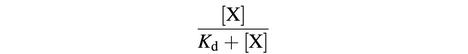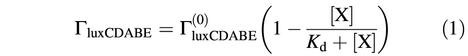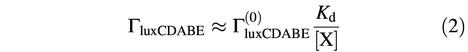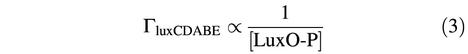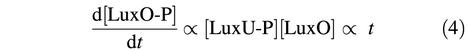Abstract
In a process called quorum sensing, bacteria communicate with one another by exchanging chemical signals called autoinducers. In the bioluminescent marine bacterium Vibrio harveyi, two different auto inducers (AI-1 and AI-2) regulate light emission. Detection of and response to the V.harveyi autoinducers are accomplished through two two-component sensory relay systems: AI-1 is detected by the sensor LuxN and AI-2 by LuxPQ. Here we further define the V.harveyi quorum-sensing regulon by identifying 10 new quorum-sensing-controlled target genes. Our examination of signal processing and integration in the V.harveyi quorum-sensing circuit suggests that AI-1 and AI-2 act synergistically, and that the V.harveyi quorum-sensing circuit may function exclusively as a ‘coincidence detector’ that discriminates between conditions in which both autoinducers are present and all other conditions.
Keywords: autoinducer/coincidence detector/quorum sensing/signal transduction
Introduction
Quorum sensing is a process that allows bacteria to communicate using secreted chemical signaling molecules called autoinducers (Nealson and Hastings, 1979; Miller and Bassler, 2001). This process enables a population of bacteria collectively to regulate gene expression and, therefore, behavior. Using quorum sensing, bacteria assess population density by detecting a particular autoinducer whose concentration is correlated with cell density (Schauder and Bassler, 2001). This ‘census-taking’ enables the group to express specific genes only at particular population densities. In general, processes controlled by quorum sensing are ones that are unproductive when undertaken by an individual bacterium but become effective when undertaken by the group. For example, quorum sensing controls bioluminescence, secretion of virulence factors, sporulation and conjugation. Thus, quorum sensing allows bacteria to act as multicellular organisms (de Kievit and Iglewski, 2000; Miller and Bassler, 2001).
Quorum sensing regulates bioluminescence (Lux) in Vibrio harveyi, a free-living Gram-negative marine bacterium. When associated with eukaryotes, V.harveyi commonly is a member of the commensal microflora; however, V.harveyi is also a potent shrimp pathogen (Alvarez et al., 1998). Most Gram-negative bacterial quorum-sensing systems are composed of a LuxI-dependent acyl homoserine lactone (HSL) signal molecule and a LuxR-type autoinducer-binding transcriptional regulator protein. In contrast, the V.harveyi quorum-sensing circuit consists of a multichannel two-component phosphorelay signal transduction pathway (Bassler et al., 1993, 1994a). This intricate circuit is proposed to facilitate intra- and interspecies cell–cell communication and to provide V.harveyi with a mechanism to monitor both the population density and species composition of the bacterial community (Bassler, 1999).
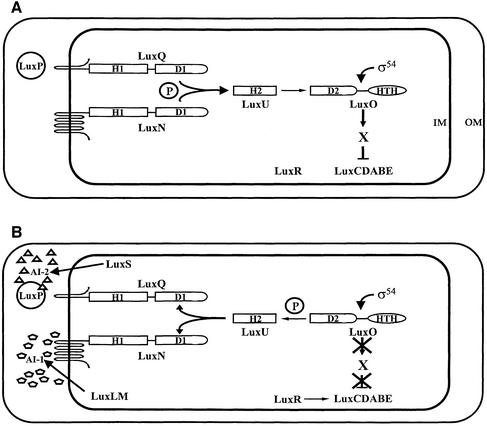
In V.harveyi, the expression of bioluminescence depends on the production and detection of two different autoinducers, AI-1 and AI-2 (Figure 1). The autoinducer synthase LuxLM produces AI-1, which is 4-hydroxyl C4 HSL (Cao and Meighen, 1989; Bassler et al., 1993). AI-2, synthesized by the LuxS enzyme, is the furanosyl borate diester 3A-methyl-5,6-dihydro-furo [2,3-D][1,3,2] dioxa borole-2,2,6,6A tetraol (Bassler et al., 1993; Surette et al., 1999; Schauder et al., 2001; Chen et al., 2002). AI-1 and AI-2 are detected via their cognate sensors LuxN and LuxPQ, respectively (Bassler et al., 1993, 1994a; Freeman et al., 2000). LuxP is homologous to the Escherichia coli periplasmic ribose-binding protein. LuxP binds AI-2 in the periplasm, and our data suggest that the LuxP–AI-2 complex interacts with LuxQ to transduce the AI-2 signal (Bassler et al., 1994a; Chen et al., 2002). LuxN and LuxQ are hybrid two-component sensor kinases, containing periplasmic sensory domains, and cytoplasmic histidine kinase and response regulator domains.
Fig. 1. The V.harveyi quorum-sensing system. The low and high cell density states of the V.harveyi quorum-sensing system are shown (A and B, respectively). The components and their putative interactions are described in the text. H, D, IM, OM and H-T-H denote histidine, aspartate, inner membrane, outer membrane and helix–turn–helix, respectively. The ‘P’ in the circle signifies that signal transduction occurs by phosphorelay. Phosphate flow in the forward direction goes from histidine (H1) to aspartate (D1) to histidine (H2) to aspartate (D2). AI-1 and AI-2 are depicted as pentagons and triangles, respectively.
At low cell density (Figure 1A), LuxN and LuxQ act as kinases and transfer phosphate to the shared phosphotransferase protein LuxU (Freeman and Bassler, 1999b). LuxU transmits the phosphate to the response regulator protein LuxO (Bassler et al., 1994b; Freeman and Bassler, 1999a). Together with the alternative sigma factor σ54, LuxO is hypothesized to activate the expression of an as yet unidentified repressor ‘X’, which negatively regulates luxCDABE (luciferase) expression, and V.harveyi does not make light (Lilley and Bassler, 2000).
At high cell density (Figure 1B), the sensors LuxN and LuxPQ detect their cognate autoinducers, AI-1 and AI-2, respectively, which convert LuxN and LuxQ from kinases to phosphatases (Freeman et al., 2000). This action reverses the flow of phosphate through the pathway, from LuxO to LuxU, and finally to LuxN and LuxQ, where the phosphoryl group is hydrolyzed. Dephosphorylation of LuxO inactivates it and terminates the expression of the repressor X. A transcriptional activator, LuxR (not similar to other LuxR-type quorum-sensing proteins) binds the luxCDABE promoter, activates transcription, and light is produced (Martin et al., 1989).
The use of two autoinducers to control quorum sensing in V.harveyi is intriguing, as one autoinducer is sufficient for density-dependent gene regulation. We suggested previously that the two autoinducers play distinct roles in V.harveyi quorum sensing. This assertion stems from two findings: (i) AI-1 is highly specific and its known production thus far is limited to V.harveyi and the closely related Vibrio species Vibrio parahaemolyticus; and (ii) AI-2 production and the AI-2 synthase, LuxS, are widespread in bacteria and to date have been shown to exist in >40 Gram-negative and Gram-positive bacterial species (Bassler et al., 1997; Surette et al., 1999; Miller and Bassler, 2001). Thus, we proposed that AI-1 and AI-2 are used by V.harveyi for intra- and interspecies communication, respectively, which allows V.harveyi to distinguish between situations when it exists predominately as a mono-culture versus situations in which it exists in a consortium (Bassler et al., 1997; Surette et al., 1999; Schauder and Bassler, 2001).
To understand further how intra- and interspecies communication is achieved in V.harveyi, in the present work, we identify genes in addition to luxCDABE that are members of the V.harveyi quorum-sensing regulon, and we determine the mechanism of their regulation. We investigate the individual roles of AI-1 and AI-2 in quorum-sensing gene regulation and show that AI-1 and AI-2 act synergistically. Because the V.harveyi quorum-sensing circuit responds to two signals, AI-1 and AI-2, there are at least four different input states: no autoinducer, AI-1 only, AI-2 only, and AI-1 + AI-2. Our measurements of light production show that the circuit has the potential to distinguish between all four states. However, under the conditions of our experiments, the V.harveyi quorum-sensing circuit appears to function primarily as a coincidence detector that differentiates between the presence of both autoinducers and all other input states.
Results
A screen for genes regulated by AI-2
We have shown that, in addition to Lux, quorum sensing regulates siderophore production and colony morphology (Lilley and Bassler, 2000). We suspected that quorum sensing controls additional behaviors and, further, that AI-1 and AI-2 could have distinct functions in cell–cell communication. To extend our definition of the quorum-sensing regulon and to investigate the precise roles of AI-1 and AI-2, we performed screens to identify genes specifically regulated by AI-1 and AI-2. Here we describe the experiments designed to identify and characterize AI-2-regulated genes; the AI-1 experiments will be reported elsewhere (Henke,J. and Bassler,B.L., in preparation).
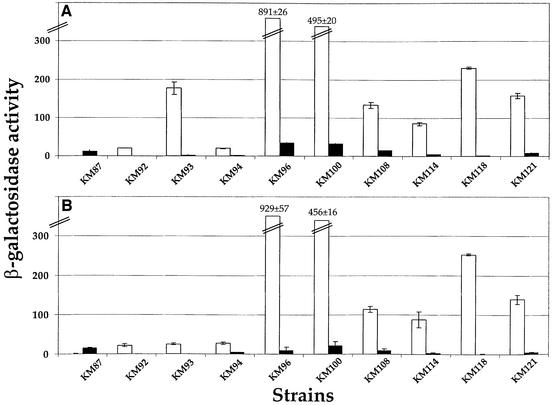
Random transposon Mini-MulacZ insertion mutagenesis was performed on the V.harveyi luxS (i.e. AI-1+, AI-2–) strain MM30. A total of 6500 insertion mutants were arrayed onto agar grids, allowed to grow into colonies and subsequently stamped to agar plate grids supplemented with 10% (v/v) cell-free culture fluids prepared from either an AI-1–, AI-2+ or an AI-1–, AI-2– V.harveyi strain. We compared the activity of the lacZ transcriptional fusions on the two sets of plates using X-gal, and identified 10 fusions that are regulated specifically by AI-2. Nine of the fusions showed reduced β-galactosidase (β-gal) activity on the Petri plates containing AI-2 compared with plates lacking AI-2.
To verify and quantify the activities of the fusions, β-gal assays were performed (Figure 2A). Prior to analysis, the strains were grown in broth containing either 10% (v/v) AI-1–, AI-2– or AI-1–, AI-2+ cell-free culture fluid (white and black bars, respectively). The fusions displayed a wide range of basal activities and extent of regulation by AI-2. Specifically, the fusion in KM87 was induced 25-fold in the presence of AI-2, and the other targets were repressed from 9- to 120-fold. To verify that the fusions were regulated by AI-2 and not by some unidentified component present in the cell-free culture fluid, the wild-type luxS gene was restored in each of the 10 insertion mutant strains. The β-gal activity of each lacZ fusion was measured in the corresponding luxS+ and luxS– strains and compared. These results (Figure 2B) show a similar pattern to that obtained by adding AI-2+ and AI-2– cell-free culture fluids.
Fig. 2. Regulation of target gene expression by AI-2. β-Gal activities for the 10 lacZ target gene fusions listed in Table I are shown. (A) Activities following addition of 10% (v/v) V.harveyi stationary phase cell-free culture fluid lacking (white bars) and containing (black bars) AI-2. (B) Activities in luxS-null (i.e. AI-2–) and luxS wild-type (i.e. AI-2+) strain backgrounds (white and black bars, respectively). β-Gal units are defined as Vmax × 0.2/[OD550 of cells × volume (ml)].
Identification of the AI-2-regulated genes
The Mini-MulacZ transposon used in the above screen contains inverted repeat sequences at its ends that interfere with PCR and preclude amplification and sequencing of the chromosome–transposon fusion junctions (Casadaban and Cohen, 1979; Metcalf et al., 1990). We therefore attempted to clone the transposon–chromosome junction from each strain, and we successfully cloned and sequenced six of the 10 fusion junctions. We have not been able to clone the other four targets so they remain unidentified. The clones are listed in Table I along with the predicted function of the genes based on database analysis. The AI-2-regulated genes encode a variety of putative functions including a secreted metalloprotease, three putative type III secretion system components and a conserved hypothetical protein. One open reading frame has no significant homology to any protein in the database. The most notable of these targets are the putative type III components as, to our knowledge, Vibrios are not known to possess such secretory systems. We have cloned additional genes encoding components of the secretory apparatus and currently are investigating their functions.
Table I. Autoinducer-regulated target genes.
| Strain | Putative function | Gene name | Accession No. | % Similarity |
|---|---|---|---|---|
| KM87 | Metalloprotease | proAC | BAA82875 | 63 |
| KM92 | Putative type III secretion proteina | NA | NA | NA |
| KM93 | No sequence homology | NA | NA | NA |
| KM94 | Type III secretion protein | pscT | AAG05080 | 82 |
| KM96 | Unknownb | |||
| KM100 | Unknownb | |||
| KM108 | Conserved hypothetical protein | VC2647c | E82049 | 87 |
| KM114 | Type III secretion protein | popB | AAC45937 | 42 |
| KM118 | Unknownb | |||
| KM121 | Unknownb |
NA = not available.
aThis gene is inferred to encode a poorly conserved type III secretion protein due to its specific location in a type III secretion operon.
bUnknown denotes that these fusions were not sequenced (see text).
cVC numbers refer to V.cholerae genome annotations.
The Lux circuit regulates the target genes
The above screen was designed to identify genes specifically controlled by AI-2. Importantly, in this experiment, we used a V.harveyi strain that was incapable of producing AI-2 but produced wild-type levels of AI-1. Therefore, this screen allowed us to identify genes whose expression remained controllable by AI-2 in the presence of high levels of AI-1. We considered two possible mechanisms that could account for this pattern of gene regulation. First, an AI-2 quorum-sensing detection–response mechanism that is distinct from, or diverges from the one we have already identified (Figure 1) could control some or all of the 10 target genes. If so, the presence of AI-1 is irrelevant for control of these genes. Secondly, the known quorum-sensing pathway (Figure 1) could control the targets. In this case, the presence of AI-1 must not cause the complete inactivation (i.e. dephosphorylation) of LuxO, or the targets could not respond to the addition of AI-2.
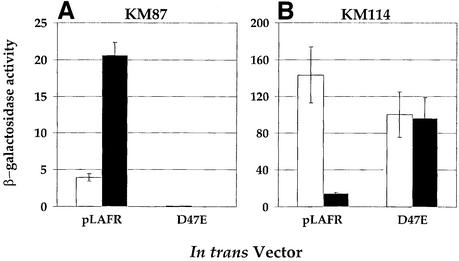
To test if the V.harveyi quorum-sensing circuit controls the targets, we introduced a dominant allele of LuxO and measured its effect on expression of the fusions. This protein, LuxO D47E, contains a missense mutation at the phosphorylation site that ‘locks’ it into a form mimicking active, LuxO-P (Freeman and Bassler, 1999a). Cells containing LuxO D47E are Lux– because the circuit is fixed in the low cell density state (Figure 1A). Additionally, the activity of LuxO D47E is not modulated by the autoinducers. If an unidentified circuit controls the 10 targets, introduction of LuxO D47E into the lacZ fusion strains should have no effect on their expression. If, on the other hand, the known quorum-sensing system controls the newly identified targets, then introduction of LuxO D47E would lock the expression of the targets into the low cell density state, and render the targets unresponsive to AI-2.
To assess whether regulation of the target fusions depends on LuxO, strains containing LuxO D47E were grown in the presence of 10% (v/v) cell-free culture fluid from an AI-1–, AI-2– strain or an AI-1–, AI-2+ strain. Subsequently, β-gal assays were performed, and Figure 3 shows the results for the AI-2 activated target gene (KM87; encoding a metalloprotease) and for one representative AI-2 repressed target gene (KM114; encoding the type III secretion PopB homolog). Introduction of LuxO D47E eliminated AI-2 regulation of the transcriptional fusions. Specifically, the left panel of Figure 3 shows that, in the presence of the vector (denoted pLAFR), lacZ transcription in strain KM87 was induced 5-fold upon addition of AI-2. However, when LuxO D47E was present (denoted D47E), transcription of lacZ was very low in both the absence and presence of AI-2. The right panel of Figure 3 shows that in the absence of AI-2, high lacZ activity was produced by strain KM114 containing the empty vector (pLAFR), and the lacZ activity decreased 10-fold when AI-2 was present. Following introduction of LuxO D47E, KM114 displayed high lacZ transcription in both the absence and presence of AI-2. The other eight negatively regulated targets behaved identically to KM114 when LuxO D47E was introduced (data not shown). We conclude that the known quorum-sensing circuit controls the fusions because each depends on LuxO for regulation. These and earlier results show that at low cell density, LuxO-P induces target genes such as those required for siderophore production (Lilley and Bassler, 2000), popB in KM114 and the eight additional targets listed in Table I. Additionally, at low cell density, LuxO-P activates expression of X that represses targets such as luxCDABE and the metalloprotease gene in KM87.
Fig. 3. The Lux signal transduction circuit controls the AI-2-regulated target genes. β-Gal activities of two representative AI-2-regulated lacZ fusions strains are shown. KM87 (A) has a fusion to a metalloprotease gene, and KM114 (B) has a fusion to a putative type III secretion popB-like gene. White and black bars denote the –AI-2 and +AI-2 conditions, respectively. AI-2 was supplied in 10% (v/v) cell-free culture fluids. pLAFR2 is the vector control, and D47E shows the result when the constitutively active luxO D47E allele was introduced on the pLAFR2 cosmid.
Both AI-1 and AI-2 control the target genes
The results of the LuxO D47E experiment were puzzling because it was not clear how the activity/phosphorylation state of LuxO could be modulated by AI-2 in the presence of high levels of AI-1. We wanted to verify that addition of AI-2 impacts LuxO activity in the presence of AI-1, and to show that the above results are not a consequence of the presence of a constitutive gain-of-function allele. Since both AI-1 and AI-2 channel information to LuxO, we reasoned that, in addition to AI-2, AI-1 should regulate the expression of the 10 targets.
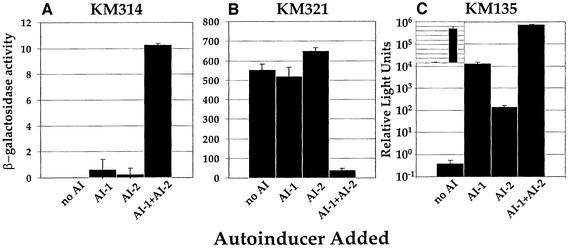
To test if AI-1 affects expression of the AI-2 regulated lacZ fusions, we needed to assay LacZ activity in V.harveyi strains that did not produce AI-1 so that we could add it exogenously. To do this, we constructed an in-frame deletion of the AI-1 synthase luxLM on the chromosome of each fusion strain. Thus, we engineered 10 strains that produced neither AI-1 nor AI-2, and each strain contained a different AI-2-regulated lacZ fusion (see Figure 2; Table I). Subsequently, we measured lacZ transcription following the addition of cell-free culture fluids containing no autoinducer, only AI-1, only AI-2, or both AI-1 and AI-2. Figure 4 shows the results for the AI-1–, AI-2– derivative of the activated fusion KM87 (this strain is called KM314, Figure 4A) and the AI-1–, AI-2– derivative of the representative repressed fusion KM114 (this strain is called KM321, Figure 4B). Again, the eight additional repressed fusions behaved identically to KM321. As expected, transcription of the fusion in KM314 was low in the absence of autoinducers and high in the presence of both autoinducers (1 and 10 U, respectively). The fusion in KM321 was regulated by the autoinducers in a reciprocal manner (550 U in the absence and 40 U in the presence of both autoinducers). However, in both cases, fluids containing only AI-1 or only AI-2 did not significantly impact expression of the lacZ fusions, showing that the autoinducers act synergistically to control gene expression.
Fig. 4. AI-1 and AI-2 act synergistically. β-Gal activities of the fusions in the luxS, luxLM derivatives of strains KM87 (KM314) and KM114 (KM321), and light production of the luxLM, luxS (AI-1–, AI-2–) strain KM135 are shown in (A), (B) and (C), respectively. The following V.harveyi cell-free culture fluids were added at 10% (v/v): MM77 (no AI), MM30 (AI-1), BB152 (AI-2), BB120 (AI-1 + AI-2). Note the logarithmic scale in (C); the inset shows the identical data plotted on a linear scale. Relative light units (RLU) are defined as c.p.m. × 103/c.f.u./ml.
The finding that the target genes are only induced or repressed when both autoinducers are present together, while striking, should be considered in light of the method used in their initial identification. As discussed earlier, the parent strain used in the screen has an AI-1+, AI-2– phenotype. The bank of insertion strains was screened for differential LacZ activity on plates with and without added AI-2. Thus, the conditions on the screening plates were AI-1+, AI-2– versus AI-1+, AI-2+. Two possible classes of target genes could be identified in this experiment: (i) genes requiring only AI-2 for regulation; and (ii) genes requiring both AI-1 and AI-2 for regulation. We only identified target genes in the latter class. This screen is not saturated, so it is possible that additional genes exist requiring both AI-1 and AI-2 for their control as well as genes requiring only AI-2 for their regulation.
In an experiment similar to those in Figure 4A and B, we measured light production in an AI-1–, AI-2– strain (KM135) following the addition of neither, one or both autoinducers. We observed (Figure 4C) <1 RLU (light production/cell) in the absence of both autoinducers, and 106 RLU when both autoinducers were added. When only AI-1 was present, 104 RLU were observed, and the addition of fluids with AI-2 alone resulted in 102 RLU. Our measurements thus distinguish four discrete levels of light output. However, these differences may not reflect distinct physiological states of V.harveyi. It is important to consider that alone, AI-1 and AI-2 stimulate only 1 and 0.01%, respectively, of the amount of light production that both autoinducers together stimulate. To depict this situation more clearly, we have plotted the Lux data on a linear scale in the inset to Figure 4C. Possibly, the low levels of light induced by AI-1 or AI-2 alone represent ‘leakage’ from a system that is designed to detect only the simultaneous presence of both autoinducers.
AI-1 and AI-2 act synergistically
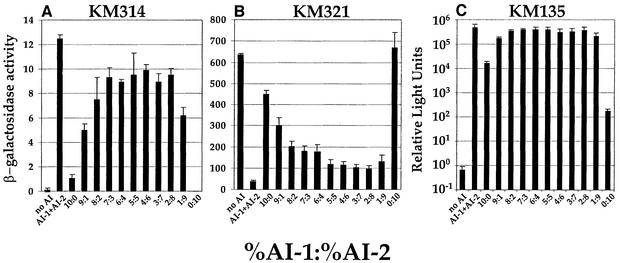
To better understand how AI-1 and AI-2 jointly regulate gene expression, we analyzed the effect of varying the levels of AI-1 and AI-2 on expression of the target genes. Our goal was to understand if the autoinducers play equivalent roles or if one autoinducer plays a major role and one a minor role in gene regulation. We varied the ratio of AI-1 to AI-2 present in the experiments by adding different mixtures of cell-free culture fluids from AI-1+, AI-2– and AI-1–, AI-2+ strains. We held constant the total amount of cell-free culture fluids supplied to the growth medium at 10% (v/v) so that we could compare these results with those in Figures 2–4. Figure 5A and B shows the results of β-gal assays for KM314 and KM321, respectively, and Figure 5C shows the luxCDABE expression data from KM135. In each panel, the two left-most bars show the controls when cell-free culture fluid containing no autoinducers (denoted no AI) and wild-type culture fluid containing both autoinducers at their native ratio (denoted AI-1 + AI-2) were added. These controls show the minimal and maximal levels of expression of the lacZ fusions and of luxCDABE. As in Figure 4, fluid with AI-1 alone or fluid with AI-2 alone did not dramatically alter gene expression, whereas every combination of the two autoinducers significantly changed transcription of the genes. In the case of the lacZ fusions, each was regulated appreciably (activated or repressed) when as little as 1% (v/v) cell-free culture fluid containing either autoinducer was added in combination with 9% (v/v) of the fluid containing the other autoinducer. In the case of luxCDABE expression, exactly as in Figure 4C, each autoinducer alone stimulated only a small amount of light production compared with both autoinducers together, although AI-1 alone had a stronger inducing effect than AI-2 alone.
Fig. 5. Non-native ratios of AI-1 and AI-2 properly regulate target gene expression. β-gal activity (KM314 and KM321) and light output (KM135) are shown, respectively, in (A), (B) and (C) (logarithmic scale), for conditions in which different mixtures of cell-free culture fluids were added at total concentrations of 10% (v/v). The AI-1-containing culture fluid was prepared from MM30, the AI-2 fluid from BB152, the AI-1 + AI-2 fluid from BB120 and the no AI fluid from MM77.
Low levels of autoinducers control gene expression
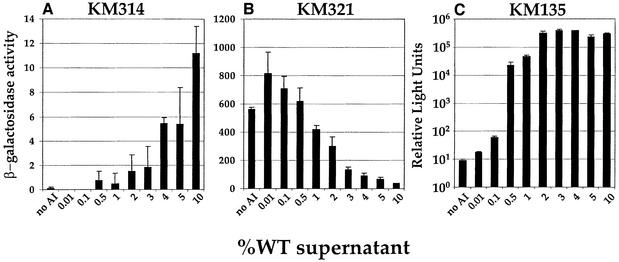
We wanted to determine how gene expression is modulated when we maintained the native ratio of AI-1:AI-2 but varied the total amount of signal present and, in so doing, to determine the amount of autoinducer required for half-maximal activation (KM314), repression (KM321) or luxCDABE expression (KM135). To do this, we added different amounts of cell-free culture fluid prepared from wild-type V.harveyi (i.e. AI-1+, AI-2+) to the various strains.
Half-maximal response was achieved when 4% (v/v) wild-type cell-free culture fluid was added to KM314 (Figure 6A), and half-maximal repression (KM321, Figure 6B) occurred following the addition of 2% (v/v) wild-type culture fluid. Less autoinducer was required for luxCDABE regulation, as significant activation occurred at 0.5% (v/v) and full activation occurred when 2% (v/v) culture fluid was added (Figure 6C). Together, the results shown in Figures 5 and 6 suggest first, that both AI-1 and AI-2 play major roles in quorum-sensing gene regulation in V.harveyi, and, secondly, that even if the amount of one signal is much lower than the other signal, or only low levels of both signals are present, the combination of the two signals is effective for quorum-sensing gene regulation.
Fig. 6. Low concentrations of the autoinducers are sufficient for gene regulation. β-Gal activity was measured for KM314 (A) and KM321 (B), and light emission for KM135 (C; logarithmic scale) following the addition of various amounts (v/v) of V.harveyi BB120 (wild-type; AI-1+, AI-2+) culture fluid.
Both AI-1 and AI-2 are required for gene regulation
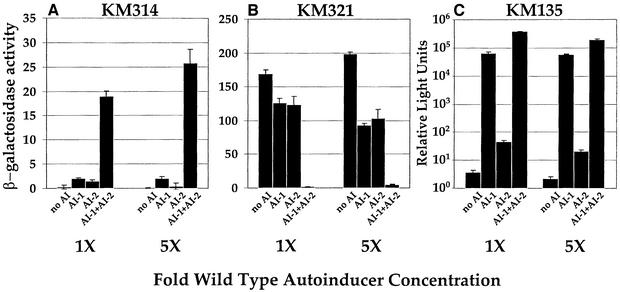
Figures 4–6 show that low levels of the autoinducers have significant effects on V.harveyi gene expression. However, in each of the above experiments, the total autoinducer added was equivalent to that present in ≤10% (v/v) V.harveyi cell-free culture fluids. Much more autoinducer could be present in 100% V.harveyi stationary phase culture fluids. We could not add 100% fluids to V.harveyi because these preparations have toxic effects, presumably from stationary phase by-products that inhibit growth. To test the effects of higher concentrations of AI-1 and AI-2 on V.harveyi gene regulation, we added AI-1 and AI-2 that we had prepared by in vitro procedures. AI-1 was prepared by the method of Cao and Meighen (1989) and AI-2 by the method of Schauder et al. (2001). AI-1 and AI-2 are estimated to be present in V.harveyi stationary phase fluids at 10–20 µM (AI-1) and 20–50 µM (AI-2). We added the autoinducers at 20 µM (1× wild-type concentration) and 100 µM (5× wild-type concentration), and monitored lacZ and luxCDABE expression (Figure 7). As observed above, individually, each autoinducer had only a minor impact on lacZ expression. Even at ∼5-fold higher concentration of autoinducer than that estimated to be present at stationary phase, neither AI-1 nor AI-2 alone dramatically modulated lacZ or luxCDABE gene transcription. Specifically, in KM314, 5-fold excess AI-1 or AI-2 had a <10% effect on the activation of lacZ transcription (Figure 7A), and in KM321, the highest concentration of autoinducer caused 50% repression of lacZ (Figure 7B). Individually, 1× or 5× AI-1 induced 33%, and 1× or 5× AI-2 induced 0.01% of the level of light production induced by the autoinducers combined (Figure 7C).
Fig. 7. High concentrations of the individual autoinducers do not regulate gene expression. (A, B and C) The β-gal activity (KM314 and KM321) and light production (KM135; logarithmic scale), respectively, in the absence and presence of AI-1 and/or AI-2 prepared by in vitro methods. 1× and 5× denote the addition of 20 and 100 µM of the specified synthetic autoinducer.
A model for Lux signal transduction
We have suggested that LuxO-P activates the expression of a putative function X, which represses (directly or indirectly) the expression of the luciferase operon luxCDABE (Figure 1). If repression of luxCDABE by X is non-cooperative, then the probability that X is bound and active as a repressor is:
where Kd is the dissociation constant for X. Thus, the rate of transcription of luxCDABE, ΓluxCDABE, is given by:
where 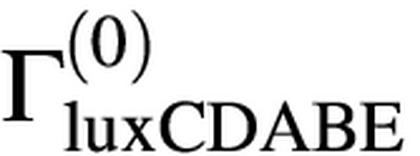 is the maximum rate of transcription of the luxCDABE operon, achieved when no repressor X is present. In the regime of strong repression, when [X]>>Kd, the rate of expression of luxCDABE is:
is the maximum rate of transcription of the luxCDABE operon, achieved when no repressor X is present. In the regime of strong repression, when [X]>>Kd, the rate of expression of luxCDABE is:
If the concentration of X is simply proportional to the concentration of LuxO-P at steady state, then luxCDABE expression is inversely proportional to the concentration of LuxO-P,
Equation 3 specifies that a large increase in light production coincides with a large decrease in the concentration of LuxO-P, which is true if cooperativity is present as well. Hence, our observation of a proportionally large increase in bioluminescence upon the sequential addition of AI-1 and AI-2 implies that the concentration of LuxO-P drops dramatically upon the addition of each autoinducer.
Discussion
Results from this and previous studies indicate that V.harveyi produces and responds to two autoinducers, AI-1 and AI-2, exclusively through the Lux circuit, depicted in Figure 1. Here we report measurements for the regulation of the expression of luxCDABE and two other genes (of 10 we identified) following the addition of a variety of different concentrations and ratios of the two autoinducers. Examination of these data gives us a means to connect the responses of V.harveyi to autoinducers with the molecular circuitry of the Lux system. Our biolumin escence measurements show that the V.harveyi quorum-sensing system can discriminate between no autoinducer, AI-1 only, AI-2 only, and AI-1 + AI-2, demonstrating that the binary information encoded in the presence or absence of one or both autoinducers is preserved by V.harveyi. Since this and previous work clearly show that all of the sensory information transmitted through the V.harveyi circuit converges on LuxO, the information supplied by the presence or absence of each autoinducer must be represented internally by four widely separated levels of LuxO-P. Hence, the observation of four distinguishable levels of bioluminescence implies that a large decrease in [LuxO-P] must occur following the addition of each autoinducer. In effect, the V.harveyi quorum-sensing system converts two binary inputs, i.e. low/high concentrations of the two autoinducers, into a four-level output, i.e. the four discrete levels of LuxO-P. This ‘digital processing’ can be accomplished if the presence of one autoinducer initiates a phosphatase activity that is much stronger than the remaining kinase activity, but not strong enough to deplete LuxO-P completely, as occurs when both autoinducers are present.
Although our bioluminescence data show that the V.harveyi quorum-sensing circuit clearly is capable of distinguishing all four test conditions, this does not necessarily imply that all four conditions are physiologically meaningful. Indeed, our results suggest that the quorum-sensing circuit could act as a coincidence detector whose function is to discriminate between the presence of both autoinducers and all other conditions. The autoinducers alone induce only 1% (AI-1) and 0.01% (AI-2) of the light production induced by the autoinducers together (Figure 4C). The responses to individual autoinducers are similar for the other target genes (Figure 4A and B).
The Lux circuit (Figure 1) has several distinguishing features that we argue could facilitate and/or enhance its operation as a coincidence detector for AI-1 and AI-2. These are: (i) the inputs from LuxN and LuxQ are combined into a single phosphotransfer pathway; (ii) both LuxN and LuxQ switch from kinase to phosphatase activity in the presence of their respective autoinducers; (iii) control of the luxCDABE operon appears to be mediated by an additional factor X, whose expression is controlled by LuxO; and (iv) the Lux pathway includes the protein LuxU, whose role is to transfer phosphate between LuxN/Q and LuxO. Here we suggest why each of these features is well suited to promote coincidence detection.
(i) Coincidence detectors function in diverse biological processes including auditory fibers in basilar membranes, neocortical pyramidal neurons, visual attention and rectifying electrical synapses (Edwards et al., 1998; Joris et al., 1998; Pena et al., 2001; Stuart and Hausser, 2001). In each case, these biological coincidence detectors integrate information from two distinct sensory detectors. In the present context, a quorum-sensing coincidence detector for AI-1 and AI-2 must combine signals emanating from two sensors. We have already demonstrated that the two autoinducer signals are detected by LuxN and LuxQ, and that this information is combined at LuxU in the Lux circuit (Bassler et al., 1993, 1994a; Freeman and Bassler, 1999b).
(ii) The LuxN and LuxQ sensors switch from kinase mode to phosphatase mode in the presence of their respective autoinducing ligands (Freeman et al., 2000). Importantly, when both autoinducers are present, LuxN and LuxQ act as phosphatases, causing dephosphorylation of LuxU-P and, at one remove, of LuxO-P. In the ‘coincidence’ state, with both autoinducers present, there is no LuxN or LuxQ kinase activity. In all other states, our results show that there is finite kinase activity, leading to an active fraction of LuxO-P. Thus, we conclude that V.harveyi’s internal representation of the presence of both autoinducers is simply the absence of LuxO-P. This design is only preferable to the alternative if the background phosphatase rate is large compared with the background kinase rate. Consistent with this idea, we have shown previously that, in a double luxN, luxQ null mutant, maximal, constitutive light production occurs (Freeman and Bassler, 1999a). Thus we know that no active LuxO-P exists under these conditions, indicating that, in the absence of the Lux sensors, no kinase exists in V.harveyi that is capable of phosphorylating LuxO. In contrast, we suggest that a scheme where LuxN and LuxQ act as kinases in the presence of the autoinducers would not produce as clear a coincidence signal, since any background phosphatase, or any intrinsic LuxU-P and/or LuxO-P decay, would lead to the presence of both LuxO and LuxO-P under all conditions.
(iii) The control of the luxCDABE operon and, by extension, other quorum-sensing targets by the putative repressor X implies a delayed temporal response to the autoinducers. According to our model, in the presence of either autoinducer, a finite amount of LuxO-P exists in the cell, and this is enough to result in production of the repressor X. Only when both autoinducers are present, is LuxO-P absent, and only then can expression of X be terminated. De-repression of luxCDABE therefore requires the presence of both autoinducers for a period of time long enough for the repressor X to degrade or to be inactivated. Thus, we suggest that a long coincidence of both autoinducers is required to stimulate the expression of bioluminescence. We therefore also expect that a temporal delay exists between the presence of both autoinducers and the expression of other target genes. Future work examining the precise timing and pattern of quorum-sensing-controlled gene expression should reveal if this is indeed the case.
(iv) Inclusion of LuxU in the circuit could serve to reduce the sensitivity to noise of this slow coincidence detection scheme. For example, a brief drop in the concentration of either autoinducer initiates the kinase activity in one sensor, leading to an increase with time of LuxU-P. The concentration of LuxU-P, starting from a negligible level, will increase linearly with time [LuxU-P ∝ t, since phosphorylation of LuxU will occur at a constant rate. However, the concentration of LuxO-P will increase more slowly with time, since the rate of phosphorylation of LuxO is itself proportional to the concentration of LuxU-P. More precisely, the concentration of LuxO-P, also starting from a negligible level, will obey:
which implies [LuxO-P] ∝ t2. This slow increase of [LuxO-P] in response to the activation of a kinase ensures that very little LuxO-P will accumulate if either kinase is briefly active. In effect, the additional step in the cascade in which phosphate is transferred through LuxU probably ‘filters out’ noise stemming from brief interruptions in the autoinducer concentrations, and thus prevents a brief drop in the concentration of one or both autoinducers from resetting the slow coincidence clock in the regulation of quorum-sensing-controlled genes.
The screen used to identify AI-2-regulated targets was performed in the presence of AI-1. We also performed an analogous screen for genes that are regulated by AI-1 in the presence of AI-2, and our results are similar to those presented here, arguing for a coincidence detector that differentiates between the simultaneous presence of both autoinducers and all other conditions (no autoinducer, AI-1 only, AI-2 only). However, Figure 4C shows that the V.harveyi quorum-sensing circuit can indeed distinguish between all four autoinducer states. It is too early to tell whether the differences in light levels visible on a log scale represent meaningful physiological states or only represent ‘leakage’ of a coincidence detector. Even if the system functions as a coincidence detector for some genes, there may be other classes of genes that are regulated differently by the four autoinducer states. For example, there could be genes that respond only to the coincidental absence of both autoinducers compared with all other conditions (i.e. AI-1 only, AI-2 only and AI-1 + AI-2). Additionally, there could be genes that respond to only the presence of one autoinducer coupled with the absence of the other autoinducer. These additional classes of genes could be identified in a lacZ reporter screen carried out in a luxLM, luxS (i.e. AI-1–, AI-2–) background. If these classes of target genes exist, we would conclude that the ability of the V.harveyi quorum-sensing circuit to distinguish between AI-1 and AI-2 (as shown in Figure 4C) is physiologically significant.
Given that V.harveyi produces both AI-1 and AI-2, what is the advantage of detecting both autoinducers? Unlike the laboratory, the natural environment may have niches in which production or accumulation of only one or the other autoinducer occurs. Independent of whether the circuit operates as a coincidence detector or by some other logic, niches with none, one or both autoinducers could be distinguished. Recent work in quorum sensing shows that production of a particular autoinducer does not necessarily coincide with its accumulation. For example, Bacillus produces an HSL hydrolase that inactivates the autoinducer of Erwinia carotovora (Dong et al., 2001). Variovorax paradoxus consumes HSLs as a source of carbon and nitrogen (Leadbetter and Greenberg, 2000). Finally, many enteric bacteria including E.coli and Salmonella typhimurium internalize AI-2 (Surette and Bassler, 1998, 1999; Taga et al., 2001). Although V.harveyi does not appear to eliminate autoinducers from the external environment, when it exists in mixed-species consortia, one or both autoinducers might be inactivated or removed. A second possibility is that the two signals serve to reduce the sensitivity of the system to noise or to ‘trickery’ by other organisms. Specifically, multisignal detection could protect the fidelity of quorum sensing from molecules that happen to be similar in structure to the autoinducers or from autoinducer mimics made by competing bacterial species or susceptible hosts.
Here we have shown that besides luxCDABE, 10 additional targets are members of the Lux regulon. It is not surprising that quorum sensing controls a large number of genes, as the transition between an individualistic existence and life as a member of a community is likely to be complex and requires alteration of many behaviors. Surprisingly, nine of the 10 genes we identified are repressed by the autoinducers. These genes probably represent a small sample of the total quorum-sensing-regulated genes, so we do not know if this pattern of regulation is significant. Among the autoinducer-repressed targets are type III secretion genes which are virulence determinants in many bacteria (Cornelis and Van Gijsegem, 2000; Lee et al., 2001). Our results suggest that, in V.harveyi, these particular virulence functions are likely to be required early in the encounter with the host prior to when V.harveyi has achieved a high cell density. This finding is consistent with recent work performed in the related species Vibrio cholerae, where we showed that the virulence regulon is expressed at low cell density and repressed by the presence of the autoinducers at high cell density (Miller et al., 2002; Zhu et al., 2002). In V.harveyi, a secreted metalloprotease is regulated in the opposite manner, i.e. it is activated at high cell density, suggesting that this virulence factor is useful in later steps in the host–bacterial pathogen interaction. This is also the case for the homologous metalloprotease in V.cholerae and other marine Vibrios (Jobling and Holmes, 1997; Shao and Hor, 2001; Zhu et al., 2002). We are currently performing shrimp infection experiments with wild-type and mutant V.harveyi strains to determine if and when quorum sensing, type III secretion and protease production are required.
We predict that many other quorum-sensing-regulated behaviors remain to be identified in V.harveyi. Experiments aimed at providing a more comprehensive definition of the Lux regulon should aid our understanding of what and how cell–cell communication influences community existence, and how communal and solitary behaviors enhance survival of V.harveyi in the different niches in which it resides.
Materials and methods
Bacterial strains and media
The strains and plasmids used in this study are listed in Table II. Vibrio harveyi was grown in HI medium at 30°C with aeration for genetic experiments, chromosomal DNA preparation and PCR analysis. β-Gal assays and luminescence assays were performed on V.harveyi strains grown in AB medium. The recipes for HI and AB have been reported previously (Bassler et al., 1994b; Freeman and Bassler, 1999a). Antibiotics were used at the following concentrations: ampicillin, 100 mg/l; kanamycin, 100 mg/l; chloramphenicol, 10 mg/l; gentamicin, 100 mg/l; tetracycline, 10 mg/l; and polymixin B, 50 mg/l.
Table II. Vibrio harveyi strains and plasmids.
| Strain | Relevant features | Source |
|---|---|---|
|
V.harveyi |
|
|
| BB120 | Wild type | Bassler et al. (1997) |
| BB152 | luxLM::Tn5 | Bassler et al. (1994a) |
| MM30 | luxS::Tn5 | Surette et al. (1999) |
| MM77 | luxLM::Tn5, luxS::CmR | This study |
| KM87 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM92 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM93 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM94 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM96 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM100 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM108 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM114 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM118 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM121 | luxS::Tn5, Mini-MulacZ CmR | This study |
| KM135 | ΔluxLM, luxS::Tn5 | This study |
| KM150 | KM87 pLAFR2 | This study |
| KM157 | KM114 pLAFR2 | This study |
| KM169 | KM108 luxS+ | This study |
| KM174 | KM87 pJAF822 | This study |
| KM181 | KM114 pJAF822 | This study |
| KM185 | KM92 luxS+ | This study |
| KM187 | KM118 luxS+ | This study |
| KM189 | KM87 luxS+ | This study |
| KM195 | KM93 luxS+ | This study |
| KM197 | KM94 luxS+ | This study |
| KM199 | KM96 luxS+ | This study |
| KM201 | KM114 luxS+ | This study |
| KM253 | KM121 luxS+ | This study |
| KM255 | KM100 luxS+ | This study |
| KM314 | KM87 ΔluxLM | This study |
| KM321 |
KM114 ΔluxLM |
This study |
| Plasmids |
|
|
| pBR322 | AmpR | Bolivar et al. (1977) |
| pLAFR2 | Broad host range; mob, TetR | Friedman et al. (1982) |
| pRK2013 | Broad host range, tra | Ditta et al. (1980) |
| pPH1JI | Broad host range, tra, mob | Beringer et al. (1978) |
| pJAF822 | pLAFR2 with luxO D47E | Freeman and Bassler (1999a) |
| pBB2929 | pLAFR2 with luxS | Surette et al. (1999) |
| pKM556 | pLAFR2 with ΔluxLM | This study |
DNA manipulations
DNA manipulations were performed as described previously in Sambrook et al. (1989). Taq DNA polymerase (Boehringer Mannheim Biochemicals) was used in PCRs. Restriction endonucleases and T4 DNA ligase came from New England Biolabs. DNA ligation reactions were transformed by electroporation into E.coli JM109 [supE Δ(lac-proAB) hsdR17 recA1 F– traD36 proAB+ lacIq lacZΔM15].
Screen for AI-2 regulated genes
Random lacZ transcriptional fusions were generated in V.harveyi strain MM30 (luxS::Tn5 KanR) using the transposon Mini-MulacZ (CmR) as described previously (Martin et al., 1989). Colonies were arrayed onto HI agar grids containing Cm, and subsequently stamped to AB, Cm plates containing 10% (v/v) cell-free culture fluid from either the V.harveyi AI-1–, AI-2+ strain BB152 (luxLM::Tn5 KanR) or from the V.harveyi AI-1–, AI-2– strain MM77 (luxLM::Tn5 KanR, luxS::CmR). Cell-free culture fluids were harvested following 14 h of growth as described previously (Bassler et al., 1993). The AB plates were supplemented with X-Gal to assay for β-gal activity (40 mg/l).
Cloning and identification of AI-2 regulated genes
Chromosomal DNA was isolated from the fusion strains as described in Murray and Thompson (1980), digested with EcoRI, and shotgun cloned into pBR322. Following electroporation into E.coli JM109, transformants were screened for β-gal activity on LB agar plates containing X-Gal. Plasmid DNA from blue colonies was isolated and digested to verify that it contained an insert of sufficient length to contain the lacZ gene and flanking V.harveyi genomic DNA. Insert DNAs were sequenced and subsequently analyzed using Sequencher 4.1 (Gene Codes) and NCBI Blast (http://www.ncbi.nlm.nih.gov/BLAST/).
Construction of V.harveyi strains
Cosmid pBB2929 was used to restore the wild-type luxS gene on the chromosome of V.harveyi strains. pBB2929 was conjugated into the luxS-null V.harveyi fusion strains (Table I), followed by allelic replacement. These methods have been described previously (Bassler et al., 1993). Gene replacement was verified by PCR and Southern blot analysis. Cosmid pJAF822 was conjugated into V.harveyi fusion strains to introduce luxO D47E. To construct AI-1–, AI-2– V.harveyi lacZ fusion strains, an in-frame deletion of luxLM was generated on the chromosome of each luxS-null strain containing a target lacZ fusion (listed in Table I). To do this, plasmid pKM556, containing the luxLM deletion adjacent to luxN in pLAFR2, was conjugated into the V.harveyi strains, and the deletion construction transferred to the chromosomes of the recipients by allelic replacement.
β-galactosidase assays
Vibrio harveyi strains were grown at 30°C with aeration for 14 h in AB medium containing autoinducers as specified. For strains with cosmids, the AB was supplemented with Tet. A 1.5 ml aliquot of culture was harvested and resuspended in 1 ml of Z buffer. β-Gal assays were performed as described previously (Slauch and Silhavy, 1991). β-gal units were calculated as (Vmax × 0.2)/[OD600 of cells × volume (ml)]. All assays were performed in triplicate.
Bioluminescence assays
The V.harveyi strains were grown 12–14 h in AB medium containing the specified concentrations of autoinducers. Bioluminescence was measured as described previously in a Wallac model 1409 liquid scintillation counter (Freeman and Bassler, 1999a). Relative light units (RLU) are defined as c.p.m. × 103/colony-forming units (c.f.u.)/ml.
Acknowledgments
Acknowledgements
We thank F.Hughson, T.Silhavy and Bassler lab members for insightful discussions. This work was supported by Office of Naval Research Grant N00014-99-1-0767, NSF grant MCB-0094447 and NIGMS grant GM65859 (B.L.B.).
References
- Alvarez J.D., Austin,B., Alvarez,A.M. and Reyes,M. (1998) Vibrio harveyi: a pathogen of penaeid shrimps and fish in Venezuela. J. Fish Dis., 21, 313–316. [DOI] [PubMed] [Google Scholar]
- Bassler B.L. (1999) How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol., 2, 582–587. [DOI] [PubMed] [Google Scholar]
- Bassler B.L., Wright,M., Showalter,R.E. and Silverman,M.R. (1993) Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol., 9, 773–786. [DOI] [PubMed] [Google Scholar]
- Bassler B.L., Wright,M. and Silverman,M.R. (1994a) Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol., 13, 273–286. [DOI] [PubMed] [Google Scholar]
- Bassler B.L., Wright,M. and Silverman,M.R. (1994b) Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol., 12, 403–412. [DOI] [PubMed] [Google Scholar]
- Bassler B.L., Greenberg,E.P. and Stevens,A.M. (1997) Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol., 179, 4043–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer J.E., Beynon,J.L., Buchanan-Wollaston,A.V. and Johnston,A.W.B. (1978) Transfer of the drug resistance transposon Tn5 to Rhizobium. Nature, 276, 633–634. [Google Scholar]
- Bolivar F., Rodriguez,R.L., Greene,P.J., Betlach,M.C., Heyneker,H.L. and Boyer,H.W. (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2, 95–113. [PubMed] [Google Scholar]
- Cao J.G. and Meighen,E.A. (1989) Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem., 264, 21670–21676. [PubMed] [Google Scholar]
- Casadaban M.J. and Cohen,S.N. (1979) Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl Acad. Sci. USA, 76, 4530–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Schauder,S., Potier,N., Van Dorsselaer,A., Pelczer,I., Bassler,B.L. and Hughson,F.M. (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature, 415, 545–549. [DOI] [PubMed] [Google Scholar]
- Cornelis G.R. and Van Gijsegem,F. (2000) Assembly and function of type III secretory systems. Annu. Rev. Microbiol., 54, 735–774. [DOI] [PubMed] [Google Scholar]
- de Kievit T.R. and Iglewski,B.H. (2000) Bacterial quorum sensing in pathogenic relationships. Infect. Immun., 68, 4839–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield,S., Corbin,D. and Helinski,D.R. (1980) Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl Acad. Sci. USA, 77, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.H., Wang,L.H., Xu,J.L., Zhang,H.B., Zhang,X.F. and Zhang,L.H. (2001) Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature, 411, 813–817. [DOI] [PubMed] [Google Scholar]
- Edwards D.H., Yeh,S.R. and Krasne,F.B. (1998) Neuronal coincidence detection by voltage-sensitive electrical synapses. Proc. Natl Acad. Sci. USA, 95, 7145–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.A. and Bassler,B.L. (1999a) A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol., 31, 665–677. [DOI] [PubMed] [Google Scholar]
- Freeman J.A. and Bassler,B.L. (1999b) Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol., 181, 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.A., Lilley,B.N. and Bassler,B.L. (2000) A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol. Microbiol., 35, 139–149. [DOI] [PubMed] [Google Scholar]
- Friedman A.M., Long,S.R., Brown,S.E., Buikema,W.J. and Ausubel,F.M. (1982) Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene, 18, 289–296. [DOI] [PubMed] [Google Scholar]
- Jobling M.G. and Holmes,R.K. (1997) Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol., 26, 1023–1034. [DOI] [PubMed] [Google Scholar]
- Joris P.X., Smith,P.H. and Yin,T.C. (1998) Coincidence detection in the auditory system: 50 years after Jeffress. Neuron, 21, 1235–1238. [DOI] [PubMed] [Google Scholar]
- Leadbetter J.R. and Greenberg,E.P. (2000) Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol., 182, 6921–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Butler,S.M. and Camilli,A. (2001) Selection for in vivo regulators of bacterial virulence. Proc. Natl Acad. Sci. USA, 98, 6889–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley B.N. and Bassler,B.L. (2000) Regulation of quorum sensing in Vibrio harveyi by LuxO and σ-54. Mol. Microbiol., 36, 940–954. [DOI] [PubMed] [Google Scholar]
- Martin M., Showalter,R. and Silverman,M. (1989) Identification of a locus controlling expression of luminescence genes in Vibrio harveyi. J. Bacteriol., 171, 2406–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W.W., Steed,P.M. and Wanner,B.L. (1990) Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J. Bacteriol., 172, 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.B. and Bassler,B.L. (2001) Quorum sensing in bacteria. Annu. Rev. Microbiol., 55, 165–199. [DOI] [PubMed] [Google Scholar]
- Miller M., Skorupski,K., Lenz,D., Taylor,R. and Bassler,B. (2002) Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell, 110, 303–314. [DOI] [PubMed] [Google Scholar]
- Murray M.G. and Thompson,W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res., 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K.H. and Hastings,J.W. (1979) Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev., 43, 496–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena J.L., Viete,S., Funabiki,K., Saberi,K. and Konishi,M. (2001) Cochlear and neural delays for coincidence detection in owls. J. Neurosci., 21, 9455–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schauder S. and Bassler,B.L. (2001) The languages of bacteria. Genes Dev., 15, 1468–1480. [DOI] [PubMed] [Google Scholar]
- Schauder S., Shokat,K., Surette,M.G. and Bassler,B.L. (2001) The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol., 41, 463–476. [DOI] [PubMed] [Google Scholar]
- Shao C.P. and Hor,L.I. (2001) Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol., 183, 1369–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch J.M. and Silhavy,T.J. (1991) cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol., 173, 4039–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart G.J. and Hausser,M. (2001) Dendritic coincidence detection of EPSPs and action potentials. Nat. Neurosci., 4, 63–71. [DOI] [PubMed] [Google Scholar]
- Surette M.G. and Bassler,B.L. (1998) Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl Acad. Sci. USA, 95, 7046–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M.G. and Bassler,B.L. (1999) Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol., 31, 585–595. [DOI] [PubMed] [Google Scholar]
- Surette M.G., Miller,M.B. and Bassler,B.L. (1999) Quorum sensing in Escherichia coli, Salmonella typhimurium and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl Acad. Sci. USA, 96, 1639–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga M.E., Semmelhack,J.L. and Bassler,B.L. (2001) The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol., 42, 777–793. [DOI] [PubMed] [Google Scholar]
- Zhu J., Miller,M.B., Vance,R.E., Dziejman,M., Bassler,B.L. and Mekalanos,J.J. (2002) Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl Acad. Sci. USA, 99, 3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]