Abstract
The preprotein translocase of the inner membrane of mitochondria (TIM23 complex) is the main entry gate for proteins of the matrix and the inner membrane. We isolated the TIM23 complex of Neurospora crassa. Besides Tim23 and Tim17, it contained a novel component, referred to as Tim50. Tim50 spans the inner membrane with a single transmembrane segment and exposes a large hydrophilic domain in the intermembrane space. Tim50 is essential for viability of yeast. Mitochondria from cells depleted of Tim50 displayed strongly reduced import kinetics of preproteins using the TIM23 complex. Tim50 could be cross-linked to preproteins that were halted at the level of the translocase of the outer membrane (TOM complex) or spanning both TOM and TIM23 complexes. We suggest that Tim50 plays a crucial role in the transfer of preproteins from the TOM complex to the TIM23 complex through the intermembrane space.
Keywords: import/mitochondria/TIM23 complex/Tim50/translocation
Introduction
Four different complexes have been discovered in mitochondria that are involved in the targeting of newly synthesized proteins to the various mitochondrial subcompartments (Koehler, 2000; Paschen and Neupert, 2001; Pfanner and Wiedemann, 2002; Stuart, 2002). The TOM complex in the outer membrane is the entry gate for mitochondrial proteins (Endo and Kohda, 2002). The inner membrane contains the TIM23 and TIM22 complexes which facilitate the import of proteins into the matrix space and into the inner membrane (Tokatlidis et al., 2000; Jensen and Dunn, 2002; Pfanner and Chacinska, 2002). The OXA1 complex mediates insertion of proteins into the inner membrane from the matrix side (Hell et al., 2001).
The TIM23 complex is responsible for the import of the vast majority of mitochondrial proteins. It was found to consist of the integral membrane components Tim23 and Tim17 (Dekker et al., 1993, 1997; Emtage and Jensen, 1993; Maarse et al., 1994; Ryan et al., 1994; Berthold et al., 1995). Both are predicted to span the inner membrane four times and are believed to constitute the protein-conducting channel for precursors containing mitochondrial matrix targeting signals (MTS) (Ryan and Jensen, 1993; Kubrich et al., 1994; Milisav et al., 2001; Truscott et al., 2001). Evidence for Tim23 acting as a component that recognizes the MTS in the intermembrane space (IMS) has been presented (Bauer et al., 1996). Tim44 is loosely associated with Tim23 and Tim17 (Berthold et al., 1995). It is attached to the inner membrane as a peripheral protein and recruits the mitochondrial heat shock protein 70 (mtHsp70) to the import site (Kronidou et al., 1994; Rassow et al., 1994; Schneider et al., 1994). Tim44 and mtHsp70 are the major components of the mitochondrial protein import motor. The nucleotide exchange factor Mge1 assists in the cyclic binding of mtHsp70 to Tim44 and precursor, which facilitates the vectorial movement of precursor polypeptide chains in the direction of the matrix (Matouschek et al., 2000; Ryan and Pfanner, 2001; Neupert and Brunner, 2002). Despite these insights into the structure and function of the TIM23 complex, many questions as to its composition, dynamics, energetics and interaction with the TOM complex remain to be answered.
Here we report on the isolation of the TIM23 complex of Neurospora crassa and, in addition to the known components Tim23 and Tim17, we have detected a novel protein component of 56 kDa molecular mass. In the yeast TIM23 complex, this protein was also present as a homolog of 50 kDa. The component was named Tim50 according to the terminology for Tom and Tim proteins (Pfanner et al., 1996). In Neurospora and yeast, Tim50 was found in substoichiometric amounts in the TIM23 translocase. Tim50 is a protein essential for the viability of yeast. Mitochondria from cells in which Tim50 was depleted by downregulation show deficiencies in the import of mitochondrial proteins using the TIM23 pathway. No evidence was obtained that the TIM22 pathway is affected. Tim50 was observed to be in close proximity to segments of preproteins present in the IMS on their way into the matrix. Even intermediates, which accumulated at the level of the TOM complex in the absence of a membrane potential, could be cross-linked to Tim50.
We propose that Tim50 plays a crucial role in the IMS by facilitating the transfer of precursor proteins from the TOM complex to the TIM23 complex.
Results
Purification of the TIM23 complex
In order to isolate the TIM23 complex from the mitochondria of N.crassa, we constructed a strain in which the gene for tim23 was inactivated by sheltered RIP (repeat induced point mutation) (D.Mokranjac and F.Nargang, in preparation). This strain was rescued with a plasmid coding for a version of Tim23 carrying a His9 tag at the N-terminus. The His-tagged Tim23 was fully functional in vivo as this strain showed normal growth behavior (data not shown). Mitochondria were isolated, solubilized in various detergents and the detergent lysates were analyzed for association of Tim17 and Tim23. Triton X-100 and dodecyl maltoside led to dissociation of the complex. In contrast, when mitochondria were dissolved in buffer containing digitonin and passed over an Ni-NTA column, both Tim components were retained. Tim44, however, was not recovered in the complex of Tim23 with Tim17 in either low or high salt conditions (data not shown).
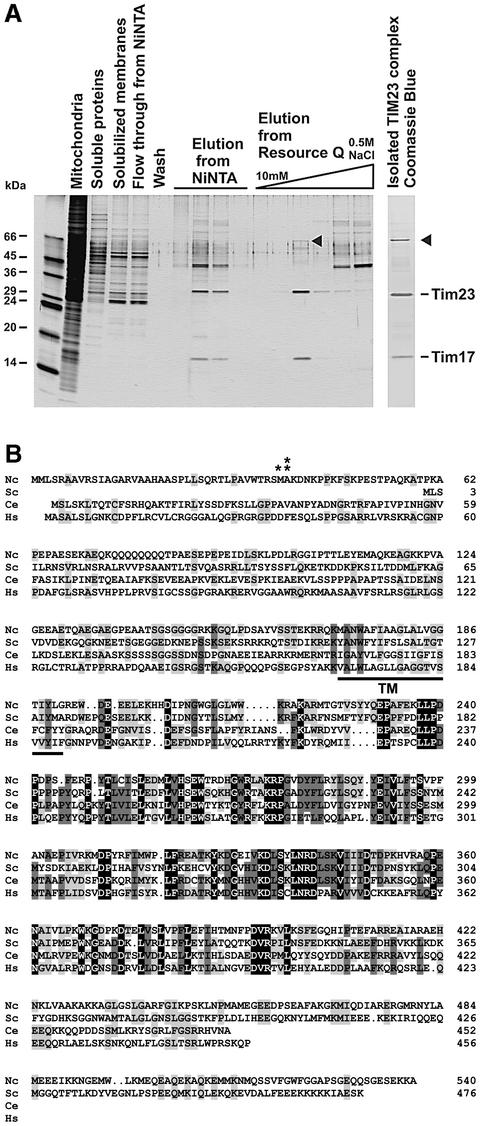
Digitonin was used in further experiments for the isolation of the TIM23 complex. Mitochondria were isolated from ∼300 g of cells, and the membrane fraction was solubilized in digitonin and passed over Ni-NTA. Bound material was eluted with imidazole-containing buffer and subjected to anion exchange chromatography on a Resource Q column. The TIM23 complex was recovered essentially in a single fraction (Figure 1A). The identities of the two major bands as Tim23 and Tim17 were confirmed by immunoblotting and mass spectrometry (data not shown). In addition, a minor band with an apparent molecular mass of ∼56 kDa was visible upon silver and Coomassie Blue staining (Figure 1A). The latter protein was isolated from the gel and analyzed by mass spectrometry (shown as Supplementary data available at The EMBO Journal Online). The peptides obtained were used to identify the corresponding cDNA from the Neurospora genome database. The cDNA encodes a protein of 540 amino acids (DDBJ/EMBL/GenBank accession No. AY188754).
Fig. 1. Identification of a 56 kDa protein present in the TIM23 complex. (A) Purification of the TIM23 complex from N.crassa. Isolated mitochondrial membranes from a Neurospora strain, which harbors a Tim23 with an N-terminal His9 tag, were solubilized in digitonin and passed over an Ni-NTA column. Bound material was eluted with an imidazole-containing buffer and applied to a Resource Q ion exchange column at low salt concentration. Elution was performed with a gradient of 10–500 mM NaCl. Fractions were analyzed by SDS–PAGE and silver staining. The TIM23 complex was eluted from the Resource Q column essentially in a single fraction (left panel). Coomassie Blue staining of such a fraction is also shown (right panel). The 56 kDa protein is indicated by arrowheads. (B) Alignment of the Neurospora 56 kDa protein with its homologs from S.cerevisiae (YPL063w), C.elegans (T25076) and H.sapiens (XM_053074). Partial and complete conservation of amino acid residues are indicated by shaded and black backgrounds, respectively. The predicted single transmembrane domain (TM) is underlined. The asterisk shows the predicted cleavage site by the mitochondrial processing peptidase for the Neurospora protein, and the double asterisk the determined N-terminal residue of the isolated protein.
The 56 kDa protein associated with the TIM23 complex is predicted to contain a mitochondrial MTS at the N-terminus. Cleavage of the targeting signal occurs either after residue 38, as the following alanine was the first residue identified by N-terminal sequencing of the purified protein, or, more probably, after residue 37, as this is a canonical mitochondrial processing peptidase (MPP) cleavage site (Gavel and von Heijne, 1990). There is a strong prediction for the presence of an α-helical hydrophobic membrane-spanning stretch at residues 171–191. These structural elements predict a topology of the mature protein with the N-terminal 133 residues in the matrix and a large hydrophilic domain of 349 residues in the IMS.
A search of databases yielded homologs in the genomes of all eukaryotic organisms inspected. Figure 1B shows an alignment of the deduced sequences of Saccharomyces cerevisiae, Caenorhabditis elegans and Homo sapiens. These homologs have the same structural organization as the 56 kDa protein; however, only the C-terminal segment following the hydrophobic stretch shows clear sequence conservation.
To verify that the 56 kDa protein is a component of the inner membrane with the predicted topology, subcellular and submitochondrial localization experiments were performed. The protein was found in the mitochondrial fraction (Figure 2A). Treatment of mitochondria with proteinase K (PK) led to degradation of the surface-exposed Tom70, but not of the 56 kDa protein (Figure 2B). Mitochondria were titrated with digitonin to open successively the outer and the inner membranes, and were treated with PK. The 56 kDa protein was degraded as soon as the outer membrane was opened, in agreement with the predicted removal of a 40 kDa IMS domain (Figure 2B). It could not be extracted by alkaline treatment of mitochondria (Figure 2C). Finally, the protein was synthesized in a reticulocyte lysate. Upon treatment with yeast mitochondrial processing peptidase, it was processed to a slightly smaller species which had the mobility of the isolated 56 kDa species. Upon incubation with isolated mitochondria in the presence of a membrane potential, a major part of the precursor was converted to this latter species, a part of which was imported completely into mitochondria, as it was resistant to treatment of the mitochondria with protease (data not shown).
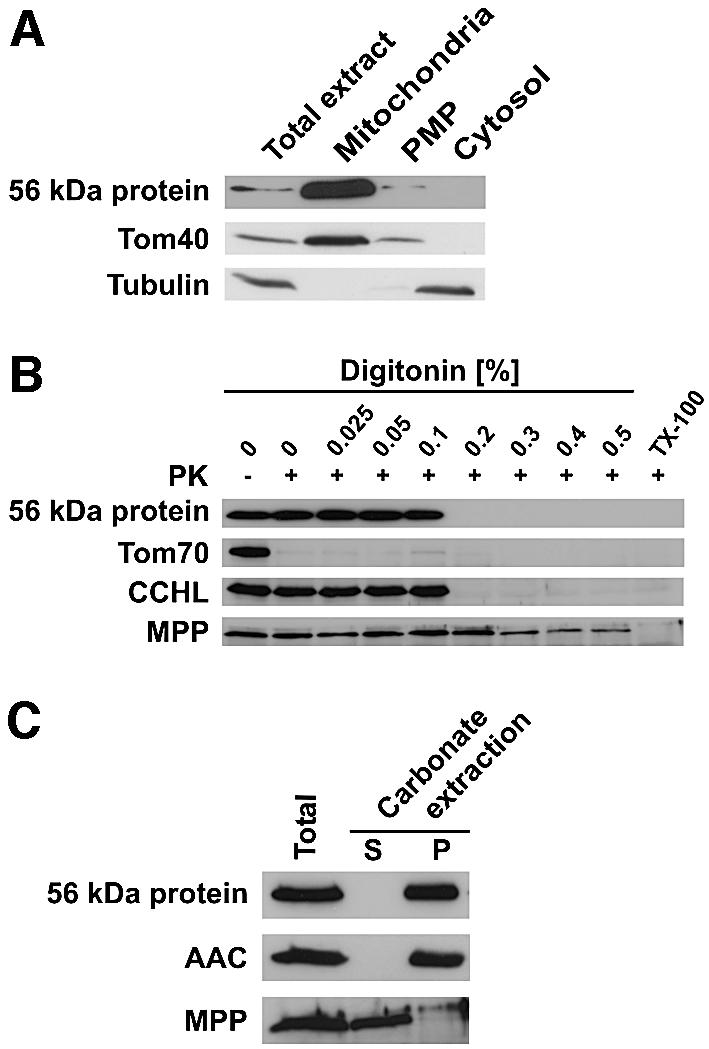
Fig. 2. Subcellular and submitochondrial localization of the 56 kDa protein in Neurospora. (A) The 56 kDa protein is located in mitochondria. Neurospora cells were fractionated into a mitochondrial fraction (mitochondria), a post-mitochondrial pellet (PMP) and a soluble fraction (cytosol). The fractions were analyzed by SDS–PAGE and immunoblotting with antibodies against the 56 kDa protein, Tom40 and tubulin, as mitochondrial and cytosolic markers, respectively. (B) Mitochondria were titrated with digitonin to open successively the outer and inner membrane. Samples were treated with proteinase K (PK) and analyzed by SDS–PAGE and immunoblotting with antibodies against the 56 kDa protein, and Tom70, cytochrome c heme lyase (CCHL) and the mitochondrial processing peptidase (MPP) as markers for outer membrane, intermembrane space and matrix, respectively. (C) Alkaline extraction of the 56 kDa protein. Mitochondria were treated with 0.1 M sodium carbonate, and soluble (S) and membrane fractions (P) were separated by centrifugation and analyzed as in (A) using antibodies against an integral membrane protein, ADP/ATP carrier (AAC), and a soluble protein, MPP, as controls.
In conclusion, the TIM23-associated 56 kDa protein is a mitochondrial inner membrane protein with a large hydrophilic domain exposed into the IMS.
Interaction of the 56 kDa protein with the TIM23 complex
Is the 56 kDa protein associated with the TIM23 complex in a specific manner? Wild-type Neurospora mitochondria were solubilized in digitonin and incubated with antibodies against either Tim23, Tim17, the 56 kDa protein or Oxa1, in amounts sufficient to achieve complete immuno precipitation of the respective protein. Immunoprecipitates and supernatants were subjected to SDS–PAGE and immunoblotting with the same antibodies, as well as antibodies against the ADP/ATP carrier (AAC) (Figure 3A). The antibody against Tim23 led to virtually complete depletion of both Tim23 and Tim17 from the supernatant. In addition, a significant portion of the 56 kDa component was found in the immunoprecipitate; yet, the larger portion of this protein was not precipitated. Essentially the same result was obtained when antibodies against Tim17 were used. Antibodies against the 56 kDa protein were able to deplete this protein from the detergent extract, but only a fraction of Tim23 and Tim17 was co-precipitated (Figure 3A). Antibodies against Oxa1 as a control only precipitated the Oxa1, but no Tim proteins. In addition, the abundant inner membrane protein, AAC, was not detected in the immunoprecipitates.
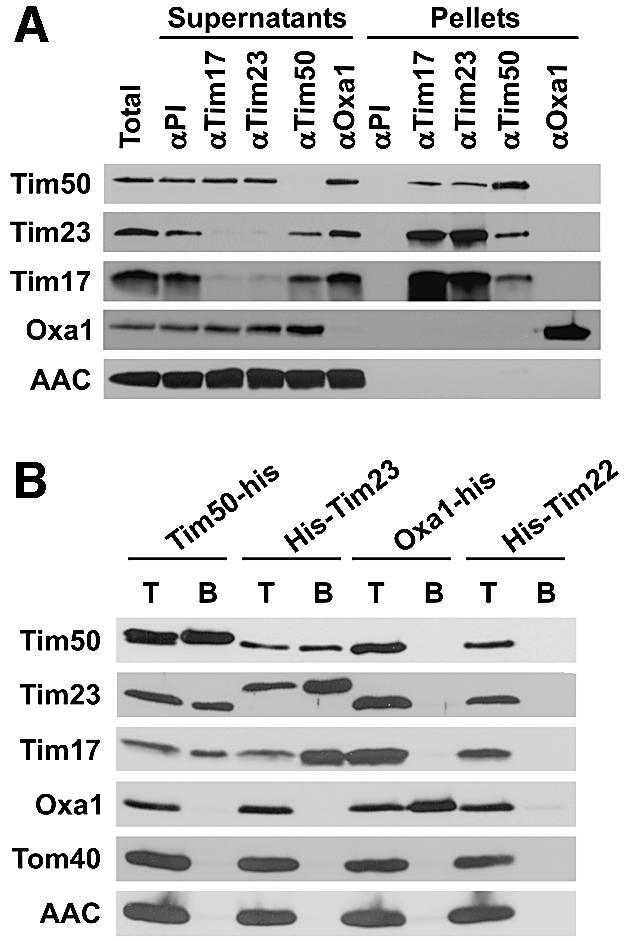
Fig. 3. Specific association of Tim50 with the TIM23 complex. (A) Co-immunoprecipitation of Tim50 with TIM23 components. Wild-type Neurospora mitochondria were solubilized with digitonin and incubated with antibodies against Tim50, Tim23, Tim17 and Oxa1, or with pre-immune IgG pre-bound to protein A–Sepharose beads. The beads were collected by centrifugation, washed, and bound proteins were eluted with Laemmli buffer. The supernatants were diluted directly in Laemmli buffer. Pellets and supernatants were analyzed by SDS–PAGE and western blotting for the antigens indicated. ‘Total’ and ‘Supernatants’ represent 25% of material present in ‘Pellets’. (B) Co-purification of Tim50 with TIM23 complex and vice versa via Ni-NTA beads. Mitochondria were isolated from Neurospora strains in which either Tim50, Tim23, Tim22 or Oxa1 carried a His tag. They were solubilized with digitonin and incubated with Ni-NTA beads. After elution with imidazole-containing buffer, proteins were analyzed by SDS–PAGE and western blotting for the indicated mitochondrial components. T, total digitonin solubilized material, representing 25% of input; B, material bound to Ni-NTA.
The association of the 56 kDa protein with Tim23 and Tim17 was also analyzed with mitochondrial extracts from cells expressing His-tagged versions of Tim23 or of the 56 kDa protein (Figure 3B). When mitochondrial extracts containing His9-Tim23 were passed over Ni-NTA, the 56 kDa protein was partly retained. Conversely, when mitochondria containing His-tagged 56 kDa protein were analyzed in the same way, Tim23 and Tim17 were retained. Again, control proteins Tom40, Oxa1 and AAC were not retained, demonstrating the specificity of the interaction. Furthermore, using extracts from control strains expressing His-tagged forms of Oxa1 or Tim22, neither Tim17, Tim23 nor the 56 kDa protein were retained on the column.
These results show that the 56 kDa protein is indeed associated specifically with the TIM23 complex. We therefore consider this protein a true TIM component. Following the established nomenclature for components of the import machinery (Pfanner et al., 1996), we refer to this protein from here on as Tim50, according to the molecular weight of the homolog in yeast.
Tim50 was not present in the TIM23 complex in stoichiometric amounts, at least not under the conditions of our experiments. To determine the percentage of total Tim50 in the TIM23 complex, N.crassa cells were metabolically labeled with [35S]sulfate. Mitochondria were isolated, co-immunoprecipitation was performed as described above, and analyzed by SDS–PAGE. The radioactive signals in the protein bands were quantified using a phosphoimager. Between 10 and 20% of total Tim50 was found to be associated with the TIM23 complex. In addition, the total amounts in mitochondria of Tim50, Tim23 and Tim17 were compared with each other by immunodepletion after lysis of isolated mitochondria with SDS-containing buffer (see Supplementary data). Considering the specific contents of methionine and cysteine, the molar ratio of Tim50:Tim23:Tim17 in mitochondria was determined to be ∼2:1:1. Thus, the ratio in the purified complex is ∼0.3:1:1, reflecting either dynamic or labile interaction of Tim50 with the other two components. It should be noted that Coomassie Blue staining of the bands in the TIM23 complex (Figure 1A, right panel) may not reflect the stoichiometry of the complex. Purified Tim50 was found to be stained relatively strongly, as revealed by a comparison of absorption at 280 nm and absorption in the Bradford assay which uses Coomassie G250 (data not shown).
We conclude that Tim50 interacts with the TIM23 complex; however, only a fraction of it is found in association with the complex when isolated in the presence of the mild detergent digitonin.
Role of Tim50 in the import of preproteins
What is the function of Tim50? We addressed this question using the yeast S.cerevisiae, as genetic analysis is straightforward in this organism. The size of the yeast protein is consistent with an MPP cleavage site after residue 35 and with a putative cleavage by the mitochondrial intermediate peptidase (MIP) after residue 43 (Figure 1B). Tim50 of yeast was confirmed to be a mitochondrial constituent and to have the same topology as the Neurospora homolog (data not shown). In co-immunoprecipitation experiments, yeast Tim50 was also found to be associated specifically with the TIM23 complex in substoichiometric amounts (data not shown). We investigated whether Tim50 is essential for the viability of yeast cells. The TIM50 gene was deleted in the diploid strain W303, and tetrad analysis was carried out. A strict 2:2 segregation showed that the gene for Tim50 is essential for cell viability (Figure 4A).
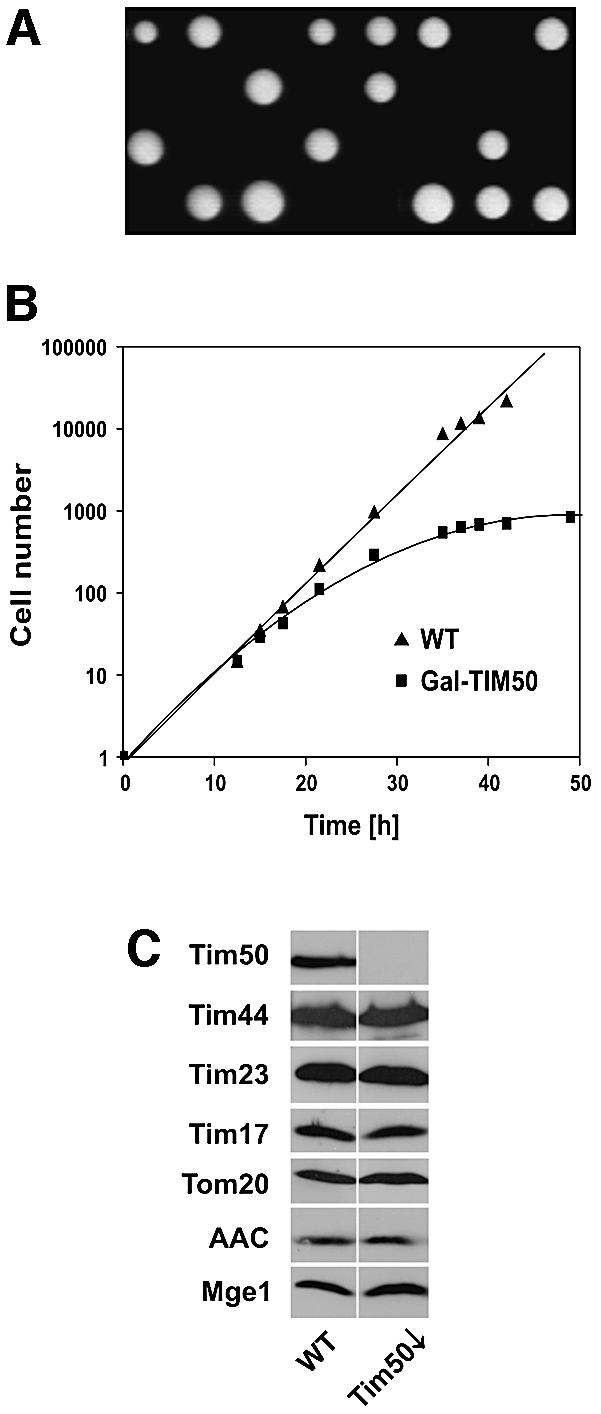
Fig. 4. Tim50 is essential for viability of yeast cells. (A) Disruption of the TIM50 gene. The TIM50 gene was disrupted in diploid cells by homologous recombination. Cells were sporulated and subjected to tetrad analysis. Only two out of four spores were viable in all tetrads analyzed, indicating that Tim50 is an essential protein in yeast. (B) Downregulation of Tim50 in cells affects their growth. Cells expressing Tim50 under the control of the GAL10 promoter (Gal-TIM50) and wild-type cells (WT) were grown in medium containing 0.1% galactose. At time point zero, they were shifted to medium containing 0.1% glucose; the cell number was set to 1. (C) Cells depleted of Tim50 (Tim50↓) show normal levels of other mitochondrial proteins. WT and Tim50↓ cells were harvested 33 h after shift to glucose. Mitochondria were isolated and analyzed by SDS–PAGE and western blotting for the indicated proteins.
For further analysis, a yeast strain was generated in which the TIM50 promoter was replaced by the GAL10 promoter (Gal-TIM50). To deplete cells of Tim50, they were shifted from lactate medium containing galactose to lactate medium containing glucose. This shift led to a reduced growth rate and to virtual arrest of growth after ∼38 h (Figure 4B).
Mitochondria were isolated from YPH499 cells (wild type) and from Gal-TIM50 cells (Tim50↓) 33 h after the shift to glucose medium, and the levels of various mitochondrial proteins were determined (Figure 4C). Tim50 was virtually absent in mitochondria after the shift. The levels of Tim17, Tim23 and Tim44 were not altered in Tim50-depleted cells. In addition, proteins of the outer membrane (Tom20), the inner membrane (AAC) and the matrix (Mge1) were present in amounts similar to wild-type. This suggests that the integrity of mitochondria was not severely affected by depletion of Tim50.
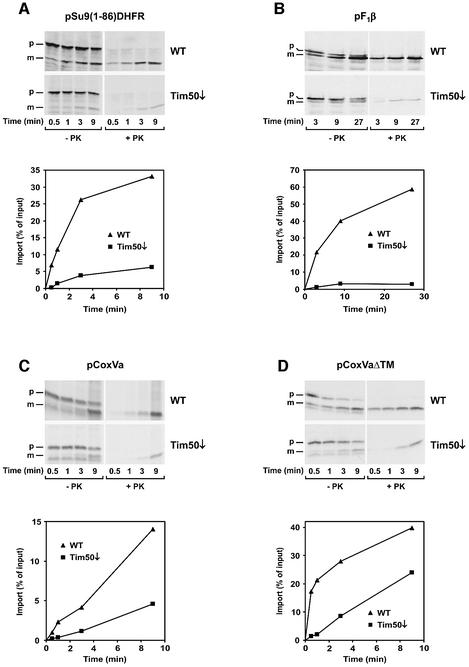
To analyze whether Tim50 functions as a translocation component, we tested the ability of mitochondria isolated from Tim50-depleted cells to import various radiolabeled precursor proteins. First, we analyzed the import of matrix and inner membrane proteins with a cleavable presequence, which are substrates of the TIM23 complex. The following precursors for matrix proteins were used: pF1β, precursor to the F1β subunit of the F1F0-ATP synthase; and pSu9(1–86)DHFR, which consists of the first 86 amino acids of the precursor of F1F0-ATP synthase subunit 9 of N.crassa fused to the complete mouse dihydrofolate reductase. In addition, the precursor of cytochrome c oxidase subunit Va (pCoxVa) was used as an example of an inner membrane protein. Import of pF1β and of pSu9(1–86)DHFR was strongly reduced in mitochondria from cells depleted of Tim50 (Figure 5A and B). Similar results were obtained for other matrix proteins, e.g. the yeast frataxin homolog, Yfh1, and the J-type co-chaperone, Jac1 (data not shown). Import of pCoxVa, which becomes localized by arrest of translocation in the inner membrane, was also affected, but to a lesser degree (Figure 5C).
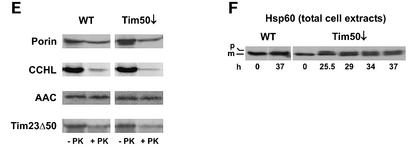
Fig. 5. Mitochondria isolated from Tim50-depleted cells are defective in import of preproteins using the TIM23 translocase but not in import of preproteins using only the TOM complex or the TIM22 translocase. Mitochondria were isolated from wild-type cells (WT) and cells depleted of Tim50 for 33 h (Tim50↓). Radiolabeled precursor proteins were synthesized in reticulocyte lysate and incubated with the mitochondria for the indicated time periods at 25°C (A, B and E) or 12°C (C and D). The import reaction was stopped by placing samples on ice and adding 1 µM valinomycin. Samples were divided, and one-half was incubated with proteinase K (+PK). Reisolated mitochondria were subjected to SDS–PAGE and autoradiography (upper panel). PK-protected material was quantified (lower panel). (A) pSu9(1–86)DHFR, a fusion protein consisting of the N-terminal 86 residues of the presubunit 9 of ATP synthase and dihydrofolate reductase. (B) pF1β, the precursor of subunit β of ATP synthase. (C) pCoxVa, the precursor of cytochrome oxidase subunit Va. (D) pCoxVaΔTM, the pCoxVa precursor with deletion of the transmembrane domain. (E) CCHL, cytochrome c heme lyase; AAC, ADP/ATP carrier; Tim23Δ50, Tim23 with residues 1–50 deleted. The indicated precursors were imported for 15 min into WT and Tim50↓ mitochondria as described above. (F) Tim50-depleted cells accumulate Hsp60 precursor in vivo. WT and Tim50↓ cells were harvested at different time points after shift to glucose-containing medium. Total cell extracts were prepared by alkaline lysis and analyzed by SDS–PAGE and immunodecoration against Hsp60. p, precursor form of Hsp60; m, mature form of Hsp60.
Notably, the observed difference between pCoxVa and the matrix proteins in their dependence on Tim50 for efficient import was not due to the particular presequence. pCoxVaΔTM, a precursor derived from pCoxVa by deletion of its transmembrane domain, becomes sorted to the mitochondrial matrix. Its initial rate of import into Tim50-depleted mitochondria was reduced more strongly than that of pCoxVa (Figure 5D). Thus, it appears that the mature part of a preprotein influences the degree of dependence on Tim50.
The activity of the TOM complex was not affected by depletion of Tim50, as porin and cytochrome c heme lyase (CCHL) were imported at the same rate in depleted and control mitochondria (Figure 5E).
Import of proteins which are inserted into the inner membrane by the TIM22 complex was also analyzed. AAC was imported as well in Tim50-depleted mitochondria as in control mitochondria, as was the precursor of Tim23Δ50, a protein derived from Tim23 by deleting the first 50 amino acid residues (Figure 5E). These results show that the TIM22 pathway is not affected by depletion of Tim50.
We determined the membrane potential ΔΨ of Tim50-depleted mitochondria and measured a decrease by 20–30% compared with wild-type mitochondria. It is very unlikely that the observed import defects of 90% of matrix-targeted precursors were due to the slightly reduced membrane potential, as the AAC was imported at a similar rate in Tim50-depleted and wild-type mitochondria. AAC and matrix-targeted precursors carrying the subunit 9 presequence were reported to require a similar threshold of ΔΨ, and a very low ΔΨ generated by K+ diffusion was able to drive import of matrix-targeted preproteins (Martin et al., 1991). The following observations support this conclusion. (i) Preproteins carrying the MTS of Su9 need less ΔΨ than pCoxVa for import (Gärtner et al., 1995), yet in Tim50-depleted mitochondria, the initial rate of import of pSu9(1–86)DHFR was much lower than the initial rate of pCoxVa import. (ii) Precursors with the same presequence should be affected to the same degree in processing and import, if ΔΨ was limiting. This was not the case, as observed for the pair pCoxVa and pCoxVa ΔTM (Figure 5C and D). Taken together, these data show that a lowered ΔΨ was not responsible for the reduced import rates of preproteins in Tim50-depleted mitochondria.
A defect in the import of preproteins in Tim50-depleted mitochondria is also suggested by observations with intact cells. Total extracts of cells depleted of Tim50 by downregulation for 25.5–37 h were analyzed for the presence of precursor and mature forms of Hsp60. This protein has been shown to accumulate in the cytosol as the precursor form under conditions of impeded import (Moczko et al., 1994; Schmitt et al., 1995). Indeed, the precursor accumulated upon growth of cells in the absence of galactose (Figure 5F). In summary, these results demonstrate that Tim50 has a distinct role in the import of proteins along the TIM23 pathway.
Interaction of Tim50 with preproteins in the IMS
To understand how Tim50 is involved in the translocation of precursor proteins into mitochondria, its interaction with preproteins in transit was analyzed by cross-linking. The chimeric protein pb2Δ19(167)DHFRK5 (Schneider et al., 1994), consisting of the first 167 amino acids of cytochrome b2 with a 19 residue deletion in the sorting signal, was imported into wild-type mitochondria under the following conditions: in the absence or presence of ΔΨ, and in the absence or presence of methotrexate (MTX). In the absence of ΔΨ, precursors do not reach the translocation channel in the inner membrane and are only partially imported across the outer membrane (Paschen and Neupert, 2001). In contrast, in the presence of ΔΨ, the precursor is imported completely into the mitochondrial matrix and processed. The DHFR moiety of the precursor can be stably folded by addition of MTX. Upon import in the presence of ΔΨ, the precursor is arrested in a topology spanning the outer membrane, the IMS and the inner membrane, and is bound by mtHsp70 in the matrix (Schneider et al., 1994). After incubating mitochondria with the precursor under these three import conditions, cross-linking was performed followed by immuno precipitation with antibodies against Tim50 and other components of the translocating machinery. We used the cross-linker 1,5-difluoro-2,4-dinitrobenzene (DFDNB), which has a particularly short spacer arm of 0.3 nm. Cross-linked products of precursor and Tim50 were observed when import was performed in the absence of ΔΨ or when the precursor was arrested by MTX, but not upon complete import into the matrix (Figure 6A). In the latter case, mtHsp70 was found in association with the preprotein. These experiments indicate that Tim50 is in contact with or in the close vicinity of incoming precursor proteins in the IMS. Interestingly, the cross-linking pattern in the absence of ΔΨ was different from that obtained with arrested precursor spanning both membranes. This may reflect different interactions of Tim50 with the preprotein. Precursor protein arrested at the level of the outer membrane and protruding into the IMS by depletion of membrane potential could be cross-linked only to Tim50, whereas cross-linking of the spanning intermediate was observed not only to Tim50, but also to Tim23, Tim44 and mtHsp70 (Figure 6A).
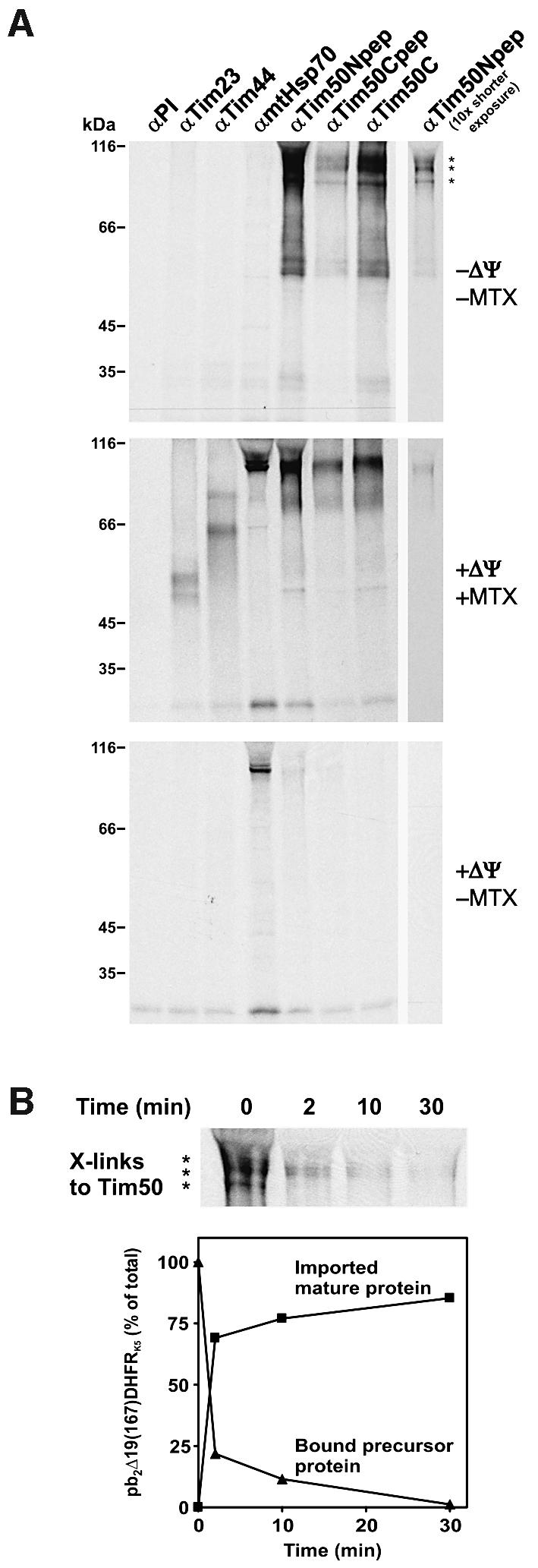
Fig. 6. Cross-linking of Tim50 to preproteins in transit. (A) Precursor pb2Δ19(167)DHFRK5 was imported into isolated wild-type yeast mitochondria under various conditions and subjected to cross-linking with DFDNB as described in Materials and methods. Mitochondria were re-isolated and solubilized in SDS-containing buffer. Immunoprecipitation was performed with the antibodies indicated. The immunoprecipitates were analyzed by SDS–PAGE and autoradiography. PI, pre-immune serum. Top panel: import in the absence of a membrane potential ΔΨ and of methotrexate (MTX). The observed cross-linked products to Tim50 are marked with asterisks. Middle panel: import in the presence of ΔΨ and MTX. Lower panel: import in the presence of ΔΨ and absence of MTX. (B) The precursor of pb2Δ19(167)DHFRK5 was bound to mitochondria in the absence of a membrane potential. Mitochondria were re-isolated, re-energized and incubated further. At the time points indicated, aliquots were withdrawn for cross-linking or for SDS–PAGE directly. Upper panel: samples which were subjected to cross-linking and immunoprecipitation with antibodies against Tim50 (αTim50Npep) were analyzed as in (A). Lower panel: samples were subjected directly to SDS–PAGE, autoradiography and quantification. The amounts of bound precursor protein and imported mature protein at various time points were related to total precursor bound at time point zero in the absence of ΔΨ, which was set to 100%.
To confirm such a dynamic interaction, the precursor was first accumulated at the level of the TOM complex in the absence of ΔΨ. Then a ΔΨ was re-established, and import and cross-linking to Tim50 were followed in a time-dependent manner. A cross-linked species was observed with the bound precursor at time zero of the experiment, but decreased upon incubation in the presence of ΔΨ (Figure 6B, upper panel). At the same time, the precursor was processed and imported (Figure 6B, lower panel). The decrease of cross-links with Tim50 corresponded to the kinetics of processing and import. Thus, Tim50 appears to interact with the precursor as soon as it reaches the trans side of the TOM complex and as long as segments of it are present in the IMS.
Discussion
We describe the isolation of the TIM23 complex of N.crassa. The complex, in addition to Tim23 and Tim17, contains a new component, referred to as Tim50. Tim50 is present in virtually all eukaryotic organisms, as homologs were found in all genomes that have been completely sequenced. Tim50 is anchored in the inner membrane with a single transmembrane segment; the N-terminal segment is exposed to the matrix, and the C-terminal segment to the IMS. Tim50 has an NIF domain (NLI-interacting factor) within its C-terminal segment. The NIF domain is found in many proteins of diverse functions, e.g. NLI-interacting factors, RNA polymerase II CTD phosphatase and FCP1 serine phosphatase, yet its function is still unclear. Tim50 in Neurospora and yeast was found not to be present in stoichiometric amounts in the TIM23 complex. The majority of Tim50 in yeast was recovered in a complex of apparent molecular mass of ∼250 kDa (S.A.Paschen and K.Hell, unpublished observation). These observations suggest that Tim50 may interact with the TIM23 complex in a dynamic fashion rather than being an integral fixed part of the TIM23 complex.
Deletion of Tim50 in yeast is lethal. Depletion of Tim50 leads to a virtually complete block of growth. Thus, this protein belongs to the relatively small group of mitochondrial proteins that are essential for viability of yeast cells. This group comprises components of the import complexes, molecular chaperones and enzymes of the biogenesis pathways of Fe/S proteins (Lill and Kispal, 2000; Voos and Rottgers, 2002). We present evidence here that Tim50 functions in protein transport into mitochondria. Mitochondria isolated from yeast cells in which Tim50 was depleted displayed a strong reduction of import in the case of all matrix proteins analyzed, and partial inhibition of import of proteins which are sorted by the TIM23 complex using the stop-transfer mechanism. Import by the TIM22 complex was virtually unaffected. Similarly, no reduction of TOM complex activity was observed, as insertion of outer membrane proteins was unaffected by depletion of Tim50.
The involvement of Tim50 in translocation of preproteins is reflected in its proximity to preprotein in transit. Cross-linking of Tim50 with precursor was achieved when precursor was accumulated as an intermediate spanning TOM and TIM23 complexes simultaneously. Cross-linking of a precursor arrested across both mitochondrial membranes to a protein of 50 kDa has been reported (Kanamori et al., 1997); it seems likely now that this component was Tim50. Remarkably, efficient cross-linking was also seen when precursor was bound to mitochondria in the absence of a membrane potential, i.e. at the level of the TOM complex. These observations argue for an interaction of Tim50 with various segments of the precursor as these are entering or crossing the IMS.
Our data allow us to put forward a working hypothesis that integrates the various findings. Tim50 has a major domain exposed to the IMS. This domain is highly conserved in evolution, in contrast to the matrix domain. It appears to have a major role in Tim50 function upon protein import. Furthermore, this domain interacts with, or at least is close to, segments of precursors which are present in the IMS. Thus, Tim50 would function by supporting the transfer of preproteins from the TOM complex to the TIM23 complex, thereby increasing efficiency of translocation. Accordingly, it may associate with both these complexes in a weak and transient manner. This would explain why only part of the Tim50 present in mitochondria is associated with the TIM23 complex at any one time. The interaction of Tim50 with the TIM23 complex might occur through the segment of the IMS domain of Tim23, which was proposed to function as a presequence receptor (Bauer et al., 1996). The similarity of the adverse effects on import into intact mitochondria of depletion of Tim50 and deletion of the IMS domain of Tim23 (Bauer et al., 1996; Ryan et al., 1998) would support this view.
Additional functions of Tim50 seem possible. This will have to be addressed in further studies. In particular, Tim50 could also have a role in the assembly of the TIM23 translocation pore or exert a more general chaperone function in the IMS.
Two papers were published recently that report the identification of Tim50 in yeast. Their results on the characterization largely agree with those presented here for Neurospora and yeast (Geissler et al., 2002; Yamamoto et al., 2002).
Materials and methods
Neurospora crassa strains
Transformation and handling of N.crassa strains were carried out as described previously (Davis and De Serres, 1970; Vollmer and Yanofsky, 1986). The wild-type strain used in this study is 74A (Fungal Genetics Stock Center No. 2489). The strain expressing the His-tagged version of Tim23 was generated essentially in the same way as reported for the strain expressing the His-tagged version of Oxa1 (Nargang et al., 2002) and will be described in detail elsewhere (D.Mokranjac and F.Nargang, in preparation). To generate the strain expressing His-tagged Tim50, the TIM50 gene was amplified from genomic DNA introducing a C-terminal His9 tag by PCR, cloned into pKS-bar-cpc1 vector and transformed into wild-type cells.
Yeast strains and cell growth
The wild-type strains YPH499, W303 and W334 were used (Hovland et al., 1989; Sikorski and Hieter, 1989; Hell et al., 2000). The diploid yeast strain W303 was used for replacement of the TIM50 gene by the HIS3 gene. The Gal-TIM50 strain, in which expression of Tim50 is under galactose control, was constructed by replacing the 100 bp upstream of the TIM50 reading frame with a GAL10 promoter-containing cassette in the wild-type strain YPH499 (see Supplementary data). For depletion of Tim50, Gal-TIM50 cultures were shifted from lactate medium containing 0.1% galactose to lactate medium containing 0.1% glucose for the indicated periods. Control wild-type cells were treated in the same way. Otherwise cells were cultured on lactate medium.
Total yeast cell extracts
Total yeast cell extracts were prepared by alkaline lysis (Kushnirov, 2000). The cell pellet of 1 ml of yeast culture (OD595nm = 2.5) was resuspended in 200 µl of 0.2 M NaOH. After incubation for 5 min at room temperature, cells were re-isolated by centrifugation, resuspended in Laemmli buffer and analyzed by SDS–PAGE and western blotting.
Purification of TIM23 complex
The TIM23 complex was isolated from 1.5 g of mitochondria of an N.crassa strain expressing the His-tagged Tim23. Mitochondria were resuspended in ice-cold buffer A [50 mM Na-phosphate pH 8.0, 300 mM NaCl, 20 mM imidazole, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF)] and sonicated with 12 pulses (Branson Sonifier, duty 80%, output 6). Membranes were collected by ultracentrifugation at 100 000 g for 45 min, and solubilized in buffer A containing 2% (w/v) digitonin. After a clarifying spin, the supernatant was loaded on an Ni- NTA–agarose column. The column was washed with 10 column volumes of buffer A containing 0.05% digitonin, followed by a three column volume wash with the same buffer but reduced salt (10 mM NaCl) prior to elution with 300 mM imidazole. Fractions containing TIM23 complex were passed over a PD-10 column (Amersham) equilibrated with 50 mM Tris–HCl pH 8.0, 10 mM NaCl, 10% glycerol, 0.05% digitonin, and then loaded on a 1 ml Resource Q column (Amersham). Bound proteins were eluted with the above buffer using a salt gradient from 10 to 500 mM NaCl. Samples were analyzed by SDS–PAGE and Coomassie Blue or silver staining (Blum et al., 1987).
In small-scale co-purification experiments, 1 mg of mitochondria containing His-tagged versions of Tim50, Tim23, Oxa1 or Tim22 were solubilized as above. After a clarifying spin, the supernatants were incubated with 50 µl of Ni-NTA–agarose beads for 1 h, beads were washed and bound material was eluted with Laemmli buffer containing 300 mM imidazole. Samples were analyzed by SDS–PAGE and immunodecoration.
Identification of TIM50
The protein band with an apparent molecular mass of ∼56 kDa, which co-purifies with the TIM23 complex, was excised from a Coomassie Blue-stained gel and analyzed by mass spectrometry (WITA Proteomics AG, Berlin, Germany). The tryptic fingerprint revealed a previously unknown open reading frame in the Neurospora database (see Supplementary data). Since the start codon assigned in the database was questionable, primers were designed for several potential start codons and used, in combination with primer annealing at the end of the gene, for the PCR amplification of the DNA from the genomic DNA and a cDNA library. A PCR product was observed with all tested 5′-primers using genomic DNA as a template, but only one primer generated a product in this PCR using the cDNA library. Therefore, the ATG present in this primer was denoted as the start codon. The sequence amplified from the cDNA library revealed the presence of a 64 bp intron in the genomic DNA. The cDNA sequence was cloned into pGem4 vector (Promega) for in vitro transcription and translation.
Antibodies to Tim50
The DNA segment coding for amino acids 189–540 of Neurospora Tim50 was cloned into the pQE-30 vector (Qiagen) and the protein expressed in Escherichia coli XL1-Blue cells. It was purified on an Ni-NTA–agarose column (Qiagen) in 6 M guanidine-HCl according to the manufacturer’s instructions, and injected into rabbits.
Antibodies against the C-terminus of yeast Tim50 were generated in rabbits by injection of a protein comprising residues 131–479 which was expressed with a C-terminal His tag in XL1-Blue cells and purified on an Ni-NTA–agarose column under native conditions. Antibodies against N- and C-terminal peptides of yeast Tim50 were raised in rabbits using the N-terminal peptide QKETKDDKPKSILTDC and the C-terminal peptide CLEKQKEVDALFEEEK. All antibodies were affinity purified as described previously (Harlow and Lane, 1988).
Cross-linking
35S-labeled precursor protein pb2Δ19(167)DHFRK5 was imported into wild-type yeast mitochondria under three different conditions. To arrest the precursor at the level of the outer membrane, mitochondria were incubated for 10 min with 6 µM oligomycin, 25 µM carbonyl cyanide m-chlorophenylhydrazone (CCCP) and 0.5 µM valinomycin to dissipate membrane potential before precursor protein was added. To arrest the precursor protein as a spanning intermediate across both membranes, precursor protein was pre-incubated with MTX and NADPH and then imported into energized mitochondria in the presence of 2 µM MTX and 5 mM NADPH. Following the import reaction, samples were chilled on ice and the cross-linker DFDNB (Pierce) was added to a final concentration of 200 µM. After 30 min, cross-linker was quenched by addition of a 1/10 volume of 1 M glycine pH 8.8. Mitochondria were re-isolated, solubilized in SDS-containing buffer, and cross-linked products were analyzed by immunoprecipitation, SDS–PAGE and autoradiography.
In the chase experiments, mitochondria were de-energized by addition of 50 µM CCCP before pb2Δ19(167)DHFRK5 was added. Mitochondria were then re-isolated and membrane potential was re-established by addition of 2 mM dithiothreitol (DTT) and 5 mM NADH. After different time periods, aliquots were removed and either directly analyzed for fully imported protein or subjected to cross-linking and immunoprecipitation with antibodies against the N-terminal peptide of Tim50 as above.
Miscellaneous methods
The following procedures were applied as published: subcellular and submitochondrial fractionation (Segui-Real et al., 1993), preparation of mitochondria (Daum et al., 1982), protein determination (Bradford, 1976), sporulation and tetrad analysis of yeast cells (Sherman et al., 1986) and immunoprecipitation (Hell et al., 1998). Protein import into mitochondria, co-immunoprecipitation and quantification of endogenous Tim50 levels are described in the Supplementary data.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Ulrike Gärtner, Marica Malesic and Heiko Germeroth for excellent technical assistance, Drs Andreas Reichert and Johannes Herrmann for critically reading the manuscript, Professor Manfred Schliwa, Munich, for the generous gift of antibody against tubulin, and Stephan Meier for providing the CoxVa constructs. This work was supported by grants from the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 594 (Teilprojekt B3, B4), the Fonds der Chemischen Industrie, the Canadian Institutes of Health Research (F.E.N.), Alberta Ingenuity Fund (S.C.H.) and a predoctoral fellowship from the Boehringer Ingelheim Fonds (S.A.P.).
References
- Bauer M.F., Sirrenberg,C., Neupert,W. and Brunner,M. (1996) Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell, 87, 33–41. [DOI] [PubMed] [Google Scholar]
- Berthold J., Bauer,M.F., Schneider,H.C., Klaus,C., Dietmeier,K., Neupert,W. and Brunner,M. (1995) The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell, 81, 1085–1093. [DOI] [PubMed] [Google Scholar]
- Blum H., Beier,H. and Gross,H.J. (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis, 8, 93–99. [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilising the principle of protein–dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Daum G., Bohni,P.C. and Schatz,G. (1982) Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem., 257, 13028–13033. [PubMed] [Google Scholar]
- Davis R.H. and De Serres,F.J. (1970) Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol., 17, 79–143. [Google Scholar]
- Dekker P.J., Keil,P., Rassow,J., Maarse,A.C., Pfanner,N. and Meijer,M. (1993) Identification of MIM23, a putative component of the protein import machinery of the mitochondrial inner membrane. FEBS Lett., 330, 66–70. [DOI] [PubMed] [Google Scholar]
- Dekker P.J., Martin,F., Maarse,A.C., Bomer,U., Muller,H., Guiard,B., Meijer,M., Rassow,J. and Pfanner,N. (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J., 16, 5408–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage J.L. and Jensen,R.E. (1993) MAS6 encodes an essential inner membrane component of the yeast mitochondrial protein import pathway. J. Cell Biol., 122, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T. and Kohda,D. (2002) Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys. Acta, 1592, 3–14. [DOI] [PubMed] [Google Scholar]
- Gärtner F., Voos,W., Querol,A., Miller,B.R., Craig,E.A., Cumsky,M.G. and Pfanner,N. (1995) Mitochondrial import of subunit Va of cytochrome c oxidase characterized with yeast mutants—independence from receptors, but requirement for matrix hsp70 translocase function. J. Biol. Chem., 270, 3788–3795. [DOI] [PubMed] [Google Scholar]
- Gavel Y. and von Heijne,G. (1990) Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng., 4, 33–37. [DOI] [PubMed] [Google Scholar]
- Geissler A. et al. (2002) The mitochondrial presequence translocase. An essential role of Tim50 in directing preproteins to the import channel. Cell, 111, 507–518. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hell K., Herrmann,J.M., Pratje,E., Neupert,W. and Stuart,R.A. (1998) Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl Acad. Sci. USA, 95, 2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Tzagoloff,A., Neupert,W. and Stuart,R.A. (2000) Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem., 275, 4571–4578. [DOI] [PubMed] [Google Scholar]
- Hell K., Neupert,W. and Stuart,R.A. (2001) Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J., 20, 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovland P., Flick,J., Johnston,M. and Sclafani,R.A. (1989) Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene, 83, 57–64. [DOI] [PubMed] [Google Scholar]
- Jensen R. and Dunn,C. (2002) Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim. Biophys. Acta, 1592, 25–34. [DOI] [PubMed] [Google Scholar]
- Kanamori T., Nishikawa,S., Shin,I., Schultz,P.G. and Endo,T. (1997) Probing the environment along the protein import pathways in yeast mitochondria by site-specific photocrosslinking. Proc. Natl Acad. Sci. USA, 94, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C.M. (2000) Protein translocation pathways of the mitochondrion. FEBS Lett., 476, 27–31. [DOI] [PubMed] [Google Scholar]
- Kronidou N.G., Oppliger,W., Bolliger,L., Hannavy,K., Glick,B.S., Schatz,G. and Horst,M. (1994) Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc. Natl Acad. Sci. USA., 91, 12818–12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubrich M., Keil,P., Rassow,J., Dekker,P.J., Blom,J., Meijer,M. and Pfanner,N. (1994) The polytopic mitochondrial inner membrane proteins MIM17 and MIM23 operate at the same preprotein import site. FEBS Lett., 349, 222–228. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. (2000) Rapid and reliable protein extraction from yeast. Yeast, 16, 857–860. [DOI] [PubMed] [Google Scholar]
- Lill R. and Kispal,G. (2000) Maturation of cellular Fe–S proteins: an essential function of mitochondria. Trends Biochem. Sci., 25, 352–356. [DOI] [PubMed] [Google Scholar]
- Maarse A.C., Blom,J., Keil,P., Pfanner,N. and Meijer,M. (1994) Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett., 349, 215–221. [DOI] [PubMed] [Google Scholar]
- Martin J., Mahlke,K. and Pfanner,N. (1991) Role of an energized inner membrane in mitochondrial protein import. Δψ drives the movement of presequences. J. Biol. Chem., 266, 18051–18057. [PubMed] [Google Scholar]
- Matouschek A., Pfanner,N. and Voos,W. (2000) Protein unfolding by mitochondria. The Hsp70 import motor. EMBO rep., 1, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milisav I., Moro,F., Neupert,W. and Brunner,M. (2001) Modular structure of the TIM23 preprotein translocase of mitochondria. J. Biol. Chem., 276, 25856–25861. [DOI] [PubMed] [Google Scholar]
- Moczko M., Ehmann,B., Gartner,F., Honlinger,A., Schafer,E. and Pfanner,N. (1994) Deletion of the receptor MOM19 strongly impairs import of cleavable preproteins into Saccharomyces cerevisiae mitochondria. J. Biol. Chem., 269, 9045–9051. [PubMed] [Google Scholar]
- Nargang F.E., Preuss,M., Neupert,W. and Herrmann,J.M. (2002) The Oxa1 protein forms a homooligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa. J. Biol. Chem., 277, 12846–12853. [DOI] [PubMed] [Google Scholar]
- Neupert W. and Brunner,M. (2002) The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol., 3, 555–565. [DOI] [PubMed] [Google Scholar]
- Paschen S.A. and Neupert,W. (2001) Protein import into mitochondria. IUBMB Life, 52, 101–112. [DOI] [PubMed] [Google Scholar]
- Pfanner N. and Chacinska,A. (2002) The mitochondrial import machinery: preprotein-conducting channels with binding sites for presequences. Biochim. Biophys. Acta, 1592, 15–24. [DOI] [PubMed] [Google Scholar]
- Pfanner N. and Wiedemann,N. (2002) Mitochondrial protein import: two membranes, three translocases. Curr. Opin. Cell Biol., 14, 400–411. [DOI] [PubMed] [Google Scholar]
- Pfanner N. et al. (1996) Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem. Sci., 21, 51–52. [PubMed] [Google Scholar]
- Rassow J., Maarse,A.C., Krainer,E., Kubrich,M., Muller,H., Meijer,M., Craig,E.A. and Pfanner,N. (1994) Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol., 127, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.R. and Jensen,R.E. (1993) Mas6p can be cross-linked to an arrested precursor and interacts with other proteins during mitochondrial protein import. J. Biol. Chem., 268, 23743–23746. [PubMed] [Google Scholar]
- Ryan K.R., Menold,M.M., Garrett,S. and Jensen,R.E. (1994) SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol. Biol. Cell, 5, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.R., Leung,R.S. and Jensen,R.E. (1998) Characterization of the mitochondrial inner membrane translocase complex: the Tim23p hydrophobic domain interacts with Tim17p but not with other Tim23p molecules. Mol. Cell. Biol., 18, 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M.T. and Pfanner,N. (2001) Hsp70 proteins in protein translocation. Adv. Protein Chem., 59, 223–242. [DOI] [PubMed] [Google Scholar]
- Schmitt M., Neupert,W. and Langer,T. (1995) Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J., 14, 3434–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H.C., Berthold,J., Bauer,M.F., Dietmeier,K., Guiard,B., Brunner,M. and Neupert,W. (1994) Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature, 371, 768–774. [DOI] [PubMed] [Google Scholar]
- Segui-Real B., Kispal,G., Lill,R. and Neupert,W. (1993) Functional independence of the protein translocation machineries in mitochondrial outer and inner membranes: passage of preproteins through the intermembrane space. EMBO J., 12, 2211–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Hicks,J. (1986) Methods in Yeast Genetics: A Laboratory Course. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart R. (2002) Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex. Biochim. Biophys. Acta, 1592, 79–87. [DOI] [PubMed] [Google Scholar]
- Tokatlidis K., Vial,S., Luciano,P., Vergnolle,M. and Clemence,S. (2000) Membrane protein import in yeast mitochondria. Biochem. Soc. Trans, 28, 495–499. [PubMed] [Google Scholar]
- Truscott K.N., Kovermann,P., Geissler,A., Merlin,A., Meijer,M., Driessen,A.J., Rassow,J., Pfanner,N. and Wagner,R. (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol., 8, 1074–1082. [DOI] [PubMed] [Google Scholar]
- Vollmer S.J. and Yanofsky,C. (1986) Efficient cloning of genes of Neurospora crassa. Proc. Natl Acad. Sci. USA, 83, 4869–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W. and Rottgers,K. (2002) Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta, 1592, 51–62. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Esaki,M., Kanamori,T., Tamura,Y., Nishikawa,S. and Endo,T. (2002) Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell, 111, 519–528. [DOI] [PubMed] [Google Scholar]