Abstract
The human snRNA genes transcribed by RNA polymerase II (e.g. U1 and U2) have a characteristic TATA-less promoter containing an essential proximal sequence element. Formation of the 3′ end of these non-polyadenylated RNAs requires a specialized 3′ box element whose function is promoter specific. Here we show that truncation of the C-terminal domain (CTD) of RNA polymerase II and treatment of cells with CTD kinase inhibitors, including DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole), causes a dramatic reduction in proper 3′ end formation of U2 transcripts. Activation of 3′ box recognition by the phosphorylated CTD would be consistent with the role of phospho-CTD in mRNA processing. CTD kinase inhibitors, however, have little effect on initiation or elongation of transcription of the U2 genes, whereas elongation of transcription of the β-actin gene is severely affected. This result highlights differences in transcription of snRNA and mRNA genes.
Keywords: CTD/pol II/RNA processing/snRNA/transcription
Introduction
In addition to protein-coding genes, RNA polymerase II (pol II) transcribes the genes for the non-coding snRNAs such as U1 and U2, which are required for processing of pre-mRNAs. These snRNA genes have specialized TATA-less promoters with a very simple structure comprising the basal proximal sequence element (PSE) and an upstream enhancer-like distal sequence element (DSE) (reviewed by Hernandez, 2001). Unlike pre-mRNAs, snRNAs are not spliced, and 3′ end formation is dependent on a conserved ‘3′ box’ rather than a polyadenylation signal or the 3′ processing signal of replication-activated histone genes (reviewed in Cuello et al., 1999). We have shown that transcription of the U2 snRNA genes continues beyond the 3′ box (Cuello et al., 1999), and this element is, therefore, likely to function directly as a 3′ end processing element in the RNA, rather than a transcription termination signal. However, the 3′ box is required for termination of transcription to occur downstream from the U2 promoter (Cuello et al., 1999). Intriguingly, the 3′ box only functions efficiently if transcription by pol II is initiated from a PSE-containing promoter (see Cuello et al., 1999), indicating that events at the snRNA gene promoter are somehow linked to downstream processing reactions.
The conserved C-terminal domain (CTD) of pol II is important for efficient transcription of some mRNA genes in vitro (reviewed by Dahmus, 1996). In addition, efficient transcription of some transfected mRNA genes is CTD dependent (Gerber et al., 1995), and there appears to be a global defect in transcription of chromosomal protein-coding genes in the absence of the CTD (Meininghaus et al., 2000). In mammals, the CTD has an unusual structure of 52 tandem heptad repeats, which can be phosphorylated in a reversible manner. CTD kinases include the cyclin-dependent kinase, CDK7, subunit of the basal transcription factor TFIIH and the CDK9 subunit of the elongation factor P-TEFb. A conserved CTD phosphatase, called Fcp1, has also been identified. Dynamic site-specific phosphorylation and dephosphorylation appear to be critical for CTD function and correct mRNA gene expression (reviewed by Prelich, 2002). For example, the phosphorylated form of the CTD interacts with capping enzymes and enhances 5′ capping of pre-mRNA, splicing and 3′ cleavage (reviewed by Bentley, 2002). Dantonel et al. (1997) have also shown that the pol II basal transcription factor TFIID can mediate interaction between polyadenylation factors and the CTD. Thus, the CTD couples transcription and RNA processing in expression of protein-coding genes.
We proposed that the CTD plays a similar role to connect initiation and 3′ end processing in expression of the U2 snRNA genes, and that specific promoter factors could help recruit the 3′ box-dependent processing machinery (Cuello et al., 1999). In agreement with this, we show here that correctly initiated transcripts are not processed properly at the 3′ end when U2 genes are transcribed by pol II lacking most of the CTD. Inhibitors of CTD kinases affect recognition of the 3′ box, causing readthrough of this signal. These inhibitors, however, have little effect on U2 transcription, although transcription of the β-actin gene is drastically affected.
These results indicate that, in common with protein-coding genes, snRNA genes require the CTD of pol II for accurate and efficient RNA processing. In addition, a phosphorylated CTD may be required for processing of the transcripts. In contrast to the β-actin gene, however, transcription of U2 snRNA genes does not require hyperphosphorylation of the CTD, pointing to differences in the molecular events associated with transcription of these two gene types.
Results
The CTD of pol II is required for high levels of steady-state RNA from human U2 snRNA gene constructs and correct 3′ end formation of the transcripts
To test the hypothesis that the CTD of pol II plays a role in 3′ end formation of transcripts from the human U2 snRNA genes, we have used the in vivo complementation system devised by Gerber et al. (1995). An α-amanitin-resistant pol II large subunit is supplied exogenously by transfection, and transcription by polymerase containing the endogenous pol II large subunit can be inhibited by addition of this drug. In this way, the effect of changes in the large subunit can be assessed. Previous experiments using this system have shown that, in mammals, the CTD of this subunit of pol II is required for transcription of some mRNA genes (Gerber et al., 1995) and for processing of pre-mRNA (McCracken et al., 1997b).
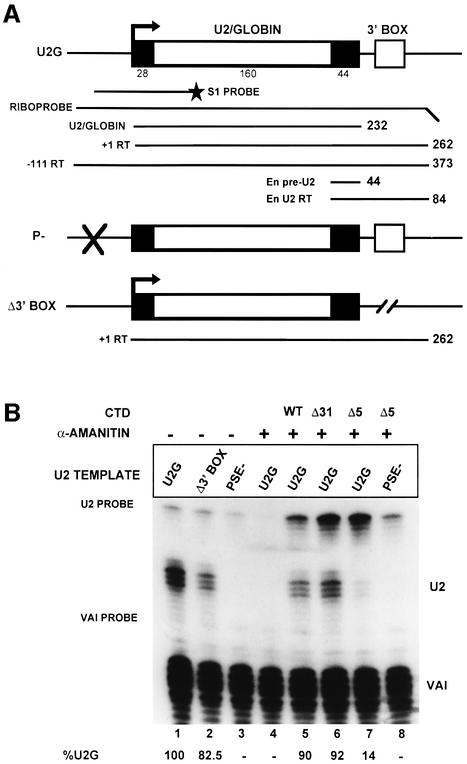
For our analysis, we have used U2 gene constructs where the majority of the region encoding the mature RNA is replaced by β-globin gene sequences (U2G; Figure 1A). This allows differentiation between the products of transfected and endogenous genes. In addition, the 3′ processed RNAs from the transfected, marked, genes are exported to the cytoplasm but are neither further 3′ end processed nor re-imported into the nucleus since the necessary signals are lacking (reviewed in Huang and Pederson, 1999). Constructs with or without the 3′ box and with a mutated PSE were transfected (Figure 1A), and RNA was analysed at the 5′ end by S1 nuclease mapping (Figure 1B) and by RNase protection (Figure 1C and D). We have used the adenovirus pol III-transcribed VAI gene as a co-transfection control, and the quantitation determined relative to this is shown at the bottom of each figure. Since several plasmids must be present together in the same cell for these experiments to work, we have used 293 human embryonic kidney cells that transfect with high efficiency.
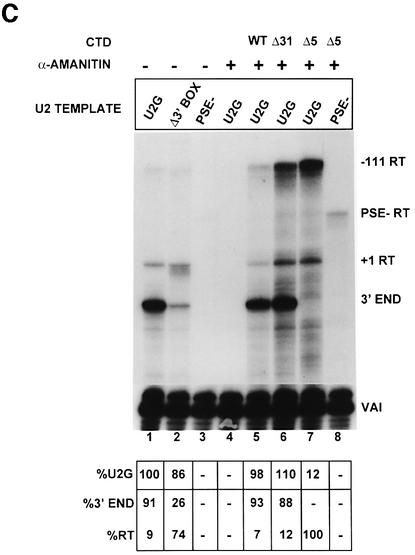
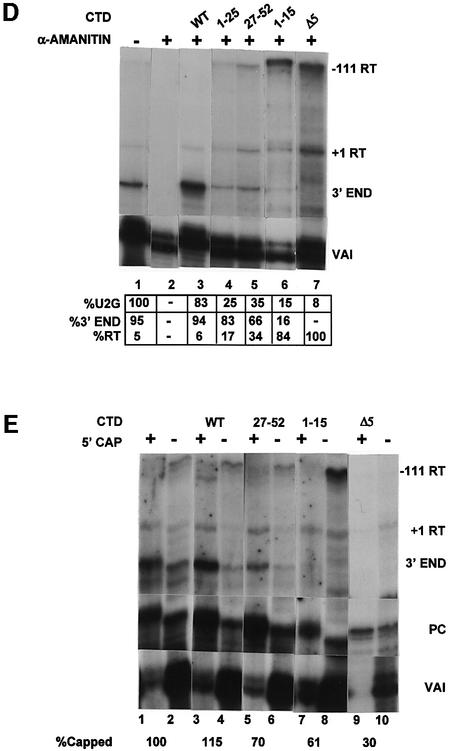
Fig. 1. The CTD of pol II is required for high steady-state levels of RNA from U2 snRNA gene constructs and correct 3′ end formation of the transcripts. (A) The structure of the U2–globin (U2G) constructs (see Materials and methods). P– is transcriptionally inactive due to a mutation in the PSE of the promoter (Cuello et al., 1999). The Δ3′ box has had the 3′ box deleted (Cuello et al., 1999). The relative positions of the S1 probe and riboprobes are shown below the diagram of the U2G construct. The size of the expected products of RNase protection analysis is also noted on the right of each construct. +1RT and –111RT are products that initiate at +1 and –111, respectively, and read through beyond the mismatch with the riboprobe. (B) The results of S1 analysis of RNA transcribed from the constructs described in (A). The U2G and CTD construct used and the addition of α-amanitin is noted above each lane. The positions of S1 products corresponding to VAI RNA (VAI) (see Materials and methods) and U2–globin RNA (U2) are noted on the right, and the position of the probes is noted on the left. The amount of properly initiated U2-specific S1 product relative to lane 1 is shown below each lane. (C) The results of RNase protection analysis of RNA transcribed from the constructs described in (A). The U2G and CTD construct used and the addition of α-amanitin is noted above each lane. The positions of the protected products are noted on the right. The amount of properly initiated U2G-specific protection products relative to lane 1 is shown beneath each lane. A breakdown of the relative amount of correct 3′ end and readthrough (RT) is also noted below each lane. (D) The result of RNase protection analysis of U2G RNA transcribed by the CTD constructs indicated above the lanes. The positions of the protected products are noted on the right. The amount of properly initiated U2G-specific protection products relative to lane 1 is shown beneath each lane. A breakdown of the relative amount of correct 3′ end and readthrough (RT) is also noted below each lane. (E) The results of 5′ cap analysis of U2G RNA transcribed by the CTD constructs indicated above the lanes. RNase protection analysis was carried out on RNA selected by GST–eIF4E (capped) or unselected (uncapped) after addition of a positive control RNA (PC). The percentage of capped RNA is noted below the lanes.
Analysis of the 5′ end using S1 nuclease (Figure 1B) indicates that removal of the 3′ box has little effect on the steady-state level of RNA quantitated relative to VAI (compare U2G, lane 1 and Δ3′ box, lane 2). Mutation of the PSE abolishes all specific transcription, as expected (PSE–, lane 3). After incubation of the cells with α-amanitin for 48 h, no RNA is detected from the U2G construct, indicating that transcription has been inhibited effectively and that RNA turnover has occurred (lane 4). Co-transfection of the α-amanitin-resistant large subunit of pol II with either a full-length CTD (WT, lane 5) or 31 repeats (Δ31, lane 6) restores the steady-state level of RNA from the U2G construct (lane 1). However, deletion of all but the last five repeats in the CTD has a drastic effect on the steady-state level of correctly initiated RNA (Δ5, lane 7), which is reduced to 14% of the level in lane 1. Importantly, the low level of properly initiated transcription is dependent on the PSE (compare lanes 7 and 8). The intensity of the U2G probe band is increased when Δ31 (lane 6) or Δ5 (lane 7) are transfected compared with the WT CTD (lane 5). This may indicate that some transcription is initiating upstream from the natural start site or that transcription continues round the vector when the CTD is truncated. This transcription appears to be largely dependent on the PSE since it is reduced when the PSE– template is used (PSE–, lane 8).
Analysis of the 3′ ends by RNase protection (Figure 1C) demonstrates that the majority of the 3′ ends are formed correctly in the presence of the 3′ box (U2G, lane 1) although some unprocessed or ‘readthrough’ transcripts are detected as a single band due to mismatch with the riboprobe (see Figure 1A). In the absence of the 3′ box (lane 2, Δ3′ BOX), the proportion of readthrough is significantly increased in accordance with previous results (Cuello et al., 1999). The readthrough is now present as a series of bands, perhaps due to inappropriate processing or non-specific degradation. The low level of residual, correct 3′ end formation suggests that sequences in the U2G construct other than the 3′ box can also direct this process, albeit inefficiently. Complementation with WT CTD results in the same proportion of correct 3′ end to readthrough (Figure 1C, lane 5) as for the U2G construct before addition of α-amanitin (lane 1). The proportion of readthrough is slightly higher when Δ31 is used (lane 6), suggesting that recognition of the 3′ box is slightly defective. However, no RNA with the correct 3′ end is detected when most of the repeats have been deleted (Δ5, lane 7). In this case, all of the correctly initiated RNA is detected as readthrough transcripts. As was noted for the 5′ end analysis (Figure 1B), the band corresponding to protection of the U2G riboprobe from the 5′ end at –111 to the site of the 3′ mismatch is stronger in lane 7 (Δ5 and U2G) than in lane 5 (WT CTD and U2G). This suggests, again, that some transcription is starting upstream from the natural initiation site or that the polymerase is reading right around the vector. The –111 readthrough band is weaker in lane 8 (Δ5 and PSE–), suggesting a role for the PSE in this ‘aberrant’ transcription. The amount of unprocessed RNA detected as starting at +1 and reading through when the Δ5 CTD construct is transfected is similar for the 5′ S1 (14%) and RNase protection (12%) analyses (Figure 1B and C, lanes 7), indicating that most of the correctly initiated transcripts are >232 nucleotides long.
Treatment of the cells with cycloheximide for 48 h had little effect on the level of steady-state U2G RNA relative to VAI and had no effect on the relative proportion of correct 3′ end to readthrough (data not shown). Furthermore, all three CTD constructs were expressed efficiently in the transfected cells (data not shown). Our results therefore indicate that the CTD of pol II is required for high steady-state levels of U2 RNA and for correct formation of the 3′ end. In addition, pol II with a truncated CTD may be reading around the vector and back through the natural site of initiation due to failure to terminate transcription.
The results of analysing the effect of additional, intermediate deletions of the CTD (Fong and Bentley, 2001) are shown in Figure 1D. The amount of steady-state RNA is reduced when only repeats 1–25 are present, and the proportion of readthrough is increased (compare WT, lane 3, 6%, and 1–25, lane 4, 17%). Repeats 27–52 support a slightly higher steady-state level of RNA but an increased proportion of readthrough (27–52, lane 5, 34%). The level of readthrough is increased further to 84% when only repeats 1–15 are present (lane 6).
The stepwise truncation of the CTD appears to cause a progressive decrease in processing, suggesting a correlation between the efficiency of processing and the number of repeats. However, deletion of repeats 25–16 causes a more drastic loss of processing than deletion of repeats 52–26. In addition, repeats 27–52 activate processing less well than the N-terminal 25 repeats, indicating that the two halves of the CTD are not completely interchangeable.
The CTD is required for the co-transcriptional capping of mRNAs (McCracken et al., 1997a), and the effect of CTD truncation may be the result of loss of the 5′ cap. We therefore have analysed the effect of CTD truncation on capping of U2G transcripts by binding to GST–eIF4E as previously described (McCracken et al., 1997a), followed by RNase protection analysis (Figure 1E). We have used a short synthetic capped U2G RNA, which gives a 160 nucleotide protection product, as a positive control, and the uncapped VAI transcripts serve as a negative control. Transcription by endogenous pol II (lanes 1 and 2) and pol II expressed from the WT CTD construct (WT, lanes 3 and 4) results in a similar level of capping when normalized to the positive control. Capping is reduced using either the 27–52 CTD construct (27–52, lanes 5 and 6, 70%) or the 1–15 construct (1–15, lanes 7 and 8, 61%) and reduced further but not abolished when only five repeats remain (Δ5, lanes 9 and 10, 30%). These results are in agreement with the CTD requirements for mRNA capping (Fong and Bentley, 2001). Thus, 3′ processing of U2 RNA is more severely affected by loss of the CTD than capping, indicating that the effect on processing is not due entirely to loss of the 5′ cap. This does not, however, rule out any role of the cap in the processing reaction.
The –111 readthrough RNA is mainly unselected, suggesting that most of it is uncapped (see lanes 7 and 8). We have noted that this RNA is relatively unstable through the cap-binding procedure and is not always recovered quantitatively (see, for example, Δ5 in lanes 9 and 10).
The CTD is required for transcription of the endogenous U2 genes
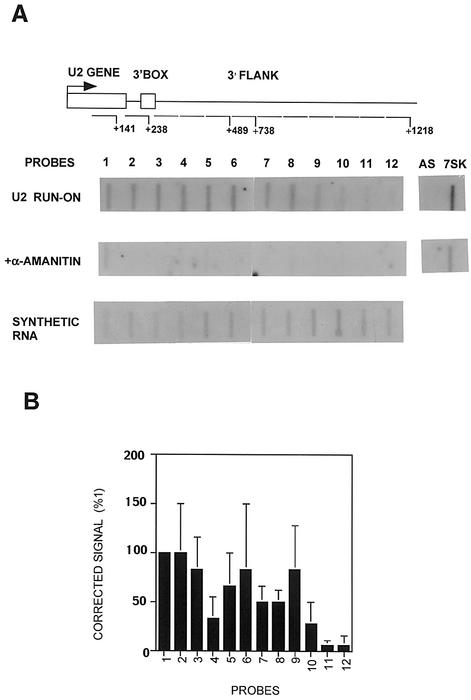
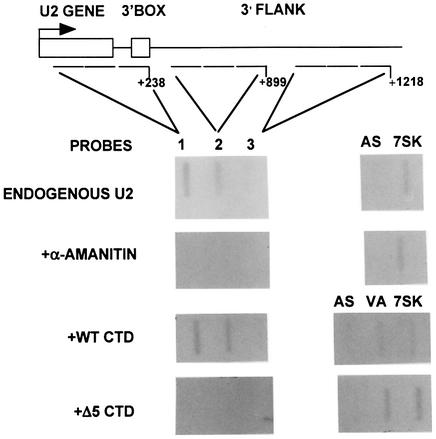
To assess the effect of CTD truncation on nascent transcription rather than steady-state levels of RNA, we have carried out nuclear run-on analysis. Analysis of nascent RNA from transfected genes is complicated by the PSE-dependent transcription across the initiation site from upstream, derived from aberrant initiation or read-around (see Figure 1). We therefore have analysed the level of nascent transcription of the endogenous U2 genes when most of the CTD is deleted. We have shown that transcription continues beyond the 3′ box of the U2 genes (Cuello et al., 1999) and we have now mapped termination to ∼1 kb downstream from the site of initiation (see Figure 5). To ensure a detectable signal in this analysis, we used sets of three oligodeoxynucleotides (oligos) in each slot corresponding to contiguous regions of 143 (oligo 1) and 240 bp (oligos 2 and 3) within the transcription unit of the U2 genes (Figure 2A). Transcription of the endogenous genes is readily detected using these U2 gene probes. Addition of α-amanitin specifically abolishes the signals over these probes, while transcription of the pol III-dependent 7SK gene is unaffected. Transfection of the WT CTD restores transcription of the U2 genes, and the profile of transcription is unchanged. However, little transcription is detected when the Δ5 construct is transfected. This suggests that the drop in specific transcription seen in the steady-state analysis of the U2G constructs reflects reduced transcription in the absence of the CTD. It also suggests that the aberrant transcription across the site of initiation detected in the steady-state analysis (see, for example, Figure 1A and C, lanes 7) is either the result of a low level of transcription or does not occur on the endogenous genes. Taken together, the results of the steady-state and nuclear run-on analyses indicate that the CTD participates in both transcription of the human U2 genes and 3′ end formation of the transcripts.
Fig. 5. Transcription of the U2 gene terminates 1 kb downstream from the site of initiation and is unaffected by CTD kinase inhibitors. (A) A diagram of the structure of the U2 gene, with the relative positions of the run-on probes marked below. The numbers noted next to the probes indicate the end of the probes used relative to the site of initiation. The results of run-on analysis, in the absence or presence of α-amanitin, and hybridization of synthetically produced RNA to the probes are shown below each probe. (B) A graphic representation of the results of the run-on analysis in (A) as a percentage of the signal over probe 1 after subtraction of α-amanitin-resistant transcription and correction for the hybridization efficiency of each probe.
Fig. 2. The CTD of pol II is required for transcription of U2 genes. A diagram of the structure of the U2 gene, with the relative positions of the probes marked below. The numbers noted next to the probes indicate the position of the probes relative to the site of initiation. The results of run-on analysis in the absence and presence of α-amanitin, and with co-transfected WT CTD or Δ5 CTD constructs, are shown below each probe. AS is a non-specific probe, and 7SK was used as an α-amanitin-resistant control in this and experiments shown in Figures 5 and 6. VA indicates that the probe is complementary to VAI transcripts.
Inhibition of CTD kinases affects recognition of the 3′ box
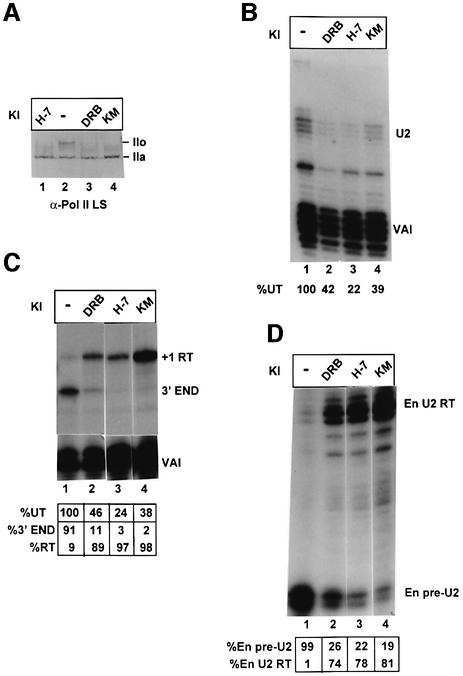
Pol II is known to exist in two alternative forms: pol II containing an unmodified form of the CTD is termed pol IIa, whereas pol II containing a hyperphosphorylated CTD is termed pol IIo. Several CDKs phosphorylate the CTD during transcription. CDK7 is a component of the pol II basal transcription factor TFIIH, CDK8 is a component of mediator that plays a role in transcriptional activation, and CDK9 is the kinase subunit of the positive transcription elongation factor, P-TEFb. Whereas CDK7 and CDK9 activate transcription (see Price, 2000), CDK8 is inhibitory (Akoulitchev et al., 2000). We have tested the effect of inhibitors of CTD kinases on transcription of U2 genes and formation of the 3′ end of the transcripts (Figure 3). HeLa cells transfected with the U2G construct (Figure 1A) were treated with 5,6-dichloro-1-β-d- ribofuranosylbenzimidazole (DRB), 1-(5-isoquino linylsulfonyl)-3-methylpiperazin (H-7) and 8-(methylthio) -4,5-dihydrothieno[3′,4′:5,6]benzoisoxazole-6-carboxamide (KM05283) (100 µM final concentration for 24 h) (see Materials and methods). All three inhibitors cause loss of hyperphosphorylation of the CTD used at this concentration in vivo (Figure 3A; Dubois et al., 1994; Lavoie et al., 2001) and inhibit CTD kinases in vitro (Serizawa et al., 1993; Mancebo et al., 1997). The steady-state level of U2 globin RNA is reduced in all cases, but is still readily detectable, and the accuracy of initiation is unaffected (Figure 3B). The effect of these inhibitors on 3′ end formation is, however, dramatic (Figure 3C). DRB causes a large proportion of transcription to read through the 3′ box, and H-7 and KM05283 cause practically complete readthrough. Since the riboprobes used have a region of complementarity to the endogenous U2 gene transcripts (see Figure 1A), we can also detect the endogenous pre-U2 (En pre-U2) and readthrough (En U2 RT) (Figure 3D). There is also a drastic change in the ratio of pre-U2 RNA to readthrough transcribed from the endogenous genes (Figure 3D).
Fig. 3. Inhibition of CTD kinases affects recognition of the 3′ box. (A) The results of western blot analysis of cells treated with 100 µM of the CTD kinase inhibitors (KI) shown above the lanes using an antibody specific for the large subunit of pol II (α-Pol II LS). The positions of the hyperphosphorylated CTD form (IIo) and the hypophosphorylated CTD form (IIa) are indicated on the right. (B) The results of S1 analysis of RNA transcribed from the U2G construct in the presence of 100 µM of the kinase inhibitors (KI) shown above the lanes. The positions of the S1 products are indicated on the right. The amount of properly initiated U2G-specific S1 product relative to the amount in untreated cells (%UT) is shown below each lane. (C) The results of RNase protection analysis of RNA transcribed from U2G in the presence of 100 µM of each kinase inhibitor (KI). The positions of the protected products are noted on the right. The amount of properly initiated U2G-specific protection products relative to the amount in untreated cells (%UT) is noted underneath each lane. A breakdown of the relative amount of correct 3′ end and readthrough (RT) is also noted below each lane. (D) The results of RNase protection analysis of endogenous U2 precursor RNA transcribed in the presence of 100 µM of each kinase inhibitor (KI). The positions of the protected products are noted on the right. A breakdown of the relative amount of correct pre-U2 3′ end and readthrough (RT) is also noted below each lane.
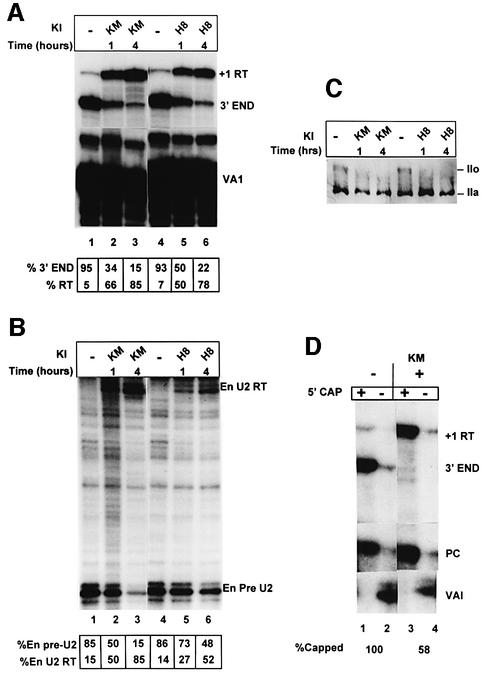
A time course was carried out using KM05283 (100 µM) and an additional inhibitor of CTD kinases, N-(2-[methylamino] ethyl)-5-isoquinolinesulfonamide hydrochloride (H-8; 200 µM) (Dubois et al., 1994) (Figure 4) to assess how rapidly the effect of these kinase inhibitors on 3′ end formation could be detected. With both inhibitors, the change in ratio of correct 3′ end to readthrough transcribed from the U2G construct is evident after 1 h (Figure 4A). The proportion of readthrough continues to increase with time, possibly due to turnover of RNA made before addition of inhibitor, and most of the RNA is detected as readthrough after 4 h treatment (lanes 3 and 6). Again, the effect is mirrored in the change in ratio of U2 precursor to readthrough transcribed from the endogenous U2 genes (Figure 4B). Both KM05283 and H-8 efficiently inhibit hyperphosphorylation of the CTD of pol II during the time course (Figure 4C).
Fig. 4. Inhibition of 3′ end processing by CTD kinase inhibitors is detectable within 1 h. (A) The results of RNase protection analysis of RNA transcribed from U2G in the presence of 100 µM of each kinase inhibitor (KI). The positions of the protected products are noted on the right. The time of incubation of cells with the inhibitors is noted above the lanes. A breakdown of the relative amount of correct pre-U2 3′ end and readthrough (RT) is also noted below each lane. (B) The results of RNase protection analysis of endogenous U2 precursor RNA transcribed in the presence of 100 µM of the kinase inhibitors (KI) shown above the lanes. The positions of the protected products are indicated on the right. The time of incubation of cells with the inhibitors is noted above the lanes. A breakdown of the relative amount of correct pre-U2 3′ end and readthrough (RT) is also noted below each lane. (C) The results of western blot analysis of cells treated with 100 µM of the CTD kinase inhibitors KM05283 and H-8 using an antibody specific for the large subunit of pol II. The positions of the hyperphosphorylated CTD form (IIo) and the hypophosphorylated CTD form (IIa) are indicated on the right. The time of incubation of cells with the inhibitors is noted above the lanes. (D) The results of 5′ cap analysis of U2G RNA from untreated and KM05832-treated cells. RNase protection analysis was carried out on RNA selected by GST–eIF4E (capped) or unselected (uncapped). The percentage of capped RNA (see Materials and methods) is noted below the lanes.
Efficient capping of mRNAs requires that the CTD is phosphorylated (McCracken et al., 1997a). It is therefore possible that inhibition of CTD kinases indirectly affects processing by inhibiting capping. To address this, we have determined the proportion of RNA selected by GST–eIF4E after treatment of cells with KM05283 for 4 h (Figure 4D). RNA selection is reduced after treatment of the cells with KM05283 (lanes 3 and 4), but a significant proportion (58%) of the, mainly readthrough, RNA is still selected and therefore capped. Identical results were obtained using RNA from H-8-treated cells (data not shown). Thus, the effect of the CTD kinase inhibitors on 3′ processing is not entirely due to inhibition of capping. Capping may be less sensitive to these drugs than 3′ processing because relatively little phosphorylation of the CTD is required to activate capping (McCracken et al., 1997a).
In an attempt to identify the CTD kinase(s) involved in the processing reaction, we have transfected constructs encoding kinase mutants of CDK7 (Makela et al., 1995), CDK8 (Akoulitchev et al., 2000) and CDK9 (Garriga et al., 1996), since CDK9 kinase mutants have been shown to have a dominant-negative effect on transcription of HIV and protein-coding genes (Mancebo et al., 1997; Kanazawa and Peterlin, 2001). Expression of these mutants had no significant effect on the ratio of readthrough to precursor of endogenous U2 gene transcripts (Supplementary data available at the EMBO Journal Online).
Transcription of the U2 genes terminates 1 kb downstream from the site of initiation and is unaffected by CTD kinase inhibitors
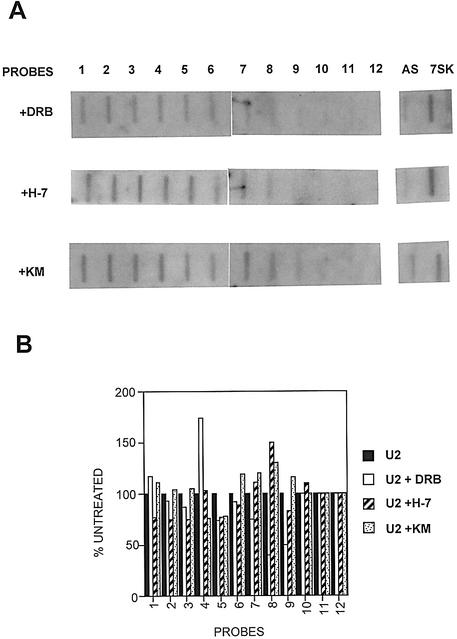
As noted above, CTD kinases play a role in initiation and elongation in transcription of protein-coding genes (see Price, 2000). We have shown that the function of the 3′ box can be linked to termination of transcription in the snRNA genes (Cuello et al., 1999). Thus, the kinase inhibitors may also affect termination of transcription. In order to test the effect of these compounds on transcription of the U2 gene, we first had to map precisely the site of termination in untreated cells. We have extended our previous run-on analysis (Cuello et al., 1999) using 47–80mer oligo probes to detect nascent transcripts (Figure 5) and have now determined that transcription continues up to ∼800 bp past the 3′ box and appears to reduce gradually rather than stop abruptly (see Figure 5B). The transcription unit of the U2 genes is, therefore, ∼1 kb in length. This is long in comparison with the much shorter transcription unit of the U1 genes (<200 bp; Cuello et al., 1999), but is much shorter than most protein-coding genes.
The transcriptional profile of the U2 genes was then analysed by nuclear run-on analysis after treating cells for 1 h with DRB, H-7 and KM05283 (Figure 6). Treatment with these kinase inhibitors has little effect on the level of transcription of the U2 genes (Figure 6A and B). In addition, and surprisingly, termination appears to be unaffected. Identical results were also obtained after treatment with kinase inhibitors for 2 h (data not shown).
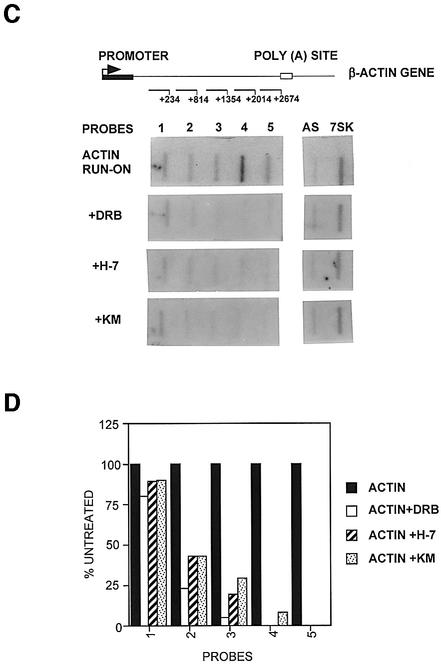
Fig. 6. Hyperphosphorylation of the CTD of pol II is not necessary for efficient transcription of the U2 gene. (A) The results of run-on analysis of the U2 genes in the presence of 100 µM CTD kinase inhibitors (indicated on the left) are shown below each probe. (B) A graphic representation of the results of the run-on analysis in (A) as a percentage of the signal over each probe relative to an untreated control (see Figure 5). (C) A diagram of the structure of the β-actin gene, with the relative positions of the probes marked below. The numbers noted next to the probes indicate the position of the end of the probes relative to the site of initiation. The results of run-on analysis in the absence or presence of 100 µM kinase inhibitors (indicated at left) are shown below each probe. (D) A graphic representation is shown of the results of the run-on analysis in (C) as a percentage of the signal over each probe relative to the untreated control.
These results indicate that inhibition of processing does not lead automatically to a loss of termination of transcription and may implicate a phosphorylation event in the coupling of the two processes.
In contrast to the U2 genes, transcription of the β-actin gene is severely affected by treatment of cells with the same concentration of inhibitors for 1 h (Figure 6C and D). In the presence of the kinase inhibitors, transcription of the β-actin gene is still quite efficient for the first 200 bp or so, indicating that initiation is occurring, and declines across the gene to very low levels after ∼1 kb.
These results suggest that hyperphosphorylation of the CTD is not required for transcription of the U2 genes. The effect of the kinase inhibitors on transcription of the β-actin gene is likely to be a direct effect of inhibition of CTD phosphorylation, although the involvement of other phosphorylated proteins in transcription of this gene cannot be ruled out.
Discussion
The results presented here demonstrate that the CTD of pol II is required for high level expression of transfected U2 snRNA gene constructs and for detectable transcription of the tandemly repeated chromosomal copies. This unusual heptad repeat structure has been shown to interact with the co-activator CRSP (Naar et al., 2002) and is required specifically for enhancer-driven transcription (Gerber et al., 1995). It is conceivable, therefore, that the CTD is required for the function of the DSE enhancer-like element located at –240 in snRNA genes. The CTD is also necessary for initiation in vitro on some TATA-less mRNA genes such as DHFR (see Akoulitchev et al., 1995), and may play a similar role in transcription of the TATA-less U2 genes. In addition, Meininghaus et al. (2000) have shown that removal of the CTD of pol II causes a global defect in transcription of mRNA genes and suggest that the CTD is necessary to access chromatin-packed templates. The failure of the CTD-less pol II to transcribe the cellular U2 genes efficiently may be due to a defect at any of these levels. We consider it likely that the effect of CTD truncation on the steady-state levels of transcripts from the transfected U2/globin constructs also reflects lower rates of initiation.
There are differences in the regulation of transcription of snRNA and protein-coding genes
Each of the 52 heptad repeats within the CTD has several potential sites of phosphorylation including serine residues at positions 2 and 5. Recent studies in living yeast cells have shown that changes in the level and position of phosphorylation of the CTD occur during the progress of the polymerase from its 5′ start site through to the 3′ end of mRNA genes (Komarnitsky et al., 2000; Schroeder et al., 2000). In mammals, pol II is recruited to protein-coding genes as a hypophosphorylated form (reviewed by Dahmus, 1996), and phosphorylation on Ser5 by CDK7 occurs at initiation (see Cho et al., 2001). Subsequent phosphorylation on both Ser2 and Ser5 by CDK9 is associated with the formation of a stable elongation complex (Kim et al., 2002). Inhibition of elongation by DRB in vitro is mediated by the DRB sensitivity-inducing factor (DSIF) comprised of the human homologues of Spt4 and Spt5 (see Price, 2000). Spt5 itself contains a motif similar to a CTD repeat and can be phosphorylated by CDK9 (see Lavoie et al., 2001). Thus, during transcription of at least some mRNA genes, the CTD of pol II undergoes a series of phosphorylation and dephosphorylation events, which may be regulated in part by phosphorylation of associated factors.
We have shown here that transcription of the β-actin mRNA gene is severely compromised by inhibitors of CTD kinases. The effect appears to be mainly on elongation, consistent with the in vitro effect of the inhibitors used on CDK9 and in agreement with previous studies showing the selective effect of DRB on transcriptional elongation of mRNA genes in vivo (reviewed by Price, 2000).
Inhibition of hyperphosphorylation of the CTD by CTD kinase inhibitors, however, does not significantly affect the level or extent of transcription of the U2 genes. This may underline a fundamental difference between snRNA genes and protein-coding genes, such as the β-actin gene (Figure 6), in the requirement for CTD hyperphosphoryl ation for production of full-length transcripts. Clearly then, the function of the CTD kinases inhibited by the drugs used is not necessary for either promoter escape or elongation in the U2 genes.
Our nuclear run-on studies have defined the transcription unit of the U2 genes as ∼1 kb, and the polymerase can therefore traverse at least this distance in the absence of CTD hyperphosphorylation. Transcription units of mRNA genes generally are much longer than this, and the polymerase may need different/additional modifications and/or associated elongation factors (e.g. P-TEFb; Price, 2000) to ensure completion of the pre-mRNA. Possibly, then, elongation of transcription of the U2 snRNA genes is ‘default’. In support of this idea, it has been shown that the human immunodeficiency virus type 1 (HIV) Tat protein can cause pol II initiated at the U2 promoter to read through the 3′ box (Ratnasabapathy et al., 1990). In transcription of HIV templates, Tat recruits CDK9 to convert the CTD to a highly phosphorylated form with enhanced processivity (Kim et al., 2002). Thus, some modifications of the CTD may both increase the elongation capacity of the polymerase and inhibit recognition of the 3′ box.
The N- and C-terminal halves of the CTD differentially activate snRNA 3′ end processing
Removal of the C-terminal half of the CTD causes a minor defect in processing the 3′ end of U2G RNA, whereas deletion of 37 repeats causes a major defect, and removal of 47 repeats abolishes detectable processing. This demonstrates that efficient processing requires at least half of the CTD. However, the two halves of the CTD do not appear to be entirely interchangeable. Repeats 27–52 are more effective in activating polyadenylation and splicing than repeats 1–25 (Fong and Bentley, 2001), whereas the reverse is the case for 3′ processing of U2G tran scripts. This suggests that different regions of the CTD are specialized for processing of pre-mRNA and pre-snRNAs.
Dephosphorylation of the CTD correlates with loss of RNA processing of U2 transcripts
CTD kinase inhibitors have a drastic effect on 3′ end processing of U2 transcripts from transfected and endogenous genes, and this cannot be attributed to an indirect effect on capping. Thus, a phospho-protein participates in recognition of the 3′ box, although the kinase responsible remains to be determined. High levels of DRB, H7, H-8 and KM05283 can inhibit both CDK7 and CDK9, in addition to other protein kinases (Serizawa et al., 1993; Dubois et al., 1994; Mancebo et al., 1997). These compounds also inhibit phosphorylation of SPT5 by CDK9 (Lavoie et al., 2001). Therefore, we cannot rule out that phosphorylated proteins additional/alternative to the phospho-CTD are required for 3′ box function. For example, 3′ end processing of a replication-activated histone mRNA is increased by phosphorylation of one of the processing factors (Dominski et al., 2002). However, the required phosphorylation event is dynamic, and there is a correlation between the loss of CTD phosphorylation and inhibition of 3′ end formation. We therefore favour the idea that phosphorylation of the CTD activates 3′ box recognition. This is an attractive hypothesis since the phosphorylated CTD is a better activator of 3′ cleavage at mammalian polyadenylation signals in vitro than the non-phosphorylated form (Hirose and Manley, 1998). Therefore, snRNA-specific processing factors may also associate directly with the phosphorylated CTD, and/or cleavage may be activated.
How could the promoter ensure recruitment of the right processing factors?
Neither the 3′ box nor a polyadenylation signal work efficiently if swapped between snRNA and protein-coding genes, highlighting the promoter dependence of 3′ end processing (see Cuello et al., 1999). The only essential element within the promoter of the U2 gene, the PSE, is recognized by PTF/SNAPc, a factor specific for transcription of snRNA genes (reviewed by Hernandez, 2001). Other basal factors include TFIIA, TFIIB, TFIIF and TFIIE that are all also required for transcription of protein-coding genes (Kuhlman et al., 1999). TATA box-binding protein (TBP) is also essential for transcription of snRNA genes (Bernues et al., 1993). However, TFIID, the complex between TBP and TBP-associated factor (TAF), cannot substitute for ‘free’ TBP for in vitro transcription from the U1 promoter (Bernues et al., 1993) and may be specific for transcription of protein-coding genes. TFIID can recruit CPSF to the CTD of pol II (Dantonel et al., 1997), affording a molecular link between promoter-bound factors and 3′ end processing. SnRNA gene-specific factors, such as PTF, are therefore good candidates for the differential recruitment of 3′ box recognition factors. However, insertion of a TATA box downstream of the PSE in the U2 promoter causes recruitment of pol III rather than pol II, and the 3′ box is no longer recognized (Lobo and Hernandez, 1989), emphasizing that PTF is not sufficient to signal 3′ end processing in the absence of pol II.
Termination of transcription of the U2 genes does not require active RNA processing
In mRNA genes, the process of transcription termination is dependent on a functional polyadenylation signal (reviewed by Proudfoot, 1989). Removal of the CTD not only affects splicing and polyadenylation but also causes loss of termination (McCracken et al., 1997b). We have shown that termination of transcription by the U1 gene terminator requires a 3′ box upstream (Cuello et al., 1999). Here we show that truncation of the CTD prevents recognition of the 3′ box and may also affect termination, causing the polymerase to read around the transfected vectors. We therefore were surprised that inhibition of 3′ end processing by the kinase inhibitors was not accompanied by a change in the profile of transcription, and in particular the site of termination. Thus, processing per se may not be necessary for termination to occur, but rather interaction of processing factors with the 3′ box somehow triggers termination downstream. Mutations in some yeast polyadenylation factors can uncouple 3′ end processing from termination (Aranda and Proudfoot, 2001; Dichtl et al., 2002), and termination can occur before 3′ end cleavage in Xenopus oocytes (Osheim et al., 1999). We can therefore envisage that inhibition of cleavage of snRNA gene transcripts may not necessarily inhibit associations required to signal downstream termination and that a phosphorylation event is necessary to couple these processes.
Materials and methods
Nuclear run-ons
Analysis was carried out as outlined in Cuello et al. (1999). For analysis of the endogenous U2 genes (Figure 5), the oligos are complementary to the U2 gene sequence from pTP18 (Pavelitz et al., 1995; DDBJ/EMBL/GenBank Accession No. U57614) taking the first base pair of the U2 coding sequence as 1: 1, 95–141; 2, 189–238; 3, 289–338; 4, 399–438; 5, 439–489; 6, 659–739; 7, 740–818; 8, 819–899; 9, 900–979; 10, 980–1057; 11, 1058–1138; 12, 1139–1218. AS is equivalent to nucleotides 1–80. The 7SK oligonucloeotide is described by Cuello et al. (1999). Synthetic RNAs were made by T7 or SP6 from sequences cloned into pGEM vectors.
For run-on analysis of endogenous genes transcribed by WT CTD or Δ5 CTD in the presence of α-amanitin, two 150 mm plates of 293 cells were transfected with 12.5 µg of CTD construct and 250 ng of VAI gene construct (see Cuello et al., 1999) using Lipofectamine (Invitrogen) according to the manufacturers instructions. α-amanitin was added to 2.5 µg/ml after 24 h, and cells were collected after a further 24 h. In this experiment, three contiguous oligos were loaded in the same slot to increase the signal. In slot 1, oligo A (Cuello et al., 1999), 1 and 2 above were loaded. In slot 2, oligos 6–8 above were loaded. In slot 3, oligos 10–12 above were loaded. The VAI oligonucleotide is complementary to bp 1–80 of the VAI coding region.
For run-on analysis of the β-actin gene (accession No. E01452) (Figure 6C), two contiguous 80mer oligos were loaded into each slot. The position of the end of the second oligo is noted on the figure.
For the experiment in Figure 6, kinase inhibitors were added to two 150 mm plates of HeLa cells to a final concentration of 100 µM. Run-on analysis was carried out after a further 1 or 2 h. H-7 and DRB were purchased from Sigma-Aldrich, and KM05283 was purchased from Maybridge.
Constructs
The U2G construct was prepared from U2 mark (Murphy, 1997) by PCR, and sequences from +28 to +169 of the U2 gene were replaced with β-globin exon 3 sequences from 151 to 7 bp upstream from the β-globin gene poly(A) site. An Acc651 site at the 5′ end and a BamHI site at the 3′ end flank the β-globin exon 3 insert. The Δ3′ box construct was made by PCR to replace the 3′ box from +19 to +41 with a C residue to create a unique MluI site. For the preparation of riboprobes, each U2G construct was amplified by PCR and recloned into the EcoRI site of pGEM4 to place the polylinker region between the T7 promoter and the 3′ end of the U2 sequence. At the same time, a mismatch with the original template was created 30 bp downstream from the expected correct 3′ end of the RNA.
Steady-state RNA analysis
A 5 µg aliquot of each U2 construct was co-transfected with 500 ng (for experiments using HeLa cells) or 15 ng (for experiments using 293 cells) of VAI using Lipofectamine as recommended by the manufacturers. To prepare total RNA after transfection, the cells were collected into phosphate-buffered saline (PBS) by scraping, centrifuged and resuspended in 100 µl of 140 mM NaCl, 1.5 mM MgCl2, 10 mM Tris pH 7.5, 1% NP-40; 100 µl of 20 mM Tris–HCl pH 7.5, 1 M NaCl, 20 mM MgCl2. Five units of DNase I were added and the mixture was incubated for 5 min at 30°C. A 100 µl aliquot of 0.4 mg/ml proteinase K, 0.4% SDS was added and the mixture was incubated at 30°C for 30 min. After H2O-equilibrated phenol/chloroform extraction and ethanol precipitation, the pellet was resuspended in 100 µl of 10 mM Tris–HCl pH 7.5, 10 mM MgCl2. Ten units of DNase I were added and the mixture was incubated for 1 h at 37°C. After H2O-equilibrated phenol/chloroform extraction and ethanol precipitation, the RNA was resuspended in water. RNase protection was carried out as outlined in Cuello et al. (1999). Riboprobes for analysis of U2G transcripts were made by T7 after digestion with Eco47III that cuts 111 bp upstream from the start of the U2 coding region. The VAI riboprobe was made by SP6 after digesting the VAI gene in pGEM4 with BamHI. S1 analysis was carried out as described by Murphy (1997) using the VAI oligo described therein and an oligo of the sequence 5′ CATTAGCCACACCAGCCACCACGGTA CCACTAGAGGATCTTAGCCAAAAGGCCGAGAAGCGATGC GCTCGCCTTC for the U2G transcripts. The nucleotides shown in bold are complementary to sequences downstream from the +1 site. The products of S1 analysis are 45 and 63 nucleotides for the VAI and U2 RNAs, respectively.
For the experiment shown in Figure 1, 5 µg of the appropriate CTD construct was co-transfected. α-amanitin was added (2.5 µg/ml final concentration) 24 h after transfection and the cells were harvested and RNA prepared after a further 48 h.
For the experiment in Figure 3, 100 µM (final concentration) of each kinase inhibitor was added to one 90 mm plate of HeLa cells 24 h after transfection. Cells were harvested and RNA prepared after a further 24 h.
For the time course shown in Figure 4, cells were incubated with kinase inhibitors (100 µM KM05283 or 200 µM H-8) for either 1 or 4 h before harvesting the cells. H-8 was from Sigma-Aldrich.
Quantitation of the results of steady-state and nuclear run-on experiments was carried out using a phosphorimager (Molecular Dynamics). Measurements were corrected against the level of expression of the VAI co-transfection control. The percentage total RNA and relative correct 3′ end:readthrough ratios were calculated by adjusting the quantitation for the number of U residues in the transcript.
Western blotting
Cells treated with kinase inhibitors were harvested, and western blotting was carried out essentially as described by Dubois et al. (1994). Anti-pol II large subunit antibody SC-9001 (Santa Cruz) was used diluted 1:500.
Capping analysis
RNA was selected by binding to GST–mouse eIF4E essentially as outlined in McCracken et al. (1997a). Bound RNA was eluted overnight at 37°C. Percentage capping was calculated by normalization to the positive control, taking the level of capping of the RNA transcribed from U2G by endogenous pol II in untreated cells as 100%.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Tori Hogan for help in making the U2G constructs, Susan McCracken for sharing preliminary data on CTD complementation experiments with U2 constructs, Andre Furger, Sasha Akoulitchev and Nick Proudfoot for helpful suggestions and for critical reading of the manuscript, J.Corden for the gift of plasmids encoding the α-amanitin-resistant pol II large subunit, J.Garriga and X.Grana for CDK9 plasmids, and Sasha Akoulitchev for CDK7 and CDK8 plasmids. J.E.M. was supported by an MRC graduate studentship. P.U. was supported by MRC Co-operative Component grant no. G9900343. A.T. was supported by MRC Co-operative Group grant no. G9826944. D.L.B. was supported by NIH grant GM58613. S.M. was supported by MRC Senior Fellowship No. G117/309.
References
- Akoulitchev S., Makela,T.P., Weinberg,R.A. and Reinberg,D. (1995) Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature, 377, 557–560. [DOI] [PubMed] [Google Scholar]
- Akoulitchev S., Chuikov,S. and Reinberg,D. (2000) TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature, 407, 102–106. [DOI] [PubMed] [Google Scholar]
- Aranda A. and Proudfoot,N.J. (2001) Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell, 7, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Bentley D. (2002) The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol., 14, 336–342. [DOI] [PubMed] [Google Scholar]
- Bernues J., Simmen,K.A., Lewis,J.D., Gunderson,S.I., Moncollin,M., Egly,J.-M. and Mattaj,I.W. (1993) Common and unique transcription factor requirements of human U1 and U6 snRNA genes. EMBO J., 12, 3573–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.J., Kobor,M.S., Kim,M., Greenblatt,J. and Buratowski,S. (2001) Opposing effects of Ctk 1 kinase and Fcp 1 phosphatase at ser2 of the RNA polymerase II C-terminal domain. Genes Dev., 15, 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello P., Boyd,D.C., Dye,M.J., Proudfoot,N.J. and Murphy,S. (1999) Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J., 18, 2867–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus M.E. (1996) Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem., 271, 19009–19012. [DOI] [PubMed] [Google Scholar]
- Dantonel J.C., Murthy,K.G.K., Manley,J.L. and Tora,L. (1997) Transcription factor TFIID recruits factor CPSF for formation of the 3′ end of mRNA. Nature, 389, 399–402. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Blank,D., Sadowski,M., Hubner,W., Weiser,S. and Keller,W. (2002) Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J., 21, 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Yang,X.C., Raska,C.S., Santiago,C., Borchers,C.H., Duronio,R.J. and Marzluff,W.F. (2002) 3′ end processing of Drosophila melanogaster histone pre-mRNAs: requirement for phosphorylated Drosophila stem–loop binding protein and coevolution of the histone pre-mRNA processing system. Mol. Cell. Biol., 22, 6648–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M.F., Nguyen,V.T., Bellier,S. and Bensaude,O. (1994) Inhibitors of transcription such as 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole and isoquinoline sulfonamide derivatives (H-8 and H-7) promote dephosphorylation of the carboxyl-terminal domain of RNA polymerase II largest subunit. J. Biol. Chem., 269, 13331–13336. [PubMed] [Google Scholar]
- Fong N. and Bentley,D.L. (2001) Capping, splicing and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev., 15, 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J., Mayol,X. and Grana,X. (1996) The CDC2-related kinase PITALRE is the catalytic subunit of active multimeric protein complexes. Biochem. J., 319, 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber H.-P., Hagmann,M., Seipel,K., Georgiev,O., West,M.A.L., Litingtung,Y., Schaffner,W. and Corden,J.L. (1995) RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature, 374, 660–662. [DOI] [PubMed] [Google Scholar]
- Hernandez N. (2001) Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem., 276, 26733–26736. [DOI] [PubMed] [Google Scholar]
- Hirose Y and Manley,J.L. (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature, 395, 93–96. [DOI] [PubMed] [Google Scholar]
- Huang Q. and Pederson,T. (1999) A human U2 RNA mutant stalled in 3′ end processing is impaired in nuclear import. Nucleic Acids Res., 27, 1025–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa S. and Peterlin,B.M. (2001) Combinations of dominant-negative class II transactivator, p300 or CDK9 proteins block the expression of MHC II genes. Int. Immunol., 13, 951–958. [DOI] [PubMed] [Google Scholar]
- Kim Y.-K., Bourgeois,C.F., Isel,C., Churcher,M.J. and Karn,J. (2002) Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immmunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol. Cell. Biol., 22, 4622–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho,E.-J. and Buratowski,S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev., 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman T.C., Cho,H., Reinberg,D. and Hernandez,N. (1999) The general transcription factors IIA, IIB, IIF and IIE are required for RNA polymerase II transcription from the human U1 snRNA promoter. Mol. Cell. Biol., 19, 2130–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie S.B., Albert,A.L., Handa,H., Vincent,M. and Bensaude,O. (2001) The peptidyl–prolyl isomerase Pin1 interacts with hSpt5 phosphorylated by Cdk9. J. Mol. Biol., 312, 675–685. [DOI] [PubMed] [Google Scholar]
- Lobo S.M. and Hernandez,N. (1989) A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell, 58, 55–67. [DOI] [PubMed] [Google Scholar]
- Makela T.P., Parvin,J.D., Kim,J., Huber,L.J., Sharp,P.A. and Weinberg,R.A. (1995) A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc. Natl Acad. Sci. USA, 92, 5174–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo H.S. et al. (1997) P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev., 11, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S. et al. (1997a) 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev., 11, 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong,N., Yankulov,K., Ballantyne,S., Pan,G.H., Greenblatt,J., Patterson,S.D., Wickens,M. and Bentley,D. (1997b) The C-terminal domain of RNA polymerase II couples messenger RNA processing to transcription. Nature, 385, 357–361. [DOI] [PubMed] [Google Scholar]
- Meininghaus M., Chapman,R.D., Horndasch,M. and Eick,D. (2000) Conditional expression of RNA polymerase II in mammalian cells. J. Biol. Chem., 275, 24375–24382. [DOI] [PubMed] [Google Scholar]
- Murphy S. (1997) Differential in vivo activation of the class II and class III snRNA genes by the POU-specific domain of Oct-1. Nucleic Acids Res., 25, 2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar A.M., Taatjes,D.J., Zhai,W., Nogales,E. and Tjian,R. (2002) Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev., 16, 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y.N., Proudfoot,N.J. and Beyer,A.L. (1999) EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell, 3, 379–387. [DOI] [PubMed] [Google Scholar]
- Pavelitz T., Rusche,L., Matera,A.G., Scharf,J.M. and Weiner,A.M. (1995) Concerted evolution of the tandem array encoding primate U2 snRNA occurs in situ, without changing the cytological context of the RNU2 locus. EMBO J., 14, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G. (2002) RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell, 1, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D. (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol., 20, 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J. (1989) How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci., 14, 105–110. [DOI] [PubMed] [Google Scholar]
- Ratnasabapathy R., Sheldon,M., Johal,L. and Hernandez,N. (1990) The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev., 4, 2061–2074. [DOI] [PubMed] [Google Scholar]
- Schroeder S.C., Schwer,B., Schuman,S. and Bentley,D. (2000) Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev., 14, 2435–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa H., Conaway,J. and Conaway,R. (1993) Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature, 363, 371–374. [DOI] [PubMed] [Google Scholar]