Abstract
RNase E isolated from Escherichia coli is contained in a multicomponent “degradosome” complex with other proteins implicated in RNA decay. Earlier work has shown that the C-terminal region of RNase E is a scaffold for the binding of degradosome components and has identified specific RNase E segments necessary for its interaction with polynucleotide phosphorylase (PNPase), RhlB RNA helicase, and enolase. Here, we report electron microscopy studies that use immunogold labeling and freeze–fracture methods to show that degradosomes exist in vivo in E. coli as multicomponent structures that associate with the cytoplasmic membrane via the N-terminal region of RNase E. Whereas PNPase and enolase are present in E. coli in large excess relative to RNase E and therefore are detected in cells largely as molecules unlinked to the RNase E scaffold, immunogold labeling and biochemical analyses show that helicase is present in approximately equimolar amounts to RNase E at all cell growth stages. Our findings, which establish the existence and cellular location of RNase E-based degradosomes in vivo in E. coli, also suggest that RNA processing and decay may occur at specific sites within cells.
Keywords: immunogold labeling, RhlB RNA helicase, RNA process, RNA degradation
RNase E is an essential Escherichia coli ribonuclease that has a key role in the degradation and/or processing of both short and long half-lived RNAs. When purified from E. coli cells, RNase E is present in a multicomponent ribonucleolytic complex (i.e., the RNA “degradosome”) that includes polynucleotide phosphorylase (PNPase), the RhlB RNA helicase, enolase, the DnaK chaperonin protein, GroEL, and polynucleotide phosphate kinase (PPK) (1–5). Specific regions required to bind certain degradosome proteins have been identified within the C-terminal half of RNase E (6), and a functionally active minimal degradosome containing only RNase E, PNPase, and helicase has been reconstituted in vitro (7).
The view that many, if not most, cellular functions are carried out in vivo by multicomponent macromolecular complexes (i.e., cellular machines) rather than by individual freely diffusable proteins has gained wide acceptance in recent years (8). Well recognized and extensively studied examples of such complexes in bacteria and higher organisms include ribosomes, replisomes, and proteasomes (9–11). However, notwithstanding the isolation of multicomponent RNase E-based complexes from E. coli (1–5, 12), there has been no direct evidence that degradosomes are present in living cells—rather than being formed in vitro as aggregates of individual proteins. The question of whether degradosomes actually exist in vivo in E. coli is especially relevant in view of evidence that truncated RNase E protein lacking the C-terminal half is sufficient for cell viability and for RNA degradation and processing in vivo in E. coli (13, 14), that RNase E homologs in certain other bacteria do not contain the scaffold region that interacts with PNPase and other degradosome proteins (15), and that purified RNase E devoid of other degradosome components is functionally active in vitro (16–19).
We set out to investigate the existence of multicomponent degradosomes in vivo and also to localize degradosome proteins. We report here that degradosomes are present in intact E. coli cells. We further show that degradosomes, whose composition appears to be dynamically regulated, are not distributed evenly throughout the cytoplasm—but instead are associated with the cytoplasmic membrane via the N-terminal region of RNase E.
Materials and Methods
Bacterial Strains and Plasmids.
The following E. coli strains were used: HB101 [supE44 hsdS20(rB−mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1], BL21(DE3) [hsdS gal(λcIts857 ind1 S am7 nin5 lacUV5-T7 gene1)], and BZ99 [his ΔtrpE5 mukB106 smbB199 (λ)] (13). HB101 and BL21(DE3) contain wild-type rne gene, and BZ99 contains a truncated rne gene that encodes amino acids 1 through 602 of the Rne protein. Plasmid pGP1–2 contains the T7 RNA polymerase gene under the control of a temperature-sensitive bacteriophage λ repressor (20). Plasmid pRE296 is identical to pRE196 (3) except that the Flag tag was replaced with a His6-Flag tag.
Degradosome Complex Purifications.
RNA degradosomes were purified as described in ref. 3 by using the Flag-epitope-tagged Rne fusion protein and a Flag-monoclonal antibody-conjugated agarose column.
Antibody Preparation.
Polyclonal antibodies were prepared as described (21). Briefly, purified degradosomes were separated by SDS/PAGE and stained with Coomassie blue. The gel slices containing individual bands of RNase E, PNPase, enolase, or RhlB helicase were frozen in liquid nitrogen, dried, and ground into a powder. The powdered gel was suspended in Freund's adjuvant and phosphate buffer (pH 7.2) used to immunize rabbits. Degradosome components in protein gels and E. coli cells were detected by using the same antibodies, except that the polyclonal antibodies against a His6-tag RhlB was received as a gift from M. Cashel (National Institutes of Health, Bethesda) and used for the detection of RhlB helicase in E. coli cells. The specificity of individual antibodies was tested by using cell lysates from E. coli cells that overexpressed each antigen.
SDS/PAGE, Western Blotting, and Quantitation.
Proteins of complexes in E. coli cell extracts were separated on 8% gels containing 0.1% SDS. Gels were stained by Coomassie brilliant blue to visualize protein bands. The ECL Western blot detection system (Amersham Pharmacia) and LAS-1000 plus (Fuji) were used to quantify proteins with serial dilutions of antigen that had been calibrated against BSA.
E. coli Cell Preparation for Immunogold Labeling.
Log-phase liquid cultures of cells containing heat-inducible Flag-Rne (3) were shifted from 32°C to 44°C for 2 h. Cells were pelletted by centrifugation at 3,000 × g for 5 min and fixed by incubation at 4°C overnight with cold 0.1 M phosphate–citrate buffer (pH 7.2) containing 1% glutaraldehyde and 1% formaldehyde. After fixation, samples were neutralized by the addition of 0.1 M ammonium chloride in phosphate–citrate buffer at 0°C, and after 30 min were dehydrated in a series of cold methanol treatments (25% at 0°C for 30 min twice, 50% at 0°C and 75% and 100% at −20°C, each for 60 min) containing 20% polyvinylpyrolidone 6000 (PVP-6000). Samples were then infiltrated serially with solutions of embedding LR-Gold reagent containing 10% PVP-6000: 25% and 50% LR-Gold (Electron Microscopy Sciences, Fort Washington, PA) at −20°C for 1 h and 2 h, respectively; 75% and 100% LR-Gold solutions at −20°C for 4 h and overnight, respectively. The embedded samples were transferred into embedding capsules (PELCO, Redding, CA), polymerized in a PELCO UVC1 Cryo chamber at −20°C for 24 h, and then continuously hardened in the PELCO UVC1 Cryo chamber at room temperature for 2–3 d. Embedded samples were stored in a dehumidifying chamber at room temperature until use.
Immunogold Single and Double Labeling Detection.
Immunogold labeling was carried out as described in refs. 22–25 with modifications. Briefly, the ultrathin sections (100 nm) of LR-Gold embedded samples were directly mounted on 200 mesh nickel grids (PELCO) covered with carbon-backed collodion film. For double labeling one hole or 100 mesh nickel grids (PELCO) were used. The individual grids were floated on 3% normal goat serum in PBS at room temperature for 15 min, and then floated on the individual primary antibodies as indicated for 15 to 30 min. After six sequential washings with 20 μl droplets of 1% normal goat serum in PBS, the grids were floated for 15 to 30 min on 5 nm gold-IgG complexes [BB International, Cardiff, U.K.; 1:100 dilution with gold-IgG solution (0.1 M potassium phosphate, pH 7.4, 4% PVP-10000, 0.01 mM polyethyleneglycol 20000, and 0.05% sodium azide)]. Grids were then washed sequentially with 6 droplets of 1% normal goat serum in PBS, followed by two 5 min washes with triple distilled water. For double labeling, immunostaining with the second primary antibody (in this case, anti-RNase E) was performed on the reverse side of the same section as described above, except that gold-IgG complexes (26) containing larger size particles (15 nm) were used. The sections were finally treated with 2% uranyl acetate and 30 mM lead citrate. Samples were examined by using a Zeiss EM109 or Philips CM100 (Philips Electron Optics, The Netherlands) electron microscope. All chemicals used for the preparation of embedded samples for immunogold labeling were obtained from either Sigma or Merck.
SDS-Digested Freeze–Fracture Replica Labeling.
SDS-digested freeze–fracture replica labeling (SDS-FRL) was carried out as described previously (27, 28) with some modifications. Briefly, samples of cells collected from cultures were prepared as above, and, supported by a gold-specimen-carrier, were quick-frozen by immersing tubes in nitrogen slush. The frozen samples were fractured at −105°C and replicated by evaporation of Pt/C from an electron-beam gun at a 45o angle, followed by carbon coating at 90o in the Freeze Etching System (BAF 400D or BAF 400F; Balzers). To release the replicas from carriers, the carriers were immersed gently in PBS. After floating off carriers, the replicas were transferred to the sample digestion buffer (2.5% SDS, 10 mM Tris⋅HCl, pH 7.2, 30 mM sucrose, 1.5 mg/ml Lysozyme) and incubated overnight at room temperature (changing the buffer at least four times). The replicas were then washed for at least 1 h with four or more changes of PBS and placed on 3% normal goat serum in PBS with two or more changes for 1 h at room temperature. Replicas were labeled with the primary antibody (i.e., anti-RNE, PNPase, Rhl B helicase, or Enolase antibody) with two or more changes for 1 h at room temperature and were then washed with 1% normal goat serum in PBS (six 5 min washes) and incubated at room temperature with 15 nm gold-IgG complexes as the secondary antibody with two changes for 1 h each. After labeling, the replicas were washed with 1% normal goat serum in PBS as above, rinsed with distilled water for 5 min twice, and picked onto grids. The samples were examined in a Zeiss EM109 or Philips CM100 electron microscope.
Results
Degradosome Components in E. coli Cells.
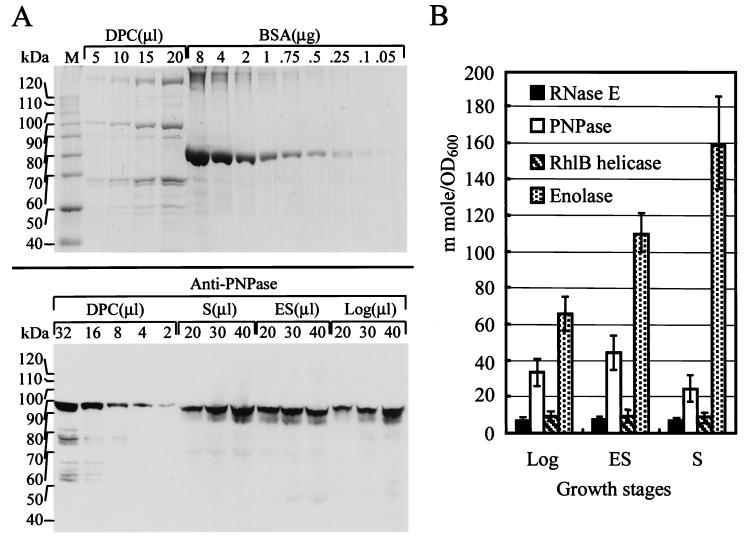
Previously, it was shown by gel filtration of partially purified RNase E that the protein was present in a complex having a molecular mass estimated by glycerol gradient sedimentation analysis to be 460 kDa (2) and by gel-filtration chromatography to be 500 kDa (4). Subsequent experiments have identified a series of proteins present in RNase E-containing complexes isolated from E. coli cells (2, 3, 5, 6) and various stoichiometric relationships between the components of these “degradosomes” have been proposed. Here, the ratios of degradosome proteins in E. coli cell extracts were determined by (i) quantitatively comparing the intensity of bands representing degradosome proteins bound to affinity purified Flag-tagged RNase E with a BSA band on Coomassie blue stained gels (Fig. 1A Upper) and (ii) serially diluting extracts from cells harvested at various growth stages and calibrating the intensity in gels of individual degradosome protein components (Fig. 1A Bottom). This enabled quantitation by Western blotting of the amount of each protein present in E. coli total cell extracts, as shown for PNPase in Fig. 1A Bottom. A histogram shows the amount of endogenous protein (in millimoles per unit of E. coli cell mass determined by measurement at OD600) determined for individual degradosome components in cells at different stages of growth (Fig. 1B). These data indicate that RNase E and the RhlB helicase were present in E. coli cells in approximately equimolar amounts throughout cell growth, but that the molar ratios of enolase and PNPase to RNase E varied with growth stage. In cells in log phase, both enolase and PNPase were present in a 5- to 10-fold excess relative to RNase E. This ratio increased through early stationary phase for both proteins and continued to increase for enolase during late stationary phase, while leveling off for PNPase. The fluctuation of the ratio of PNPase and enolase to RNase E and RhlB helicase suggests a dynamic regulation of degradosome composition.
Figure 1.
Quantitation of degradosome proteins in E. coli cell extracts. Coomassie blue stained SDS/PAGE gels containing different amounts of BSA, as indicated, for quantitation of degradosome protein components (A Upper). By using a known amount of degradosome protein component deduced from the upper panel and anti-PNPase antibody, the amount of endogenous cellular PNPase was detected in log phase (Log), early stationary phase (ES), and late stationary phase (S) by Western blotting (A Lower). (B) Quantitation of individual proteins in mmol/OD600 of cell culture. Cell lysates were prepared from cells in logarithmic phase at OD600 about 0.5, in early stationary phase at OD600 about 1.0, and in late stationary phase at OD600 about 2.3. Molecular weights (kDa) of individual protein standards are shown. Proteins were quantified by integration of band density area by using LAS-1000 plus (Fuji) and calibration curves, which were constructed from known amounts of BSA and degradosome proteins. M, protein molecular weight standards; DPC, degradosome protein components.
Location of RNase E-Complexes in Ultrathin E. coli Cell Sections.
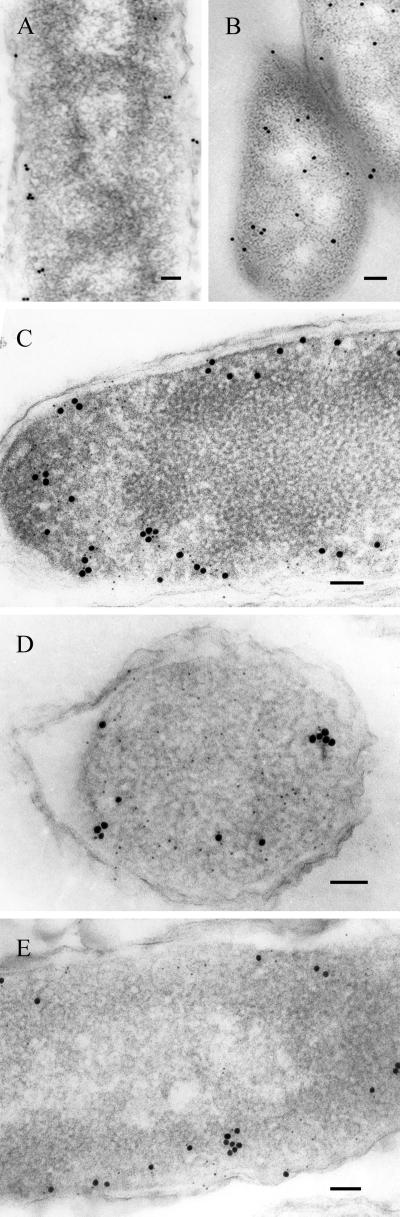
RNase E degradosomes isolated from E. coli cells contain at least three other major proteins in addition to RNase E (i.e., PNPase, RhlB helicase, and enolase) and some lesser components that include DnaK and polynucleotide phosphate kinase (PPK). Immunogold double labeling analysis of ultrathin sections of E. coli cells expressing the Flag-Rne polypeptide showed that antibodies against both RNase E and RhlB helicase were localized predominantly in regions near the inner cytoplasmic membrane. Fifteen nm immunogold particles indicate the location of RNase E (Fig. 2 A, C, D, and E) and 5 nm immunogold particles localize the RhlB helicase (Fig. 2C). The location of enolase also was not random; it was found in our immunogold analyses to be present preferentially in the vicinity of the cytoplasmic membrane in E. coli, as has been reported previously for other microorganisms (29, 30). However, the fraction of enolase not seen in the vicinity of the membrane was about twice that observed for RNase E and RhlB (Table 1, 19.15% vs. 8.9% and 10.84%, respectively). In contrast, antibody against PNPase detected protein distributed evenly throughout the cytoplasm (Fig. 2D), as was found previously (4), as well as in the vicinity of RNase E. Control experiments showed that β-galactosidase was distributed evenly throughout the cell (Fig. 2B), providing evidence of the specificity of localization of immunogold particles detecting RNase E and RhlB helicase to the cytoplasmic region near the inner membrane.
Figure 2.
Subcellular localization and colocalization of individual degradosome proteins by using immunogold double labeling. Ultrathin sections of E. coli cells singly labeled with anti-RNase E (A) or anti-β-Gal control antibody (B). (C) Double labeling of cells with anti-RNase E (15 nm gold-IgG complexes) and anti-RhlB helicase (5 nm gold-IgG complexes). (D and E) The 5 nm gold-IgG complexes labeled PNPase and enolase, respectively, are shown, whereas labeling of RNase E was the same as in C. Log phase cultured cells were used for the studies. (Bars = 100 nm.)
Table 1.
Subcellular distribution of RNase E, PNPase, Rhl B helicase and Enolase in E. coli cells detected by immunogold labeling
| Antibody | Cells examined | Immunogold particles examined | Percentage
of total immunogold particles
|
||
|---|---|---|---|---|---|
| On membrane | Within 100 nm* near membrane cytoplasmic area | Other cytoplasmic region | |||
| Rne | 124 | 2248 | 10.41 | 80.65 | 8.94 |
| PNP | 77 | 2051 | 3.66 | 27.50 | 68.84 |
| RhlB | 114 | 2232 | 12.14 | 77.02 | 10.84 |
| Eno | 112 | 1384 | 9.54 | 71.32 | 19.15 |
| β-Gal | 150 | 1589 | 1.75 | 30.90 | 67.34 |
E. coli cell cross section diameters ranged from 700 to 800 nm.
The observed colocalization of RNase E and RhlB helicase at or near the cytoplasmic membrane by electron microscopy argues strongly that degradosome complexes containing these proteins exist in living cells; such colocalization offers a level of resolution approximately 1,000-fold greater than what is seen by immunofluorescence analysis with standard confocal microscopy (22).
The information obtained by examination of randomly selected immunogold-labeled cells is presented quantitatively in Table 1. As seen, 70–80% of RNase E, RhlB, and enolase molecules detected by immunogold-labeled antibodies were found at or near the cytoplasmic membrane, whereas most PNPase molecules were located elsewhere.
Freeze–Fracture Analysis.
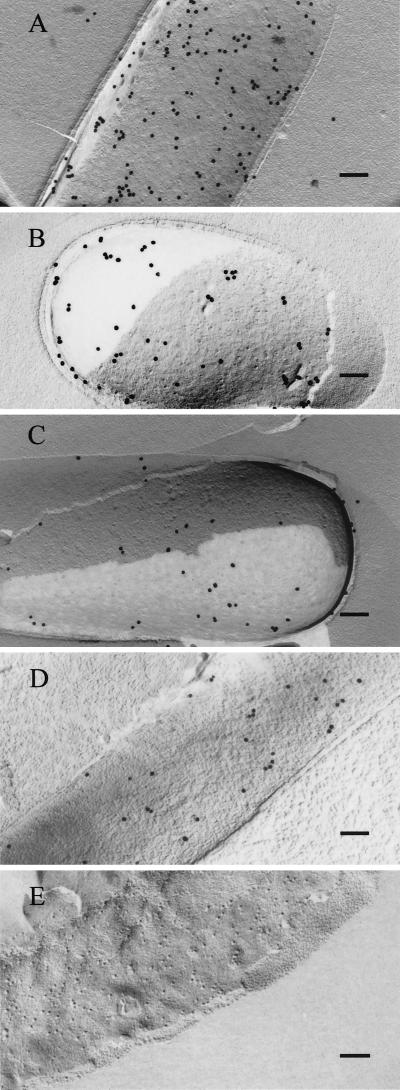
The freeze–fracture technique enables visualization of protein components of biological membranes by immunogold labeling (27, 28). This procedure, which involves the fracture of unfixed quick-frozen cells and stabilization by platinum/carbon followed by thawing, detergent treatment, and labeling of replicas by immunogold particles, as described in Materials and Methods, allowed us to identify degradosome proteins specifically attached to the E. coli cytoplasmic membrane; whereas none of the β-galactoside control protein, which is known to be cytoplasmic in location, was detected on membranes visualized by the freeze–fracture technique (Fig. 3E). Like two bona fide membrane associated proteins, TolA and Tol R (31, 32), which we detected in freeze–fracture prepared membranes (data not shown), all four of the degradosome proteins we assayed were present in freeze–fracture membrane preparations (Fig. 3 A, B, C, and D)—confirming that these degradosome proteins are membrane components.
Figure 3.
Freeze–fracture immunogold labeling of E. coli cells. Shown are SDS-digested freeze–fracture replicas of E. coli cells labeled with anti-RNase E (A), anti-PNPase (B), anti-RhlB helicase (C), anti-enolase (D), or anti-β-Gal (E) antibodies. Same growth phase for cells used in Fig. 2. (Bars = 100 nm.)
Degradosomes Associate with the Cytoplasmic Membrane Via the N-Terminal Region of RNase E.
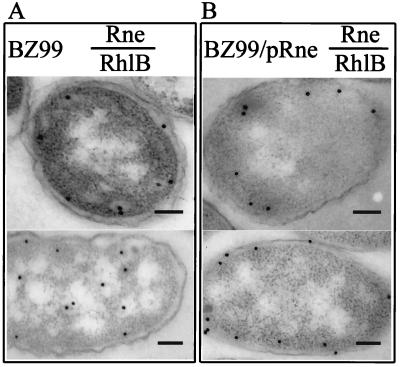
As seen in Fig. 4, ultrathin section immunogold EM analysis of a chromosomally encoded truncated mutant form of RNase E containing only the first 602 amino acids of the enzyme (i.e., lacking amino acids 603 to 1061) showed the presence of the truncated protein at the cytoplasmic membrane, indicating that the amino-terminal region of RNase E is sufficient for membrane localization (Fig. 4A Upper). However, the RhlB helicase was not increased in concentration at or near the membrane in cells producing the truncated RNase E protein (Fig. 4A Bottom), which lacks the C-terminal segment that serves as a scaffold for the binding of this helicase and other degradosome components. When full length RNase E protein was overexpressed in the mutant strain concurrently with truncated RNase E, both RNase E and the RhlB helicase were found in the vicinity of the cytoplasmic membrane (Fig. 4B Upper and Lower, respectively). Together, these results show that the N-terminal region of RNase E is sufficient to bind RNase E to the cytoplasmic membrane and suggest that this protein takes along the helicase (and presumably also other components of degradosomes) when RNase E contains the C-terminal scaffold region that interacts with these components. These results further confirm the existence of RNA degradosomes in vivo.
Figure 4.
Effect of RNase E truncation on localization of degradosome proteins. Subcellular localization of RhlB helicase and RNase E in mutant BZ99, which produces an Rne protein lacking the C-terminal region (A), or in BZ99 cells that also contain the pRne plasmid, which encodes the full length Rne protein (B). Ultrathin sections of E. coli cell labeled with anti-RNase E antibody are shown in Top. Cells labeled with anti-RhlB helicase antibody are shown in Bottom. Log phase cultured cells were used for the studies. (Bars = 100 nm.)
Discussion
The existence of degradosomes has been widely inferred from data demonstrating that macromolecular complexes containing RNase E-bound proteins can be isolated from E. coli (2–5, 12), and from evidence of RNase E interaction with other proteins by yeast two-hybrid analysis and immunoprecipitation (2, 4–7). Our results establish that degradosome complexes are in fact present in living cells and further demonstrate that these macromolecular structures associate with the E. coli cytoplasmic membrane via the N-terminal region of RNase E. As this region encodes the enzyme's ribonucleolytic activity (17, 19), our findings raise the possibility that RNA decay and processing take place at the membrane—as has been speculated previously by Miczak et al. (33) who detected enzymatic activities of several RNA-processing enzymes (i.e., RNase III, RNase E, and RNase P) in inner membrane fractions isolated from E. coli cells. As the MreE/CafA/RNaseG protein has significant homology with the N-terminal region of RNase E (34–39), this ribonuclease may also localize to the cytoplasmic membrane and carry out its actions there.
The antibodies used to identify degradosome components were not of uniform titer and thus the cellular density of immunogold particles detecting these components does not reflect the corresponding amount of protein as determined biochemically. However, the distribution of individual degradosome components within E. coli cells can be assessed by our analysis, and our data indicate that only a small fraction of PNPase is bound to RNase E in degradosomes. Although larger fractions of enolase and RhlB helicase appear to be included in membrane bound degradosomes, significant amounts of both proteins, and also of RNase E itself, remain separate from the membrane. Our current studies do not indicate whether membrane binding affects the actions of RNase E or of any of the proteins bound to RNase E in vivo. However, as RNA decay and processing can occur following the deletion of RNase E sequences essential for degradosome formation, degradosome proteins are presumed to be active in unbound form.
Acknowledgments
We thank Dr. M. Cashel for providing antibodies against RhlB helicase used for all electron microscopy studies, Dr. R. E. Webster for antibodies against TolA and TolR for freeze–fracture replica labeling, and Drs. M. Cashel, P. Knight, K. McDowall, L. Shapiro, and R. Simons for comments on the manuscript. This study fulfilled in part the requirements for the Ph.D. thesis of G.-G.L. at the National Taiwan University. The work was supported by a research grant award for Frontier Sciences from the National Science Council (to S.L.-C.), by an intramural fund from the Academia Sinica of the Republic of China (to S.L.-C.), and by National Institutes of Health Grant GM 54158 (to S.N.C.).
Abbreviation
- PNPase
polynucleotide phosphorylase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011535498.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011535498
References
- 1.Blum E, Py B, Carpousis A J, Higgins C F. Mol Microbiol. 1997;26:387–398. doi: 10.1046/j.1365-2958.1997.5901947.x. [DOI] [PubMed] [Google Scholar]
- 2.Carpousis A J, Van Houwe G, Ehretsmann C, Krisch H M. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 3.Miczak A, Kaberdin V R, Wei C-L, Lin-Chao S. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Py B, Causton H, Mudd E A, Higgins C F. Mol Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 5.Py B, Higgins C F, Krisch H M, Carpousis A J. Nature (London) 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 6.Vanzo N F, Li Y S, Py B, Blum E, Higgins C F, Raynal L C, Krisch H M, Carpousis A J. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coburn G A, Miao X, Briant D J, Mackie G A. Genes Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 9.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 10.Baumeister W, Walz J, Zuhl F, Seemuller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 11.Green R, Noller H F. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 12.Bessarab D A, Kaberdin V R, Wei C-L, Liou G-G, Lin-Chao S. Proc Natl Acad Sci USA. 1998;95:3157–3161. doi: 10.1073/pnas.95.6.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. J Bacteriol. 1996;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez P J, Marchand I, Joyce S A, Dreyfus M. Mol Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- 15.Kaberdin V R, Miczak A, Jakobsen J S, Lin-Chao S, McDowall K J, Von Gabain A. Proc Natl Acad Sci USA. 1998;95:11637–11642. doi: 10.1073/pnas.95.20.11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaberdin V R, Chao Y-H, Lin-Chao S. J Biol Chem. 1996;271:13103–13109. doi: 10.1074/jbc.271.22.13103. [DOI] [PubMed] [Google Scholar]
- 17.McDowall K J, Cohen S N. J Mol Biol. 1996;255:349–355. doi: 10.1006/jmbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 18.McDowall K J, Kaberdin V R, Wu S-W, Cohen S N, Lin-Chao S. Nature (London) 1995;374:287–290. doi: 10.1038/374287a0. [DOI] [PubMed] [Google Scholar]
- 19.Taraseviciene L, Bjork G R, Uhlin B E. J Biol Chem. 1995;270:26391–26398. doi: 10.1074/jbc.270.44.26391. [DOI] [PubMed] [Google Scholar]
- 20.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: a laboratory manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 53–137. [Google Scholar]
- 22.Bozzola J J, Russell L D. Electron Microscopy Principles and Techniques for Biologists. London: Jones and Bartlett Publishers; 1999. [Google Scholar]
- 23.Hyatt A D. In: Electron Microscopy in Biology. Harris J R, editor. New York: Oxford Univ. Press; 1991. pp. 59–81. [Google Scholar]
- 24.Lin N-S, Chen C-C, Hsu Y-H. J Histochem Cytochem. 1993;41:1513–1519. doi: 10.1177/41.10.8245409. [DOI] [PubMed] [Google Scholar]
- 25.Lin N-S, Liou G-G, Chen C-C, Chang B-Y. Zool Stud. 1995;34:142–143. [Google Scholar]
- 26.Lin N-S, Langenberg W G. J Ultrastruct Res. 1983;84:16–23. doi: 10.1016/s0022-5320(83)90082-5. [DOI] [PubMed] [Google Scholar]
- 27.Fujimoto K. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto K. Histochem Cell Biol. 1997;107:87–96. doi: 10.1007/s004180050092. [DOI] [PubMed] [Google Scholar]
- 29.Angiolella L, Facchin M, Stringaro A, Maras B, Simonetti N, Cassone A. J Infect Dis. 1996;173:684–690. doi: 10.1093/infdis/173.3.684. [DOI] [PubMed] [Google Scholar]
- 30.Edwards S R, Braley R, Chaffin W L. FEMS Microbiol Lett. 1999;177:211–216. doi: 10.1111/j.1574-6968.1999.tb13734.x. [DOI] [PubMed] [Google Scholar]
- 31.Levengood S K, Webster R E. J Bacteriol. 1989;171:6600–6609. doi: 10.1128/jb.171.12.6600-6609.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derouiche R, Benedetti H, Lazzaroni J-C, Lazdunski C, Lloubes R. J Biol Chem. 1995;270:11078–11084. doi: 10.1074/jbc.270.19.11078. [DOI] [PubMed] [Google Scholar]
- 33.Miczak A, Srivastava R A K, Apirion D. Mol Microbiol. 1991;5:1801–1810. doi: 10.1111/j.1365-2958.1991.tb01929.x. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Pandit S, Deutscher M P. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDowall K J, Hernandez R G, Lin-Chao S, Cohen S N. J Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tock M R, Walsh A P W, Carroll G, McDowall K J. J Bacteriol. 2000;275:8726–8732. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- 37.Wachi M, Doi M, Ueda T, Ueki M, Tsuritani K, Nagai K, Matsuhashi M. Gene. 1991;106:135–136. doi: 10.1016/0378-1119(91)90578-y. [DOI] [PubMed] [Google Scholar]
- 38.Wachi M, Umitsuki G, Nagai K. Mol Gen Genet. 1997;253:515–519. doi: 10.1007/s004380050352. [DOI] [PubMed] [Google Scholar]
- 39.Wachi M, Umitsuki G, Shimizu M, Takada A, Nagai K. Biochem Biophys Res Commun. 1999;259:483–488. doi: 10.1006/bbrc.1999.0806. [DOI] [PubMed] [Google Scholar]