Abstract
One of the longest standing problems in DNA repair is how cells relax chromatin in order to make DNA lesions accessible for global nucleotide excision repair (NER). Since chromatin has to be relaxed for efficient lesion detection, the key question is whether chromatin relaxation precedes lesion detection or vice versa. Chromatin accessibility factors have been proposed but not yet identified. Here we show that p53 acts as a chromatin accessibility factor, mediating UV-induced global chromatin relaxation. Using localized subnuclear UV irradiation, we demonstrate that chromatin relaxation is extended over the whole nucleus and that this process requires p53. We show that the sequence for initiation of global NER is as follows: transcription-associated lesion detection; p53-mediated global chromatin relaxation; and global lesion detection. The tumour suppressor p53 is crucial for genomic stability, a role partially explained by its pro-apoptotic capacity. We demonstrate here that p53 is also a fundamental component of DNA repair, playing a direct role in rectifying DNA damage.
Keywords: chromatin/DNA damage/nucleotide excision repair/p300/p53
Introduction
Nucleotide excision repair (NER) is the DNA repair mechanism responsible for the removal of bulky DNA adducts produced by UV light and by a number of environmental carcinogens (Friedberg et al., 1995). Its failure leads to increased cancer incidence, as observed in primary NER deficiencies such as xeroderma pigmentosum (XP) in humans and in mice deficient for the homologues of human NER genes (for recent reviews see Benhamou and Sarasin, 2000; Friedberg, 2001; van Steeg, 2001). NER comprises two pathways: transcription-coupled repair (TCR), generally the faster of the two, which removes DNA damage in transcribed regions of the genome, and global genomic repair (GGR), which eliminates DNA damage in the rest of the chromatin (Friedberg et al., 1995). Both pathways involve a number of common steps. Upon lesion detection, DNA is melted and a repair ‘bubble’ is created by the XPB and XPD helicases; this step is followed by the excision of the damaged DNA stretch (through the XPF and XPG endonucleases cutting 5′ and 3′ of the lesion, respectively) and re-synthesis of the removed stretch (de Laat et al., 1999). It is thought that damage detection in TCR is provided by the arrest of RNA polymerase II (RNA Pol II) caused by bulky DNA lesions (Friedberg, 2001; van Steeg, 2001). In GGR, the XPA protein and the XPC–hHR23B and UV-damaged DNA binding (UV-DDB) complexes have been proposed to be responsible for lesion detection (van Steeg, 2001; Stary and Sarasin, 2002). XP complementation group C patients have impaired GGR and a high cancer propensity, which is not observed in Cockayne syndrome (a primary TCR deficiency), demonstrating the importance of the repair of lesions to the global chromatin in cancer development.
The detection of bulky DNA lesions is dependent upon ‘opening’ of the chromatin (Smerdon and Thoma, 1998; Friedberg, 2001; Green and Almouzni, 2002). In the case of TCR, chromatin accessibility appears to be granted by the transcription process (Friedberg, 2001), and lesion detection appears to be processive, with a rate equal for all types of UV lesions and limited only by the pace of RNA Pol II (Tijsterman et al., 1999). In GGR, however, the rate of removal of lesions depends on their relative accessibility (Tijsterman et al., 1999), and chromatin accessibility has to be provided to enable efficient lesion detection (Friedberg, 2001; Green and Almouzni, 2002). In fact, highly accessible lesions, generally those causing a large DNA distortion, can be removed by GGR as quickly and efficiently as TCR (see for example van Oosterwijk et al., 1996). Thus, histones are acetylated following UV irradiation (Ramanathan and Smerdon, 1986), and stabilization of acetylated histones by inhibition of histone deacetylases (HDACs) enhances the rate of excision repair (Smerdon et al., 1982). NER lesion detectors such as XPA, and the XPC and UV-DDB complexes have been proposed to render the chromatin accessible for lesion detection, i.e. to act as ‘accessibility factors’, but recent work has questioned their capacity to perform this function, and the identity of the putative accessibility factor(s) remains unknown (see Hara et al., 2000; and references therein). Moreover, when damaged naked DNA is folded into nucleosomes by addition of histones, NER is not observed even when attempted with extracts from NER-proficient cells (Wang et al., 1991). This suggests that NER-associated chromatin relaxation requires nuclear com ponents or structures more complex than those present in cell extracts. Thus, the initiation of GGR poses an unresolved paradox: detection of global lesions requires chromatin relaxation, while global chromatin relaxation appears to be triggered by damage detection. Under standing this step of GGR is important for two reasons. First, failure in the factor(s) responsible for this step in GGR potentially could facilitate tumour development, and, secondly if, as mentioned above, HDAC inhibitors can increase the efficiency of NER, then chromatin accessibility offers the potential for pharmacological modulation of DNA repair efficiency.
The p53 protein is regarded as a tumour suppressor on the basis of its capacity to induce apoptosis as well as cell cycle arrest in response to a number of cellular stresses, mainly DNA damage (Levine, 1997). In addition to this, a body of evidence indicates that p53 is required for efficient NER, and that this property plays a role in the protection against the tumorigenic effects of UV light and chemicals that form bulky DNA adducts (Hanawalt, 2001). Ford and Hanawalt (1995, 1997) have shown that p53 is required for efficient GGR of UV lesions, in particular the most frequent ones, cis–syn cyclobutane pyrimidine dimers (CPDs), with little or no effect on pyrimidine (6–4) pyrimidon photoproducts (6–4PPs), and with no effect on TCR. However, the mechanism by which p53 enhances NER is not understood. For instance, p53 has been shown to bind and modulate the activities of the NER-associated helicases XPB and XPD (Wang et al., 1995), suggesting a direct participation in NER, but in vitro NER systems have shown no requirement for p53 (Léveillard et al., 1996). Furthermore, p53 can sustain the expression of the p48 component of the UV-DDB complex (Hwang et al., 1998, 1999) and thus participates at least indirectly in NER. As a consequence, the issue of the direct versus indirect participation of p53 in NER remains unresolved.
The effects of UV light on cancer-prone XP patients (Friedberg, 2001) as well as on mice lacking certain NER genes and/or p53 (van Steeg, 2001) support the concept that p53 prevents tumour promotion at least in part by sustaining efficient NER. Interestingly, the presence of functional p53 can protect cells against UV-induced apoptosis in vitro (El-Mahdy et al., 2000; McKay et al., 2000), probably due to its capacity to sustain efficient GGR, which prevents cells from entering apoptosis. This suggests that the balance between the abilities of p53 to promote NER and to induce apoptosis is crucial in the oncogenic process.
Putting together the effects of p53 on NER reported in the literature, we noticed that they resemble the requirements of a chromatin accessibility factor. As mentioned above, p53 is required for GGR of CPDs (Ford and Hanawalt, 1995, 1997). Since these lesions can be positioned within nucleosomes (as opposed to 6–4PP, which can only be accommodated in the inter-nucleosome linker; Thoma, 1999), their repair is more dependent on chromatin relaxation. p53 has the biochemical potential for promoting chromatin relaxation, since it can recruit the histone acetyltransferase (HAT) p300 to chromatin and thus facilitate histone acetylation (Espinosa and Emerson, 2001).
In the present work, we asked whether p53 could be a chromatin accessibility factor for NER. Since p53 can transactivate NER genes (see above), we first had to determine whether p53 also has to be present at the time of NER, a crucial requirement for the model. We demonstrate that this is so by inhibiting endogenous wild-type p53 in human normal diploid fibroblasts (NDFs) by microinjection of an anti-p53 antibody, and assaying NER proficiency by unscheduled DNA synthesis (UDS). Using a chromatin relaxation assay, we demonstrate that p53 is required for UV-induced global chromatin relaxation. We further show that this requirement for p53 can be bypassed by HDAC inhibitors, which can restore NER efficiency. Importantly, we show that localized UV irradiation causes p53-dependent chromatin relaxation over the whole cell nucleus, demonstrating that p53 links local and global events in NER. Finally, we show that UV-induced chromatin relaxation is achieved by p53-mediated histone acetylation and that p53 recruits p300 to sites of NER.
These observations indicate that p53 fulfils the requirements for a chromatin accessibility factor for NER, thus solving the accessibility paradox outlined above. This new role for p53 indicates that it participates directly in the maintenance of genomic stability, in a manner that is independent from induction of apoptosis or cell cycle arrest.
Results
p53 directly participates in NER
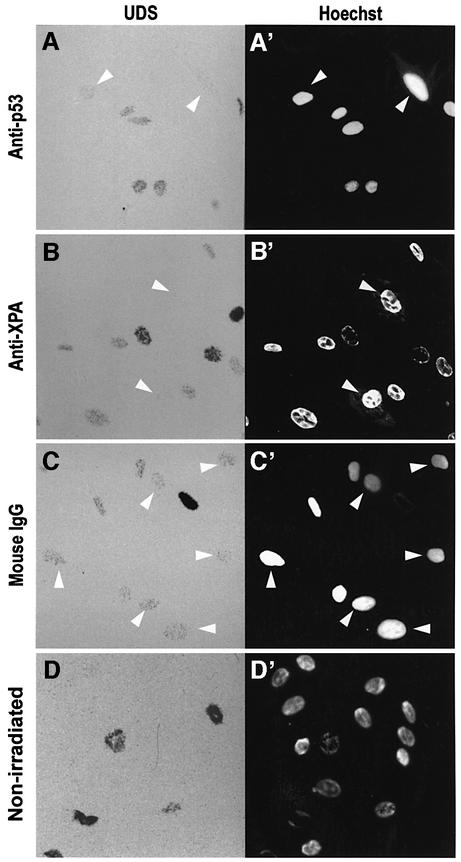
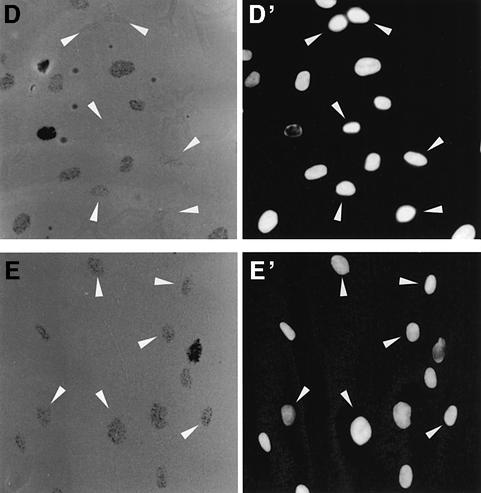
To determine whether p53 is required during NER, we blocked its availability by nuclear microinjection of an anti-p53 antibody followed by a fast NER assay such as UDS (Cleaver and Thomas, 1981). We microinjected DO-1, a mouse monoclonal antibody against the N-terminus of human p53, successfully used by others to block p53 function (Gire and Wynford-Thomas, 1998). Human NDFs were irradiated at 20 J/m2 and incubated in [3H]thymidine for 4 h. The resulting autoradiographs are shown in Figure 1A–C. The strongest [3H]thymidine incorporations correspond to S phase DNA synthesis, while intermediate densities reveal UDS. Control non-irradiated cells show only S phase incorporation of [3H]thymidine (Figure 1D and D′).
Fig. 1. Antibody blocking of p53 inhibits UDS. Human NDFs were microinjected in the nucleus with an anti-p53 monoclonal antibody (DO-1) (A), an anti-XPA antibody (B) or purified mouse IgG (C). (D) Non-irradiated control. After UV irradiation at 20 J/m2, cells were assayed for UDS. Arrows indicate cells that received the micro injections. (A′–D′) are reference images of Hoechst 33258 staining.
When p53 was blocked by nuclear microinjection of DO-1, UDS was reduced to almost undetectable levels (Figure 1A and A′, arrowed cells). The XPA protein is absolutely required for NER (de Laat et al., 1999), and cells microinjected with an anti-XPA antibody show complete inhibition of UDS, confirming that the assay specifically detects NER (Figure 1B and B′). Control cells microinjected with mouse IgG show normal UDS levels (Figure 1C and C′), demonstrating that the stress of nuclear microinjection does not affect NER. Overall, these results indicate that the presence of p53 is required for efficient NER.
The requirement for p53 in NER can be compensated by histone hyperacetylation
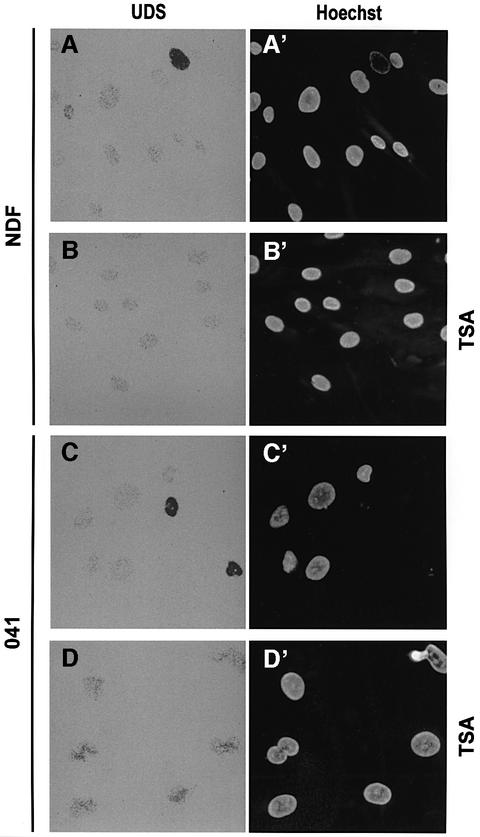
To test the hypothesis that p53 is a chromatin accessibility factor for NER, we asked whether the requirement for p53 in NER (Figure 1) can be compensated by histone hyperacetylation. In NDF controls, we show that stabili zation of acetylated histones by pre-treatment with trichostatin A (TSA, a histone deacetylase inhibitor; Marks et al., 2000) has little, if any, effect on the levels of UDS (compare Figure 2A and B).
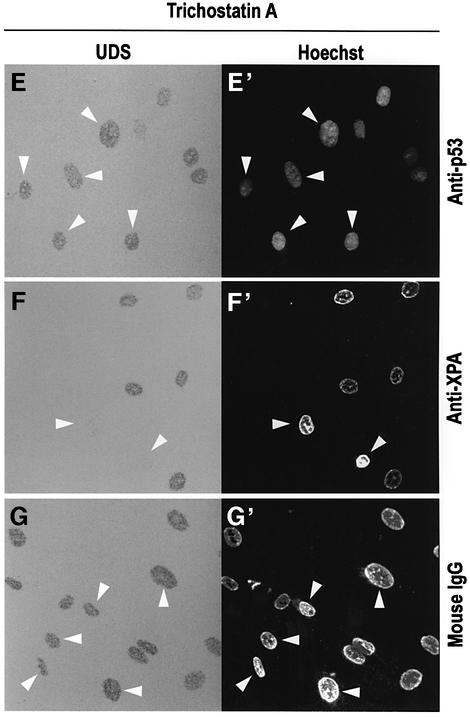
Fig. 2. Trichostatin A overcomes the effect of p53 deficiency in NER. (A–D) TSA preferentially enhances NER in p53-deficient cells. (A and B) UDS of NDF incubated for 3 h in [3H]thymidine following UV irradiation either untreated (A) or pre-incubated for 20 h with 200 ng/ml TSA (B). (C and D) The same experiment with 041 p53-deficient fibroblasts, either untreated (C) or pre-incubated with TSA (D). (E–G) Human NDFs were micro injected with anti-p53, anti-XPA or control antibodies exactly as in Figure 1, except that 200 ng/ml TSA was added to cultures 20 h prior to UV irradiation. Figure labels are as in Figure 1.
Li–Fraumeni-derived 041 fibroblasts have suffered a frameshift mutation in both p53 alleles and are therefore p53 deficient (Yin et al., 1992). This renders them deficient in GGR of CPDs (Ford and Hanawalt, 1995, 1997). TSA treatment causes a marked increase in UDS of UV-irradiated 041 cells relative to untreated cells (compare Figure 2C and D), indicating that p53 deficiency can be compensated by histone hyperacetylation.
The reduction in UDS caused by anti-p53 antibody (Figure 1) can also be compensated by histone hyperacetylation (Figure 2E). As expected, blocking of XPA, an intrinsic NER component, cannot be compensated similarly (Figure 2F). These observations support the idea that p53 can function as a chromatin accessibility factor, probably at the level of histone acetylation.
UV-induced chromatin relaxation requires p53
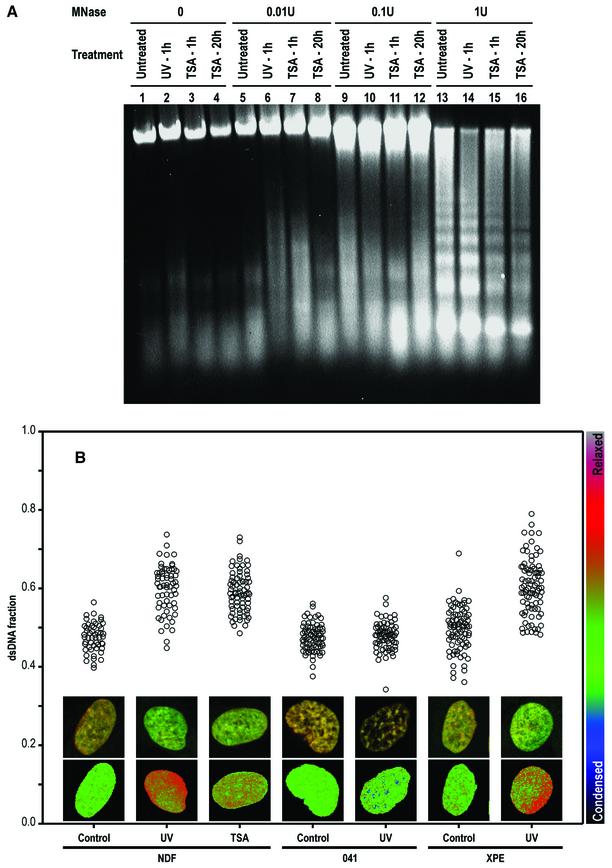
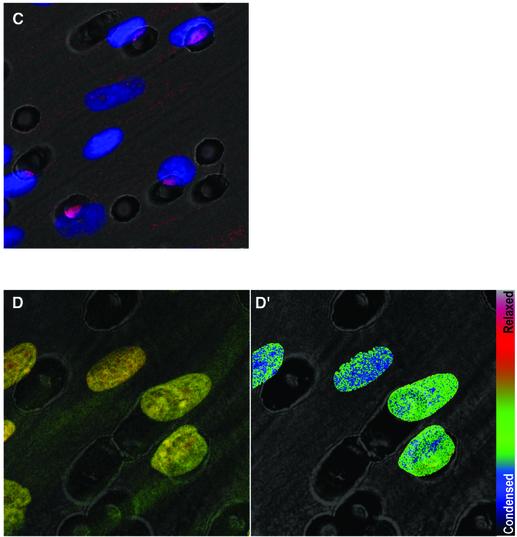
The first evidence that chromatin modifications were caused by the NER process came from the observation that histones were acetylated rapidly following UV irradiation (Ramanathan and Smerdon, 1986). Subsequently, the ‘access–repair–restore’ model was proposed and is now widely accepted (for a review see Green and Almouzni, 2002). Here, we confirmed chromatin relaxation induced by both UV irradiation and TSA, employing mild digestion with micrococcal nuclease (Figure 3A). To look directly at chromatin decondensation induced by DNA damage, we chose the partial HCl denaturation/acridine orange (HCl/AO) technique of Darzynkiewicz and co-workers (for its application to confocal microscopy, see Dobrucki and Darzynkiewicz, 2001) since it allows for single cell analysis and for microscopic study of the distribution of relaxed chromatin. This technique was applied to human NDFs either untreated, or after 1 h of treatment with UV irradiation or TSA. UV irradiation induces chromatin relaxation and, as expected, so does TSA (Figure 3B). Chromatin relaxation is shown in the image inserts of typical cells showing green and red fluorescence [double-stranded (ds)DNA, associated with relaxed chromatin, and single-stranded (ss)DNA, with condensed chromatin, respectively] and pseudo-colour images of the calculated dsDNA fraction, as well as the dsDNA fractions plotted for large numbers of cells.
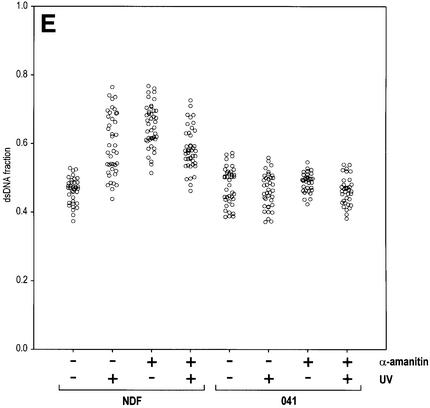
Fig. 3. UV-induced, p53-mediated global chromatin relaxation. (A) Micrococcal nuclease sensitivity as a validation of the HCl/AO assay, confirming the effect of UV irradiation and TSA on chromatin. (B) DNA denaturation sensitivity measured by the HCl/AO assay applied to human NDFs, p53-null 041 fibroblasts and XPE fibroblasts. Points indicate the extent of chromatin relaxation as the fraction of dsDNA fluorescence (green). Inserts show red/green AO images and pseudo-colour (dsDNA fraction) images. UV irradiation (4 J/m2) was assayed after 1 h, and TSA (200 ng/ml) was applied for 1 h. (C) Cells grown on Isopore filters, ‘back-irradiated’ with UV light, labelled with an anti-CPD antibody and counterstained with Hoechst 33258. (D and D′) Combination of the techniques used in (B) and (C). (E) Human NDFs and 041 fibroblasts treated as in (B), with or without the addition of 20 µg/ml α-amanitin 15 min prior to UV irradiation, and maintained throughout.
Importantly, p53-deficient 041 fibroblasts cannot undergo UV-induced chromatin relaxation (Figure 3B). This is the first demonstration that UV-induced chromatin relaxation requires p53.
Other NER proteins such as XPA, XPC and UV-DDB have been proposed as chromatin accessibility factors (see Introduction). UV-DDB has been considered a particularly strong candidate for this function, since its p48 subunit has the capacity to bind p300 (Datta et al., 2001). However, XPE fibroblasts deficient in p48 (Nichols et al., 2000) show normal UV-induced chromatin relaxation (Figure 3B), ruling out p48 as an accessibility factor. We also tested the requirements for XPA and XPC proteins, and found that XPA and XPC fibroblasts also show normal chromatin relaxation (data not shown). Thus, our data rule out UV-DDB, XPA and XPC as chromatin relaxation factors. Nevertheless, a role in chromatin remodelling (a chromatin modification different from relaxation) may still be possible. For p48, such a role is suggested by its sequence similarities to chromatin-remodelling ATPases (Hwang et al., 1998).
Local DNA damage induces global chromatin relaxation
We next addressed the long-standing problem of whether chromatin relaxation precedes global lesion detection and initiation of GGR. To do this, we studied whether a focus of localized intranuclear DNA damage would trigger chromatin relaxation over the whole cell nucleus. To introduce localized DNA damage, we modified the technique of Volker et al. (2001), based on UV irradiation of cells through filter membranes harbouring defined pores. The original technique involves laying filter discs on top of cells, irradiation and subsequent removal of the filters. This requires reference labelling to mark the localization of DNA damage. To dispense with reference labelling, incompatible with our HCl/AO protocol, we modified the technique by culturing cells directly on the filter discs and ‘back-irradiating’ with UV light. As validation, Figure 3C shows that human NDFs irradiated under such conditions exhibit DNA damage exactly coincident with the position of the filter holes (damaged DNA is labelled with an anti-CPD antibody, shown in red). This indicates that the position of a hole is a sufficient reference for the localization of the DNA damage.
By combining the above techniques, we show that localized irradiation produces global chromatin relaxation (Figure 3D and D′). This indicates that the relaxation of any particular region of global chromatin is triggered regardless of whether DNA lesions are local or distant. Since chromatin relaxation is p53 dependent (see Figure 3B), we conclude that p53 elicits relaxation throughout the nucleus and that this function can be triggered by DNA damage at a single focus within the nucleus.
Inhibition of transcription elongation as a trigger for chromatin relaxation
A potential trigger for chromatin relaxation is detection of lesions in active genes, where accessibility is provided by the transcription machinery and thus no dedicated chromatin relaxation is required. The induction of a p53 response to UV irradiation seems to be signalled by blockage of transcription elongation, which can be generated either by lesions on the DNA template or by poisoning of the RNA Pol II (Ljungman et al., 1999). Interestingly, RNA Pol II stalling at DNA lesions is believed to signal lesion detection for TCR (see Introduction; Friedberg, 2001; van Steeg, 2001). More over, we have already shown that p53 is associated with transcription sites in non-stressed cells (Rubbi and Milner, 2000). We thus reasoned that lesion-induced blockage of RNA Pol II transcription might trigger p53-dependent global chromatin relaxation. To test this hypothesis, we treated cells with α-amanitin, a specific RNA Pol II inhibitor. α-amanitin induces chromatin relaxation to levels comparable with those induced by UV irradiation (Figure 3E). Again, the effect is p53 dependent, since no relaxation was observed in p53-deficient 041 cells (Figure 3E). Note that the effects of α-amanitin and UV are not additive, in agreement with a common mechanism. In addition, we found that Cockayne syndrome fibroblasts, incapable of performing TCR, have normal induction of chromatin relaxation following UV irradiation (data not shown). This result is exactly as expected, since Cockayne syndrome cells have normal GGR and since α-amanitin treatment impairs TCR (Christians and Hanawalt, 1992); it also demonstrates that signalling for global chromatin relaxation occurs upstream of TCR and is independent of a proficient TCR machinery.
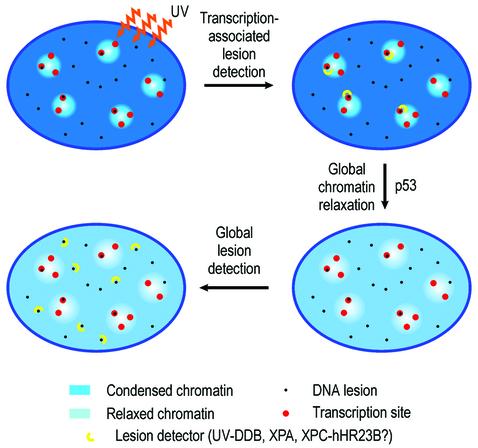
Taking the overall results of Figures 2 and 3, we conclude that p53 mediates the following pathway for global lesion accessibility: transcription-associated lesion detection (blockage of transcription elongation)→global chromatin relaxation→global lesion detection.
p53 is required for histone acetylation following UV irradiation
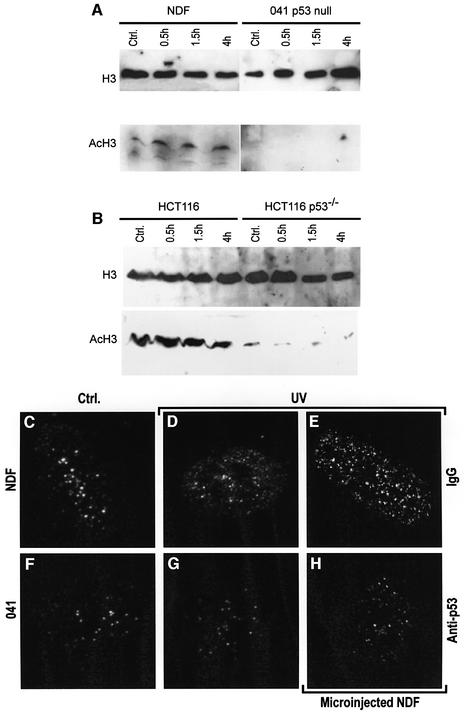
Since p53 can interact with HATs and induce chromatin relaxation, we reasoned that it could be the mediator of UV-induced histone acetylation. To test this, we followed the protocol used by Ramanathan and Smerdon (1986) to demonstrate UV-dependent induction of histone acetylation. We detected histone H3 acetylated at Lys9 (AcH3) by western blot. The requirement for wild-type p53 was studied by comparison of NDFs and 041 fibroblasts, as well as wild-type p53 and p53–/– HCT116 cells (Bunz et al., 1998). Both NDFs and HCT116 wild-type p53 cells showed a small but significant increase in AcH3 following UV irradiation (Figure 4A and B). The time profile of induction of AcH3 agreed with that reported for bulk UV-mediated histone acetylation (Ramanathan and Smerdon, 1986). In contrast, both 041 p53-deficient and p53–/– HCT116 cells showed no induction of AcH3 (Figure 4A and B). This indicates that p53 is required for UV-dependent induction of histone acetylation.
Fig. 4. p53 is required for UV-mediated histone acetylation. Human NDFs and p53-null 041 fibroblasts (A), and HCT116 human colon carcinoma cells either expressing wild-type p53 or p53–/– (B) were assayed by western blot for total histone H3 and Lys9-acetylated histone H3 (AcH3) at different times following UV irradiation at 4 J/m2. (C–H) Confocal images of cells fluorescently labelled for Lys9-acetylated histone H3. (C and D) Untreated NDFs and NDFs 4 h post-UV irradiation, respectively. (F and G) The same as (C and D) for 041 fibroblasts. NDFs microinjected in the nucleus with either purified mouse IgG (E) or an anti-p53 antibody (DO-1) (H), both UV irradiated and labelled for AcH3.
We also found that UV irradiation caused a change in the pattern of nuclear distribution of fluorescently labelled AcH3. Figure 4C and D shows confocal images of NDFs, stained for AcH3, untreated and 4 h after UV irradiation, respectively. It can be seen that the normal pattern of AcH3 (Figure 4C) consists of bright dots on a very low staining background, while irradiation (Figure 4D) changes this pattern into a more diffuse nucleoplasmic staining [image entropy analysis confirmed a significant (P = 0.01, n = 5) change in the pattern; data not shown]. As expected, p53-deficient 041 fibroblasts showed weak AcH3 fluorescence, forming a dotted pattern (Figure 4F) which did not change after UV irradiation (Figure 4G). To study whether UV-induced histone acetylation requires the presence of p53, we again used antibody blocking of endogenous p53. As seen in Figure 4H, 4 h after UV irradiation, NDFs microinjected with anti-p53 antibody had no AcH3 response, and resembled UV-irradiated 041 fibroblasts (Figure 4G). Controls microinjected with purified mouse IgG (Figure 4E) behaved as non-injected UV-irradiated NDFs (Figure 4D). These results indicate that p53 is directly involved in UV-mediated histone acetylation.
p53-mediated recruitment of p300 to sites of NER
Since p53 is not known to have intrinsic HAT activity, the model developed so far requires that a p53-related HAT (and most probably also p53) should be spatially associated with sites of NER. As a likely candidate, we chose p300, since recent work has revealed that p53 can target p300 to its consensus site in chromatin and induce histone acetylation (Espinosa and Emerson, 2001).
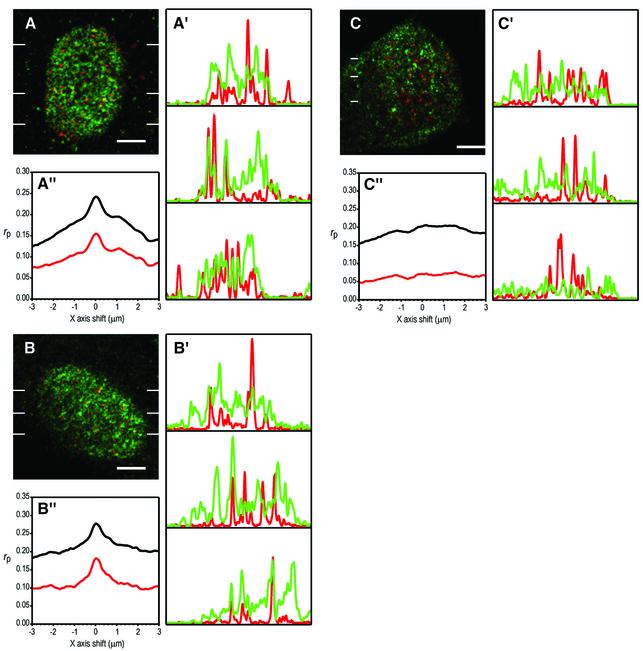
To detect sites of NER at any time after UV irradiation, we used detection of transient ssDNA stretches generated at the opening of the NER ‘bubble’ (Rubbi and Milner, 2001). Figure 5A shows a single confocal secion of a NDF nucleus stained for p53 (green) and sites of NER (red) 20 min after UV irradiation, where co-localization spots appear as yellow-orange. The co-localization of p53 and repair sites is expected to be partial since p53 may participate in other processes. For these reasons, we also monitored co-localization as line profiles (Figure 5A′), where the coincidence of intensity peaks can be observed, even for large differences in intensity. To confirm that the co-localizing yellow-orange spots immersed in an extended green signal, as well as the coincident peaks in the line profiles are statistically significant, we calculated the Pearson’s correlation coefficients at different shifts of one image with respect to the other (for equations see Rubbi and Milner, 2000). If there is no correlation between images, coefficients will oscillate around a constant background value at all shifts. If images correlate (or anti-correlate), the coefficients will depart positively (or negatively) from the background value when images are perfectly aligned. Figure 5A′′ shows Pearson’s correlation coefficients for x-axis shifts of the green image with respect to the red. The peak at 0 shift indicates that images correlate. These data indicate that p53 co-localizes with NER sites.
Fig. 5. p53 co-localizes with sites of NER, while for p300 this co-localization is p53 dependent. Single confocal sections of nuclei of human NDFs double labelled for p53 or p300 (green) and NER sites detected through transient ssDNA (red), 20 min after UV irradiation at 20 J/m2. (A and B) NDFs labelled for NER sites and for p53 and p300, respectively. (C) p53-null 041 fibroblasts labelled for NER sites and p300. All bars are 5 µm. (A′), (B′) and (C′) are line profiles of red and green images taken at the positions indicated by lines in the confocal sections. (A”), (B”) and (C”) are plots of Pearson’s correlation coefficients at a range of x-axis shifts of green images with respect to red; black and red lines correspond to plots of whole and thresholded images, respectively. (D and E) NDFs microinjected with anti-p300 purified polyclonal IgG or with non-specific IgG, respectively, and assayed for UDS as in Figure 1. (D′ and E′) Hoechst images showing the positions of nuclei. Arrows indicate injected cells.
Similarly, a clear co-localization was also found between p300 and NER sites 20 min after UV irradiation (Figure 5B, B′ and B′′). Microinjection of an anti-p53 antibody abolished the co-localization of p300 and sites of NER (data not shown). Importantly, p300 and NER sites did not co-localize in p53-deficient 041 fibroblasts (Figure 5C, C′ and C′′). From the work of Ford and Hanawalt (1995, 1997) on the NER characteristics of this cell line, we predict that the majority of the NER sites visualized in Figure 5C correspond to TCR and GGR of 6–4PPs.
To confirm the direct involvement of p300 in NER, we took advantage of the fact that p300 can be inhibited efficiently in vivo by antibody microinjection (Ait-Si-Ali et al., 2000). Figure 5D reveals that microinjection of anti-p300 causes inhibition of NER ranging from partial to complete (control microinjection of IgG in Figure 5E). This suggests that p300 is a major HAT in the NER process.
The p300 inhibition data of Figure 5D together with our data on chromatin accessibility and p53 (Figures 2–4) provide an explanation for the p53-dependent co-localization of p300 and sites of NER. Chromatin relaxed by p300 recruitment is available for lesion surveillance: if lesions are detected, NER complexes are assembled in those sites.
Discussion
Early work on the behaviour of chromatin in NER defined a strong correlation between chromatin decondensation and the onset of NER (for a review see Smerdon, 1989). However, these studies could not resolve the issue of whether chromatin decondensation is induced prior to lesion detection or whether it is provided by the NER machinery itself during or after lesion detection. The present work addresses this question. Crucially, we demonstrate UV-induced chromatin relaxation in regions where no DNA damage is present (Figure 3D and D′), i.e. upon detection of DNA damage, cells globally relax chromatin irrespective of whether the regions being relaxed actually harbour DNA lesions. Thus, the main conclusion in relation to NER is that global chromatin relaxation is prior to (and independent of) global lesion detection. Our assumption that transcription-associated lesion detection is the initiator of the process appears to be correct, since blockage of transcription elongation triggers chromatin relaxation (Figure 3E). The model is summarized in Figure 6. Interestingly, the radiomimetic drug neocarzinostatin (Povirk, 1996) did not induce chromatin relaxation (data not shown). This suggests that global chromatin relaxation is only induced by those DNA lesions that have to be repaired by NER (see below).
Fig. 6. Proposed mechanism. Chromatin relaxation events in the initiation of GGR suggested from the results in Figures 1–3. See text.
The earliest indication for a role for p53 in NER came from the observation that p53 can bind and modulate NER helicases (Wang et al., 1995), suggesting that it might be an intrinsic component of the NER machinery. However, it was shown later that p53 is not required for NER in vitro (Léveillard et al., 1996). Further detailed work by Ford and Hanawalt (1995, 1997) established that p53 is required mainly for global repair of CPDs. Thus, p53 does not appear to be required for TCR, is not necessary for the repair of naked DNA in vitro, but is required in vivo for the global repair of lesions that can be accommodated in nucleosomes (CPDs), with little effect on those which are placed in linker DNA (6–4PPs). All these characteristics led us to suspect that the role of p53 in NER could be to provide chromatin accessibility.
In Figure 2, we demonstrate that the requirement for p53 for efficient NER can be by-passed by stabilization of histone acetylation, consistent with the premise that p53 is a chromatin accessibility factor for NER. Our data from Figure 3B indicate that p53 is absolutely required for UV-induced chromatin relaxation, while the other putative global lesion detectors (XPA, XPC and UV-DDB) appear to be dispensable.
Once we identified the chromatin relaxation factor, we could explore the molecular mechanism of lesion accessibility, which would establish the model further. In Figure 4, we show that p53 plays a major part in acetylation of histone H3 and we confirm that p53 has to be present during the NER response for UV-induced histone H3 acetylation to occur. In Figure 5, we show that p53 appears to recruit p300 to global chromatin, where NER sites are to be formed. Clearly, other HATs and possibly also HDACs are likely to be involved.
Several thousands of putative p53 consensus sequences have been found in the human genome (Wang et al., 2001). In addition, recent work has revealed specific p53 binding to microsatellite sequences of limited similarity to the p53 consensus sequence (Contente et al., 2002), which may significantly increase the number of genomic p53-binding sites. It is possible that these sites may support p53-mediated HAT recruitment to chromatin following detection of DNA lesions. Even if global chromatin relaxation starts at only a limited number of sites, this may still account for the accessibility requirements of GGR, since not all of the genome appears to be equally relaxed and efficiently repaired. As Smerdon et al. (1978) demonstrated, during the first 2–3 h after UV irradiation, repair synthesis in nuclease-sensitive regions is twice as fast as in the rest of the chromatin, and this relaxed chromatin accounts for ∼30% of the genome. Importantly, DNA damage-induced binding of p53 to its consensus site does not always result in transcriptional activation, as recently demonstrated by Szak et al. (2001). We can thus speculate that a large proportion of p53-binding sites in the genome have the role of providing chromatin deconden sation origins for genome surveillance, independent of transcriptional activation. p53-binding sites could therefore act as genome ‘inspection hatches’ opened when the actively scanning transcription machinery detects DNA damage. As a chromatin accessibility factor, the tumour suppressor p53 is therefore directly involved in the protection against DNA damage. This new role is entirely independent of its ability to transactivate stress response genes or to induce apoptosis of damaged cells or their arrest in G1.
Ironically, chromatin relaxation carries increased risk of certain types of DNA damage, and a body of evidence indicates that compact chromatin is crucial for the protection against agents causing double strand DNA breaks and oxidative DNA damage. This protection is reduced following chromatin decondensation (see for example Ljungman and Hanawalt, 1992; and references therein). More specifically, UV-induced global chromatin relaxation may render DNA more susceptible to a number of DNA-damaging agents (Ljungman, 1989). Here we show that the global chromatin relaxation required to initiate GGR is tightly controlled by p53 and activated only for specific types of DNA damage (see above), demonstrating the remarkable selectivity of this process.
Upon detection of DNA damage, the cellular decision between undergoing DNA repair or committing to apoptosis is likely to be crucial in both oncogenesis and DNA damage-based anti-cancer therapy (van Steeg, 2001). In the case of bulky DNA damage, this decision appears to be particularly important, since initiation of GGR is accompanied by an increased susceptibility to other genotoxic agents, as outlined above. The identification of p53 as an accessibility factor for NER now allows for the experimental modulation of NER efficiency, and will thus be useful in understanding this decision process.
Materials and methods
Cell culture and treatment
Human NDFs (Cat. GM00038B) and XPE fibroblasts (Cat. GM01389) were obtained from Coriell Repositories (Camden, NJ) and cultured in minimal essential medium α (MEMα) + 15% FCS, as recommended. HCT116 human colon carcinoma cells expressing either wild-type p53 or p53–/– (Bunz et al., 1998; a gift from Dr Bert Vogelstein) were cultured in DMEM + 10% FCS. p53-null 041 human fibroblasts (Yin et al., 1992), were cultured in MEMα + 10% FCS. For microscopy analyses, 1–3 × 104 cells were loaded onto 13 mm diameter coverslips in 24-well plates and cultured for at least 24 h. For transient ssDNA detection, 30 µM BrdU (Sigma Chemical Co., Poole, Dorset, UK) was added 20 h later (human fibroblasts) followed by overnight release in BrdU-free medium (see Rubbi and Milner, 2001). When required, TSA (Sigma) was added at 200 ng/ml. For irradiation, cells were rinsed in PBS, exposed to a UV-C germicide tube at a fluency of 2 W/m2 for the appropriate times and immediately returned to culture medium.
Microinjection
Mouse monoclonal antibodies DO-1 and anti-XPA (NeoMarkers, Fremont, CA), rabbit anti-N-terminal fragment of human p300 (Santa Cruz, Santa Cruz, CA) and mouse and rabbit IgG (Sigma) were obtained in purified form and injected at 2 mg/ml with the addition of 1 mg/ml fluorescein isothiocyanate (FITC)–dextran 150 kDa (Sigma). Samples were brought to final concentrations by centrifugation in 10 kDa molecular weight cut-off Microcon tubes (Millipore, Watford, UK). Due to the difficulty of retaining microinjected NDFs on etched coverslips, we worked on normal HCl-cleaned glass coverslips and identified injected cells by co-injection of FITC-labelled dextran. On the day of use, Femtotips II (Eppendorf, Cambridge, UK) were scratched against the glass until a bubble pressure of ∼500 hPa in ethanol was obtained, thus ensuring sharpness and a reasonable diameter for nuclear microinjection. This was performed using an Eppendorf 5170 microinjector and 5242 micromanipulator. Coverslips with cells were placed in 6 cm Petri dishes in HBSS:MEM + 15% FCS 4:1, and returned to 24-well plates in full medium immediately after injection. Injections of antibodies were nuclear and were followed by 30–40 min incubation before irradiation.
Unscheduled DNA synthesis
After UV irradiation at 20 J/m2, cells were incubated in medium supplemented with 10 µCi/ml [3H]thymidine (Amersham Biosciences, Little Chalfont, UK) for 3 or 4 h. Next, coverslips were washed once in PBS and fixed in cold methanol for 20 min. Then, cells were dehydrated with 70, 90 and 100% ethanol, air dried and the coverslips were attached to glass slides. Slides were then coated with Hypercoat EM-1 autoradiographic emulsion (Amersham Biosciences). Following incubation at 4°C, the emulsion was developed as indicated by the manufacturer, with the addition of a final rinse with 2 µg/ml Hoechst 33258 before drying. All wide-field observations were performed using a Carl Zeiss Axiovert 135 microscope with a ×40 oil immersion Plan-NeoFluar phase contrast objective.
Chromatin relaxation
Micrococcal nuclease (Roche Diagnostics, Lewes, UK) sensitivity was assayed as described previously by Smith et al. (1998). For denaturation sensitivity (HCl/AO assay), we followed the protocol of Dobrucki and Darzynkiewicz (2001). Briefly, following UV irradiation at 4 J/m2, cells were incubated for 1 h, washed and fixed with 1% paraformaldehyde for 30 min, and then incubated in PBS, 1% BSA, 10 U/ml RNase A (Sigma) at 37°C for 1 h. Cells were then denatured for 30 s with 0.1 M HCl, stopped with 100 µg/ml acridine orange (Molecular Probes, Eugene, OR) in 0.1 M phosphate/citrate buffer pH 2.6, and mounted in the same medium, with the addition of DABCO and Mowiol (see below). Samples were scanned using 488 nm argon ion laser excitation and dual detection through HQ525/50 and HQ640LP filters (Chroma Technology Corp., Brattleboro, VT) for green (dsDNA) and red (ssDNA) fluorescence, respectively. The fraction of dsDNA was calculated as FdsDNA = G/(G + R) and displayed either numerically or as colour-coded images.
Localized irradiation
The technique for localized irradiation was adapted from Volker et al. (2001). Isopore filters 13 mm diameter, with 8 µm holes (Millipore) were glued on the edges onto 13 mm discs of UV-C-transparent 0.2 mm thick Clear Clarex plastic (Charvo, Skipton, UK), dried, sterilized with 50% ethanol and placed as inserts in 24-well culture dishes. Once cells attached to the filters, they were back-irradiated at a dose of 20 J/m2. UV-C fluency through Clear Clarex was measured separately, and irradiation times were set accordingly. Cells were fixed and processed for HCl/AO or immunofluorescence.
Western blot
HCT116 cells, both wild-type p53 and p53–/–, NDFs and 041 cells were incubated in 10 cm Petri dishes. To enhance the NER-associated AcH3 signal, replicative DNA synthesis (and thus any possible replication-associated H3 acetylation) was inhibited by adding 2 mM hydroxyurea 1 h prior to irradiation. Dishes were washed once with PBS, exposed to UV-C light at 4 J/m2, returned to culture and, at the specified times, cells were trypsinized, washed in PBS and lysed with 100 µl of SDS–PAGE sample buffer. Lysates were sonicated in order to lower the viscosity. SDS–15% polyacrylamide gels were run and proteins were electrotransferred to nitrocellulose membranes. Membranes were probed either with anti-histone H3 antibody (Santa Cruz), followed by horseradish peroxidase (HRP)-labelled secondary antibody, or with anti-Lys9-acetylated histone H3 (NeoMarkers), followed by biotinylated secondary antibody (Dako, Ely, UK) and HRP–extravidin (Sigma). Blocking and chemiluminescence solutions (Roche) were used as indicated by the manufacturer.
Staining for immunofluorescence
Cells were rinsed twice in PBS and fixed with cold methanol for 20 min followed by dipping in cold acetone and transfer to PBS-T (PBS + 0.2% Tween 20). Fixed cells were blocked in PBS-T-S (PBS-T + 10% normal serum of the same species of the secondary antibody) for 20 min. All antibodies were diluted in PBS-T-S. Antibodies and dilutions used were 1:100 for ICR1 (rat anti-BrdU; Harlan Sera-Lab, Loughborough, UK); 1:100 for DO-1 (mouse anti-p53; Oncogene, San Diego, CA); 1:100 for anti-CPD mouse monoclonal (Kamiya Biomedical, Seattle, WA); and 1:300 for rabbit anti-p300 (Santa Cruz). Rat- and rabbit-absorbed biotinylated donkey anti-mouse IgG, mouse- and rabbit-absorbed Cy3-donkey anti-rat IgG, mouse- and rat-absorbed Cy2-donkey anti-rabbit IgG anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) and FITC–rabbit anti-goat IgG (Sigma) were used diluted 1:100. Biotin was detected using streptavidin conjugated with either dichlorotriazinyl aminofluorescein (DTAF) or Cy3 (Jackson ImmunoResearch) as required. Coverslips were incubated on 20 µl drops of antibody on parafilm, for 1 h at room temperature in a humid chamber, with four 2 min washings in PBS-T. Before mounting, if required, nuclei were stained with 1 µg/ml Hoechst 33258. Samples were mounted using a Mowiol 4-88-based medium with 100 mg/ml DABCO.
Confocal microscopy and image analysis
For confocal microscopy, we used a LSM 410 system (Carl Zeiss, Welwyn Garden City, Herts, UK) equipped with Ar ion and He/Ne lasers and a ×63 1.4NA PlanApochromatic objective. Three-dimensional images were collected at a Z interval of 0.3 µm. Line profiles were obtained using built-in functions of the Carl Zeiss software. Pearson’s correlation coefficients were calculated over whole three-dimensional stacks of confocal sections spanning a whole cell nucleus, using our own software as described previously (Rubbi and Milner, 2000).
Acknowledgments
Acknowledgements
We are grateful to Dr Bert Vogelstein for providing the HCT116 p53–/– cell line, Dr Michael A.Tainsky for making available the 041 fibroblasts, Professor Alan Lehmann for providing Cockayne syndrome fibroblasts, Meg Stark and Peter Crosby for helpful advice on autoradiography, and Drs James Ford and Andrei Okorokov for discussions. This work was supported by a Yorkshire Cancer Research programme grant to J.M.
References
- Ait-Si-Ali S. et al. (2000) CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene, 19, 2430–2437. [DOI] [PubMed] [Google Scholar]
- Benhamou S. and Sarasin,A. (2000) Variability in nucleotide excision repair and cancer risk: a review. Mutat. Res., 462, 149–158. [DOI] [PubMed] [Google Scholar]
- Bunz F., Dutriaux,A., Lengauer,C., Waldman,T., Zhou,S., Brown,J.P., Sedivy,J.M., Kinzler,K.W. and Vogelstein,B. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science, 282, 1497–1501. [DOI] [PubMed] [Google Scholar]
- Christians F.C. and Hanawalt,P.C. (1992) Inhibition of transcription and strand-specific DNA repair by α-amanitin in Chinese hamster ovary cells. Mutat. Res., 274, 93–101. [DOI] [PubMed] [Google Scholar]
- Cleaver J.E. and Thomas,G.H. (1981) Measurement of unscheduled DNA synthesis by autoradiography. In Friedberg,E.C. and Hanawalt,P.C. (eds), DNA Repair: A Laboratory Manual of Research Procedures. Vol I, part B. Marcel Dekker, New York, NY, pp. 277–287.
- Contente A., Dittmer,A., Koch,M.C., Roth,J. and Dobbelstein,M. (2002) A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet., 30, 315–320. [DOI] [PubMed] [Google Scholar]
- Datta A., Bagchi,S., Nag,A., Shiyanov,P., Adami,G.R., Yoon,T. and Raychaudhuri,P. (2001) The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat. Res., 486, 89–97. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers,N.G. and Hoeijmakers,J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- Dobrucki J. and Darzynkiewicz,Z. (2001) Chromatin condensation and sensitivity of DNA in situ to denaturation during cell cycle and apoptosis—a confocal microscopy study. Micron, 32, 645–652. [DOI] [PubMed] [Google Scholar]
- El-Mahdy M.A., Hamada,F.M., Wani,M.A., Zhu,Q. and Wani,A.A. (2000) p53-degradation by HPV-16 E6 preferentially affects the removal of cyclobutane pyrimidine dimers from non-transcribed strand and sensitizes mammary epithelial cells to UV-irradiation. Mutat. Res., 459, 135–145. [DOI] [PubMed] [Google Scholar]
- Espinosa J.M. and Emerson,B.M. (2001) Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell, 8, 57–69. [DOI] [PubMed] [Google Scholar]
- Ford J.M. and Hanawalt,P.C. (1995) Li–Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc. Natl Acad. Sci. USA, 92, 8876–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J.M. and Hanawalt,P.C. (1997) Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J. Biol. Chem., 272, 28073–28080. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C. (2001) How nucleotide excision repair protects against cancer. Nat. Rev. Cancer, 1, 22–33. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Gire V. and Wynford-Thomas,D. (1998) Reinitiation of DNA synthesis and cell division in senescent human fibroblasts by microinjection of anti-p53 antibodies. Mol. Cell. Biol., 18, 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.M. and Almouzni,G. (2002) When repair meets chromatin. EMBO rep., 3, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt P.C. (2001) Controlling the efficiency of excision repair. Mutat. Res., 485, 3–13. [DOI] [PubMed] [Google Scholar]
- Hara R., Mo,J. and Sancar,A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol., 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B.J., Toering,S., Francke,U. and Chu,G. (1998) p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol., 18, 4391–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B.J., Ford,J.M., Hanawalt,P.C. and Chu,G. (1999) Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA, 96, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léveillard T., Andera,L., Bissonnette,N., Schaeffer,L., Bracco,L., Egly,J.M. and Wasylyk,B. (1996) Functional interactions between p53 and the TFIIH complex are affected by tumour-associated mutations. EMBO J., 15, 1615–1624. [PMC free article] [PubMed] [Google Scholar]
- Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- Ljungman M. (1989) Pretreatment with UV light renders the chromatin in human fibroblasts more susceptible to the DNA-damaging agents bleomycin, γ radiation and 8-methoxypsoralen. Carcinogenesis, 10, 447–451. [DOI] [PubMed] [Google Scholar]
- Ljungman M. and Hanawalt,P.C. (1992) Efficient protection against oxidative DNA damage in chromatin. Mol. Carcinog., 5, 264–269. [DOI] [PubMed] [Google Scholar]
- Ljungman M., Zhang,F., Chen,F., Rainbow,A.J. and McKay,B.C. (1999) Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene, 18, 583–592. [DOI] [PubMed] [Google Scholar]
- Marks P.A., Richon,V.M. and Rifkind,R.A. (2000) Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl Cancer. Inst., 92, 1210–1216. [DOI] [PubMed] [Google Scholar]
- McKay B.C., Chen,F., Perumalswami,C.R., Zhang,F. and Ljungman,M. (2000) The tumor suppressor p53 can both stimulate and inhibit ultraviolet light-induced apoptosis. Mol. Biol. Cell, 11, 2543–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols A.F., Itoh,T., Graham,J.A., Liu,W., Yamaizumi,M. and Linn,S. (2000) Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem., 275, 21422–21428. [DOI] [PubMed] [Google Scholar]
- Povirk L.F. (1996) DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat. Res., 355, 71–89. [DOI] [PubMed] [Google Scholar]
- Ramanathan B. and Smerdon,M.J. (1986) Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis, 7, 1087–1094. [DOI] [PubMed] [Google Scholar]
- Rubbi C.P. and Milner,J. (2000) Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene, 19, 85–96. [DOI] [PubMed] [Google Scholar]
- Rubbi C.P. and Milner,J. (2001) Analysis of nucleotide excision repair by detection of single-stranded DNA transients. Carcinogenesis, 22, 1789–1796. [DOI] [PubMed] [Google Scholar]
- Smerdon M.J. (1989) DNA excision repair at the nucleosome level of chromatin. In Lambert,M.W. and Laval,J. (eds), DNA Repair Mechanisms and their Biological Implications in Mammalian Cells. Plenum Press, New York, NY, pp. 271–294.
- Smerdon M.J. and Thoma,F. (1998) Modulations in chromatin structure during DNA damage formation and DNA repair. In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Damage and Repair. Volume II: DNA Repair in Higher Eukaryotes. Humana Press, Totowa, NJ, pp. 199–222.
- Smerdon M.J., Tlsty,T.D. and Lieberman,M.W. (1978) Distribution of ultraviolet-induced DNA repair synthesis in nuclease sensitive and resistant regions of human chromatin. Biochemistry, 17, 2377–2386. [DOI] [PubMed] [Google Scholar]
- Smerdon M.J., Lan,S.Y., Calza,R.E. and Reeves,R. (1982) Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J. Biol. Chem., 257, 13441–13447. [PubMed] [Google Scholar]
- Smith M.L., Bortnick,R.A., Sheikh,M.S. and Fornace,A.J. (1998) Chromatin relaxation by overexpression of mutant p53, HPV16-E6, or cyclin G transgenes. Exp. Cell Res., 242, 235–243. [DOI] [PubMed] [Google Scholar]
- Stary A. and Sarasin,A. (2002) The genetics of the hereditary xeroderma pigmentosum syndrome. Biochimie, 84, 49–60. [DOI] [PubMed] [Google Scholar]
- Szak S.T., Mays,D.H. and Pietenpol,J.A. (2001) Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol., 21, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F. (1999) Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J., 18, 6585–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M., de Pril,R., Tasseron-de Jong,J.G. and Brouwer,J. (1999) RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6–4) photoproduct and cyclobutane pyrimidine dimer repair in yeast. Mol. Cell. Biol., 19, 934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterwijk M.F., Filon,R., Kalle,W.H., Mullenders,L.H. and van Zeeland,A.A. (1996) The sensitivity of human fibroblasts to N-acetoxy-2-acetylaminofluorene is determined by the extent of transcription-coupled repair and/or their capability to counteract RNA synthesis inhibition. Nucleic Acids Res., 24, 4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steeg H. (2001) The role of nucleotide excision repair and loss of p53 in mutagenesis and carcinogenesis. Toxicol. Lett., 120, 209–219. [DOI] [PubMed] [Google Scholar]
- Volker M. et al. (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell, 8, 213–224. [DOI] [PubMed] [Google Scholar]
- Wang L. et al. (2001) Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J. Biol. Chem., 276, 43604–43610. [DOI] [PubMed] [Google Scholar]
- Wang X.W. et al. (1995) p53 modulation of TFIIH-associated nucleotide excision-repair activity. Nat. Genet., 10, 188–195. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Wu,X.H. and Friedberg,E.C. (1991) Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes. J. Biol. Chem., 266, 22472–22478. [PubMed] [Google Scholar]
- Yin Y., Tainsky,M.A., Bischoff,F.Z., Strong,L.C. and Wahl,G.M. (1992) Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell, 70, 937–948. [DOI] [PubMed] [Google Scholar]