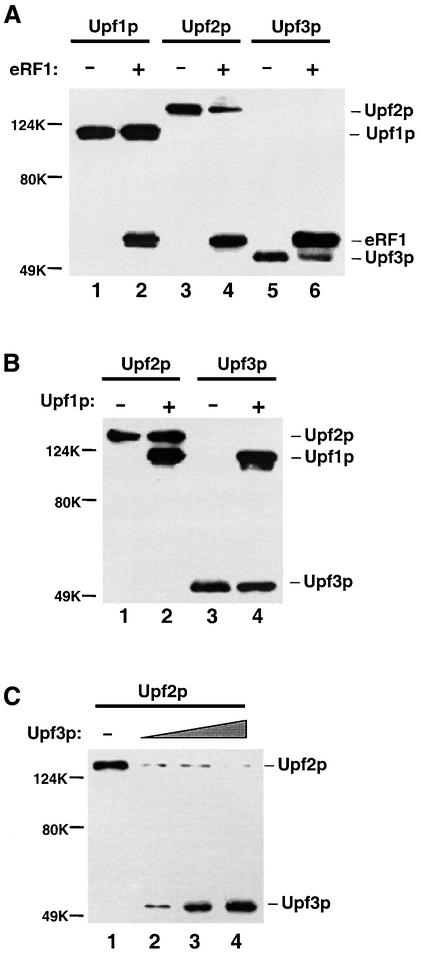
Since the Upf proteins and eRF1 bind to similar sites on eRF3, we reasoned that they might compete with each other for binding to eRF3. To test this hypothesis, a series of competition experiments was performed using the GST pull-down method. The interactions between the FLAG-tagged Upf proteins and GST–eRF3 in the absence or presence of the FLAG-eRF1 protein were first determined (Figure 7A). The results showed that the presence of eRF1 had no apparent effect on the amount of Upf1p bound to GST–eRF3 (compare lanes 1–2), but significantly reduced the amount of Upf2p (lanes 3–4) and Upf3p (lanes 5–6) bound to GST–eRF3. These results suggest that eRF1 competes with Upf2p and Upf3p, but not with Upf1p, for binding to eRF3. These results also indicate that Upf2p and Upf3p can only bind to eRF3 when it is not in complex with eRF1, but that Upf1p has the potential to associate with the eRF1–eRF3 complex. We also showed that, under high salt conditions, GST–eRF1 was unable to pull-down the FLAG-Upf1p unless eRF3 was present, providing direct evidence that the Upf1p can bind to the eRF1–eRF3 complex (data not shown).
Fig. 7. Upf2p, Upf3p and eRF1 competed with each other, but not with Upf1p, for binding to eRF3. (A) Analysis of the interactions between FLAG-Upf1p (1.0 µg), -Upf2p (1.0 µg), -Upf3p (0.5 µg) and GST–eRF3 (0.5 µg) in the absence or presence of FLAG-eRF1 (0.5 µg) by GST pull-down experiments. (B) Analysis of the interactions between FLAG-Upf2p (1.0 µg), -Upf3p (0.5 µg) and GST–eRF3 (0.5 µg) in the absence or presence of FLAG-Upf1p (1.0 µg) by GST pull-down experiments. (C) Analysis of the interactions between FLAG-Upf2p (1.0 µg) and GST–eRF3 (0.5 µg) in the absence (lane 1) or presence of increasing amounts of FLAG-Upf3p (lanes 2–4, 0.5, 1.0 and 2.0 µg) by GST pull-down experiments.
Using a similar approach, we showed that the presence of Upf1p had little or no effect on the amount of Upf2p and Upf3p bound to GST–eRF3 (Figure 7B). This indicates that eRF3 can interact with Upf1p–Upf2p or Upf1p–Upf3p at the same time. In contrast, increasing amounts of Upf3p competed away the binding of Upf2p to GST–eRF3 (Figure 7C), indicating that Upf2p and Upf3p compete with each other for binding to eRF3. Interestingly, in the reactions containing approximately equivalent concentrations of Upf2p and Upf3p, the amount of Upf2p bound to GST–eRF3 was greatly reduced, while the amount of Upf3p bound to GST–eRF3 only increased moderately (compare Figure 7A, lane 4, and 7C, lane 2). This suggests that a Upf2p–Upf3p complex was efficiently formed and this complex was unable to bind to eRF3.
Taken together, these results demonstrate that Upf2p, Upf3p and eRF1 competed with each other, but not with Upf1p, for interaction with eRF3. This conclusion is consistent with the findings that Upf2p, Upf3p and eRF1 bind to similar sites on the eRF3 protein, while the Upf1p binds to a more N-terminal site on eRF3. The competitive nature of the interaction between Upf2p, Upf3p, eRF1 and eRF3 suggests that these three factors bind to eRF3 sequentially, whereas Upf1p can bind to eRF3 together with any of these factors.
Discussion
Upf2p and Upf3p are components of the surveillance complex
In this report we showed that both upf2Δ and upf3Δ strains demonstrate a nonsense suppression phenotype. We also showed that Upf2p and Upf3p interact with the release factor eRF3. Furthermore, mutational analysis showed that the ability of Upf2p and Upf3p to complement the nonsense suppression phenotype correlated with their ability to interact with eRF3. However, unlike Upf1p, Upf2p and Upf3p do not interact with either eRF1 or the eRF1–eRF3 complex, suggesting that neither Upf2p nor Upf3p is likely to modulate the rate of peptidyl-tRNA bond hydrolysis directly. It has been proposed that during the translation termination process, a surveillance complex, which consists of at least the Upf proteins and the release factors, is assembled and searches 3′ of the termination codon for specific signals that target the transcript for rapid degradation (Czaplinski et al., 1998). Upf2p and Upf3p interact with eRF3; deleting either gene will lead to both inactivation of the NMD pathway and a nonsense suppression phenotype. These results provide evidence that the Upfps are components of the surveillance complex. Interestingly, our in vitro competition experiments showed that Upf2p, Upf3p and eRF1 compete with each other for binding to eRF3, indicating that more than one kind of complex can be formed between the Upf proteins and release factors, or that the surveillance complex is dynamic.
A sequential surveillance complex assembly model
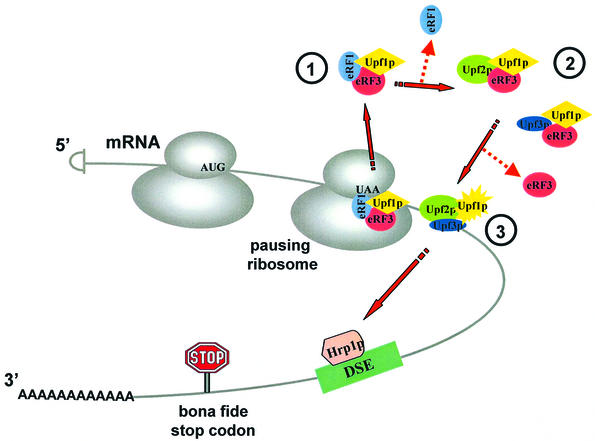
Given these observations, we propose a general model for how the surveillance complex may assemble and affect the suppression of nonsense-containing transcripts (Figure 8). Surveillance complex assembly starts when a translating ribosome pauses at a stop codon. This signals the eRF1–eRF3 complex to bind to the A site of the ribosome. During the termination process, Upf1p also becomes associated with the eRF1–eRF3 complex. After hydrolysis of the peptidyl-tRNA bond, eRF1 dissociates from the ribosome. Dissociation of eRF1 allows Upf2p (or Upf3p) to join the surveillance complex through interactions with eRF3. Subsequently, a rearrangement of the complex results in the association of Upf3p (or Upf2p) and the release of eRF3. The affinity between Upf2p and Upf3p might play an important role in this transition. The dissociation of eRF3 also activates the ATPase/helicase activity of Upf1p (Czaplinski et al., 1998). Finally, a complex forms that consists of, at a minimum, Upf1p, Upf2p and Upf3p. Since Upf1p, Upf2p and Upf3p are the only factors identified in yeast that affect both NMD and nonsense suppression, this complex may represent the mature surveillance complex. The mature surveillance complex can then initiate the subsequent steps in NMD that ultimately lead to the rapid degradation of the mRNA (Czaplinski et al., 1999; Hentze and Kulozik, 1999). Although the assembly of the surveillance complex is depicted as though it occurs while not associated with the ribosome (Figure 8), it is feasible that assembly of the complex proceeds while associated with a ribosome or ribosomal subunits.
Fig. 8. Model for the sequential surveillance complex assembly pathway. (1) The translating ribosome pauses at a premature termination codon and signals the eRF1–eRF3 complex to bind to its A site. The Upf1p becomes associated with the eRF1–eRF3 complex during the termination process. (2) After hydrolysis of the peptidyl-tRNA bond, eRF1 dissociates from the ribosome. Dissociation of eRF1 allows either Upf2p or Upf3p to bind the eRF3–Upf1p complex. (3) Rearrangement of the complex: Upf3p (or Upf2p) joins the complex and displaces eRF3 to form the mature surveillance complex.
This model implies that the assembly of the surveillance complex is part of the premature translation termination event. In the absence of any Upf proteins, the assembly of the surveillance complex is blocked. There are at least three possibilities as to how a failure in surveillance complex assembly leads to a nonsense suppression phenotype. The first possibility is that the successful assembly of the surveillance complex is needed for the release and/or regeneration of the active form of the release factors. Preventing assembly of the surveillance complex would impair the recycling of these factors and result in reduced translation termination efficiency.
A second alternative takes into account the fact that the expression levels of all three Upf proteins are significantly lower compared with the levels of release factors and ribosomes (Atkin et al., 1997; Maderazo et al., 2000), suggesting that the surveillance complex assembly might occur only during the first round of translation. Actually, the first round of translation would be sufficient to evoke NMD in all eukaryotic systems according to any current models for NMD (reviewed in Hentze and Kulozik, 1999). In support of this alternative, there is an emerging view that the first round of translation plays a distinct role in determining the translation efficiency and stability of a transcript (Fortes et al., 2000; see below). One hypothesis is that the failure in surveillance complex assembly during the first round of translation will result in an unusual mRNP structure that affects future rounds of translation, including the termination efficiency, in cis.
A third possibility is that the nonsense suppression phenotype results from a specific increase in the translation of nonsense-containing mRNAs. This possibility is based on the hypothesis that once the surveillance complex determines that a premature termination has occurred it will not only accelerate the degradation of the nonsense-containing mRNA but also repress further translation of the mRNA. In the absence of a functional surveillance complex, the nonsense-containing mRNAs are continuously translated, thus resulting in the synthesis of more full-length products and a nonsense suppression phenotype. Interestingly, the protein/mRNA ratio of a nonsense-containing mRNA was found to be three times higher in a upf1Δ strain than in a wild-type strain (Muhlrad and Parker, 1999b).
Multiple lines of evidence help support the sequential surveillance complex assembly model. In prokaryotes, it is known that the release factor RF3 stimulates the activity of RF1/RF2 by promoting their dissociation from the A site of the ribosome after hydrolysis of the peptidyl-tRNA bond (Freistroffer et al., 1997; Karimi et al., 1999). Although less is understood about the release factors in eukaryotes, it is reasonable to expect that eRF1 also dissociates from the ribosome after the hydrolysis of the peptidyl-tRNA bond. Secondly, previous results have shown that the ATPase/helicase activity of Upf1p is essential for its activity in the NMD pathway (Weng et al., 1996a) and that the Upf1p–eRF3 interaction blocks the ATPase/helicase activity of Upf1p (Czaplinski et al., 1998). These results indicate that eRF3 must be dissociated from Upf1p before Upf1p can function in the NMD pathway. In addition, we have shown that a DSE would not promote mRNA decay when positioned right after the stop codon (Zhang et al., 1995). One explanation for this finding is that the functional surveillance complex requires time to assemble. Finally, this model suggests that all the Upf proteins are involved in a unified process, and thus explains why single or multiple deletions of the UPF genes result in similar levels of nonsense-containing mRNA stabilization and nonsense suppression activity (He et al., 1997; Maderazo et al., 2000; this study).
The model presented here suggests two crucial steps in the assembly of the surveillance complex. The first is the dissociation of eRF1, which allows the binding of either Upf2p or Upf3p to the eRF3–Upf1p complex; the second is the dissociation of eRF3, which allows the binding of both Upf2p and Upf3p to Upf1p, and activates the ATPase/helicase activity of the Upf1p. Currently, it is not clear what leads to these two transitions. It is possible that the post-translational modifications play an important role in these processes. For example, the GTP or GDP forms of eRF3 may determine which factor(s) it can bind. Alternatively, it has been suggested that the UPF1 homolog in C.elegans, smg-2, undergoes a phosphorylation/dephosphorylation cycle (Page et al., 1999). Since Upf1p can bind to eRF3 together with any of the other three factors, it is possible that different phosphorylation states of Upf1p can determine which other factor(s) can become part of the complex. Future experiments are required to determine how the assembly of the surveillance complex is regulated.
Epistatic relationships of Upf1p, Upf2p and Upf3p
The role of the Upf proteins in translating nonsense-containing transcripts has also been investigated (Maderazo et al., 2000). The results showed that a upf1Δ strain demonstrates a slightly higher nonsense suppression activity than that observed in a upf2Δ (or upf3Δ) strain, and a upf1Δupf2Δ (or upf1Δupf3Δ) double deletion strain demonstrates the same level of nonsense suppression as the upf1Δ strain (Maderazo et al., 2000). Based on these results, it was proposed that Upf1p is epistatic to Upf2p and Upf3p. Furthermore, it was suggested that Upf2p and Upf3p modulate the translation termination efficiency by regulating the activity of Upf1p (Maderazo et al., 2000). Our in vitro competition experiments suggest that Upf1p assembles into the surveillance complex before Upf2p and Upf3p (Figure 8), consistent with the hypothesis that Upf1p is epistatic to Upf2p and Upf3p. The data presented here, however, do not support the idea that Upf2p and Upf3p function through Upf1p. Using an NMD-independent β-gal reporter system, we showed that single or multiple deletions of the UPF genes result in similar levels of suppression. The differences observed by the other group were relatively moderate and some were not consistent with their model. More importantly, we demonstrate that all the mutant Upf proteins that lack the ability to interact with eRF3 lose their ability to complement the nonsense suppression phenotype. Meanwhile, a mutant Upf2p that lacks the entire Upf1p-interacting domain is still >70% active. In addition, no interaction between Upf1p and Upf3p has ever been documented (He et al., 1997). Taken together, these results strongly suggest that Upf2p and Upf3p modulate the translation of nonsense-containing transcripts through interactions with eRF3, instead of the Upf1p.
The potential role of the surveillance complex at bona fide termination codons
The results presented here and elsewhere indicate that surveillance complex assembly occurs during the premature translation termination process. Although not shown directly, we anticipate that assembly of the surveillance complex also occurs at normal termination codons. This is because we have hypothesized that assembly of the surveillance complex is a prerequisite for determining whether a termination codon is normal or premature. Interestingly, recent results from a number of groups have suggested that after the first round of translation the 5′ and 3′ ends of the mRNA may interact to form a ‘closed loop’ structure through interactions between the translation initiation factor eIF4G and the poly(A)-binding protein PABP (Imataka et al., 1998). This interaction would enable the terminating ribosomes to migrate around and reinitiate at the coding region of the mRNA, and therefore would be needed for both the stability and translation efficiency of the transcript (reviewed in Sachs, 2000). Recently, it was shown that eRF3 can interact with PABP and that this interaction has an effect on the cooperative binding of PABP to the poly(A) tail (Hoshino et al., 1999). One intriguing possibility is that when the ribosome terminates at a bona fide termination codon, the surveillance complex assembly may be an integral part of the formation of the proposed ‘loop’ structure.
Materials and methods
Yeast strains and plasmids
The yeast strain KC2 (MATa ura3-52 trp1 leu2-2 tyr7-1 his4-38 met14) was created by crossing strain PLY146 with PLY22 (Leeds et al., 1991), sporulating, and screening spores for the desired markers. Deletions of the UPF genes were performed as described previously (Cui et al., 1995) and the disruptions were confirmed by Southern blotting. Yeast transformations were performed by the lithium acetate method (Philippsen et al., 1991).
A yeast 2µ plasmid, pG-1, was used as the vector in this study (Weng et al., 1996a). The FLAG-UPF1 allele has been described previously (Czaplinski et al., 1995). The 5′ portion of the UPF2 DNA sequence up to the unique HindIII site was created by PCR amplification. A FLAG epitope (sequence Met-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) was added to the N-terminus of UPF2 sequence. The 3′ portion of the UPF2 coding region was taken from a HindIII–XhoI fragment from the original UPF2 clone (Cui et al., 1995). The two DNA fragments were ligated together and cloned into the pG-1 vector to generate the FLAG-UPF2 allele. The FLAG-UPF3 and FLAG-SUP45 (eRF1) alleles, each harboring a FLAG epitope at its N-terminus, were constructed by PCR amplification using S.cerevisiae genomic DNA as template. All the constructs were confirmed by DNA sequencing.
Nonsense suppression assays
The KC2 strain was used as the wild-type strain in all nonsense suppression assays. It contains a UAG nonsense codon in the coding region of tyr7-1 and is unable to grow in medium lacking tyrosine (–tyr). To monitor the growth of wild-type and upfΔ strains, cells were grown in synthetic complete (SC) medium to an OD600 of 1.0. Three 10-fold serial dilutions were made with –tyr medium, and 10 µl of each dilution were spotted on both SC and –tyr plates. Growth was monitored at 30°C. It generally takes 1–2 days for the spots to develop on an SC plate and 2–3 days for the spots to develop on a –tyr plate.
Quantitative analysis of the nonsense suppression activities in strains harboring single or multiple UPF gene deletions was performed using a β-gal reporter construct. Yeast strains were transformed with either a wild-type LacZ gene (LacZwt) or a LacZ gene containing an early premature termination codon (LacZPTC). The LacZ genes are under the control of PGK promoter and 3′ UTR in a centromere pYCplac22 vector. For each transformant, a 20 ml culture was grown at 30°C in appropriate medium to an OD600 of 0.7–0.8, and the cells were harvested. RNA was extracted from 10 ml of the culture, and the abundance of LacZ mRNA and U3 snRNA was analyzed by northern blotting and quantified by PhosphorImaging (Molecular Dynamics). Yeast crude cytoplasmic extracts were prepared from the remaining 10 ml culture. ONPG (2-nitrophenyl-β-d-galactopyranoside) was used as substrate to measure the β-gal activity. The β-gal activity was normalized to the amount of proteins in the crude extract. The nonsense suppression activity in a particular strain was determined by calculating the ratios of β-gal activities in cells harboring the LacZPTC to cells harboring the LacZwt genes. At least three individual yeast transformants were assayed in triplicate and the ratio did not vary by >20%.
Protein purification
The proteins were purified as described previously (Czaplinski et al., 1998). The FLAG-tagged proteins were purified from yeast using anti-FLAG M2 beads (Sigma). The GST and GST fusion proteins were purified from E.coli using glutathione–Sepharose 4B beads (Pharmacia). The purified fractions were dialyzed in storage buffer [25 mM Tris pH 7.5, 50 mM KCl, 1 mM dithiothreitol (DTT) and 20% glycerol], aliquotted, and frozen at –70°C.
Immunoprecipitation
For immunoprecipitation experiments, the cells harboring FLAG-tagged UPF alleles were lysed in LB buffer (20 mM HEPES pH 7.5, 100 mM KOAc, 20% glycerol, 5 mM EDTA) with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 2 µg/ml protease inhibitors (pepstatin A, leupeptin and aprotinin). The extract (100 µg) was mixed with anti-FLAG antibody immobilized on protein A–Sepharose beads and incubated at 4°C for 4 h with constant rocking. The beads were then washed five times with 1 ml of cold LB buffer. The proteins that remained associated with the beads were released by boiling in the loading buffer and subjected to SDS–PAGE followed by immunoblotting with an anti-eRF3 antibody. The rabbit anti-eRF3 polyclonal antibody was generated using purified GST–eRF3 protein (Pocono Rabbit Farm & Laboratory).
GST pull-down experiment
Purified GST or GST fusion protein (1.0 µg) was immobilized on 10 µl of glutathione–Sepharose 4B beads and combined with the indicated amount of purified FLAG-tagged proteins in 0.5 ml of Ipp150 buffer (10 mM Tris pH 8.0, 150 mM NaCl and 0.1% NP-40). The mixtures were incubated at 4°C for 1.5 h with constant rocking. The beads were then washed three times with 1 ml of cold Ipp150 buffer. The FLAG-tagged proteins that remained associated with the beads were released by boiling in the loading buffer and subjected to SDS–PAGE followed by immunoblotting with an anti-FLAG M2 monoclonal antibody (Sigma).
For the competition experiments, a limited amount of GST–eRF3 fusion protein (0.5 µg) was incubated with two kinds of FLAG-tagged proteins at the same time. The amounts of FLAG-tagged proteins used were as indicated in the figure legends. The mixtures were incubated at 4°C for 3 h with constant rocking. The rest of the assay was performed as described above.
Acknowledgments
Acknowledgements
We thank Drs Carlos I.Gonzalez, Maria J.Ruiz-Echevarria and all the members of the Peltz laboratory for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (GM48631) and an American Heart Association Established Investigator Award given to S.W.P.
REFERENCES
- Atkin A.L., Schenkman,L.R., Eastham,M., Dahlseid,J.N., Lelivelt,M.J. and Culbertson,M.R. (1997) Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem., 272, 22163–22172. [DOI] [PubMed] [Google Scholar]
- Cui Y., Hagan,K.W., Zhang,S. and Peltz,S.W. (1995) Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev., 9, 423–436. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Weng,Y., Hagan,K.H. and Peltz,S.W. (1995) Purification and characterization of the Upf1p protein: a factor involved in translation and mRNA degradation. RNA, 1, 610–623. [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Gonzales,C.I. and Peltz,S.W. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. BioEssays, 21, 685–696. [DOI] [PubMed] [Google Scholar]
- Fortes P., Inada,T., Preiss,T., Hentze,M.W., Mattaj,I.W. and Sachs,A.B. (2000) The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell, 6, 191–196. [PubMed] [Google Scholar]
- Freistroffer D.V., Pavlov,M.Y., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J., 16, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L., Le Goff,X., Zhouravleva,G., Davydova,E., Philippe,M. and Kisselev,L. (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA, 2, 334–341. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C.I., Ruiz-Echevarria,M.J., Vasudevan,S., Henry,M.F. and Peltz,S.W. (2000) The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell, 5, 489–499. [DOI] [PubMed] [Google Scholar]
- He F., Brown,A. and Jacobson,A. (1997) Upf1p, Nmd2p and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol., 17, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hoshino S., Imai,M., Kobayashi,T., Uchida,N. and Katada,T. (1999) The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem., 274, 16677–16680. [DOI] [PubMed] [Google Scholar]
- Imataka H., Gradi,A. and Sonenberg,N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. and Peltz,S.W. (2000) Destabilization of nonsense-containing transcripts in Saccharomyces cerevisiae. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 827–847.
- Karimi R., Pavlov,M.Y., Buckingham,R.H. and Ehrenberg,M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Lee B.S. and Culbertson,M.R. (1995) Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc. Natl Acad. Sci. USA, 92, 10354–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P., Peltz,S.W., Jacobson,A. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translation termination codon. Genes Dev., 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Maderazo A.B., He,F., Mangus,D.A. and Jacobson,A. (2000) Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol., 20, 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.T., Medghalchi,S.M., Lake,R.G., Noensie,E.N. and Dietz,H.C. (2000) Novel upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol., 20, 8944–8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1999a) Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA, 5, 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1999b) Recognition of yeast mRNAs as ‘nonsense containing’ leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol. Biol. Cell, 10, 3971–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Ito,K. and Isaksson,L. (1996) Emerging understanding of translation termination. Cell, 87, 147–150. [DOI] [PubMed] [Google Scholar]
- Page M.F., Carr,B., Anders,K.R., Grimson,A. and Anderson,P. (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol., 19, 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S.W., Brown,A.H. and Jacobson,A. (1993) mRNA destabilization triggered by premature translational termination depends on three mRNA sequence elements and at least one trans-acting factor. Genes Dev., 7, 1737–1754. [DOI] [PubMed] [Google Scholar]
- Perlick H.A., Medghalchi,S.M., Spencer,F.A., Kendzior,R.J.,Jr, and Dietz,H.C. (1996) Mammalian orthologues of a yeast regulator of nonsense-transcript stability. Proc. Natl Acad. Sci. USA, 93, 10928–10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Stotz,A. and Scherf,C. (1991) DNA of Saccharomyces cerevisiae. Methods Enzymol., 194, 169–182. [DOI] [PubMed] [Google Scholar]
- Pulak R. and Anderson,P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev., 7, 1885–1897. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J. and Peltz,S.W. (1996) Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J., 15, 2810–2819. [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B. (2000) Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 447–465.
- Serin G., Gersappe,A., Black,J.D., Aronoff,R. and Maquat,L.E. (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae upf2 protein and upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol., 21, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley R.L., Lelivelt,M.J., Schenkman,L.R., Dahlseid,J.N. and Culbertson,M.R. (1998) A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci., 111, 3129–3143. [DOI] [PubMed] [Google Scholar]
- Song H., Mugnier,P., Das,A.K., Webb,H.M., Evans,D.R., Tuite,M.F., Hemmings,B.A. and Barford,D. (2000) The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Stansfield I. et al. (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J., 14, 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Kushnirov,V.V., Dagkesamanskaya,A.R., Didichenko,S.A., Chernoff,Y.O., Inge-Vechtomov,S.G. and Smirnov, V.N. (1993) Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol., 7, 683–692. [DOI] [PubMed] [Google Scholar]
- Welch E.M., Wang,W. and Peltz,S.W. (2000) Translation termination: it’s not the end of story. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 467–485.
- Weng Y., Czaplinski,K. and Peltz,S.W. (1996a) Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol., 16, 5477–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y., Czaplinski,K. and Peltz,S.W. (1996b) Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol., 16, 5491–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ruiz-Echevarría,M.J., Quan,Y. and Peltz,S.W. (1995) Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol., 15, 2231–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova,L., LeGoff,X., LeGuellec,R., Inge-Vechtomov,S., Kisselev,L. and Phillippe,M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]