Abstract
Mimicry of host chemokines and chemokine receptors to modulate chemokine activity is a strategy encoded by beta- and gammaherpesviruses, but very limited information is available on the anti-chemokine strategies encoded by alphaherpesviruses. The secretion of chemokine binding proteins (vCKBPs) has hitherto been considered a unique strategy encoded by poxviruses and gammaherpesviruses. We describe a family of novel vCKBPs in equine herpesvirus 1, bovine herpesvirus 1 and 5, and related alphaherpesviruses with no sequence similarity to chemokine receptors or other vCKBPs. We show that glycoprotein G (gG) is secreted from infected cells, binds a broad range of chemokines with high affinity and blocks chemokine activity by preventing their interaction with specific receptors. Moreover, gG also blocks chemokine binding to glycosaminoglycans, an interaction required for the correct presentation and function of chemokines in vivo. In contrast to other vCKBPs, gG may also be membrane anchored and, consistently, we show chemokine binding activity at the surface of cells expressing full-length protein. These alphaherpesvirus vCKBPs represent a novel family of proteins that bind chemokines both at the membrane and in solution.
Keywords: chemokine/glycosaminoglycan/herpesvirus/immune evasion/inflammation
Introduction
Chemokines are chemoattractant cytokines that regulate trafficking and effector functions of leukocytes, and play an important role in host defence against invading microbes and in the pathogenesis of inflammatory diseases (Baggiolini, 1998). Chemokines are basic 8–10 kDa proteins classified into four subfamilies: C, CC, CXC and CX3C. Seven-transmembrane-domain chemokine receptors are expressed in different cell subsets, thus determining the leukocyte subtype that predominates in different types of inflammation. The interaction of chemokines with glycosaminoglycans (GAGs) is required for endothelial transcytosis and correct presentation of chemokines to leukocytes and function in vivo (Middleton et al., 1997; Cinamon et al., 2001).
Viruses interact with crucial immune molecules to modulate their activity (Alcami and Koszinowski, 2000; Tortorella et al., 2000). The critical role of chemokines in antiviral defence has been highlighted by the discovery that poxviruses and herpesviruses encode proteins that mimic chemokines or chemokine receptors and secreted chemokine binding proteins (vCKBP) (Lalani et al., 2000; Murphy, 2001). Myxoma virus (MV) MT-7 or vCKBP-1 is an interferon-γ (IFN-γ) receptor that was later shown to interact with a broad range of C, CC and CXC chemokines (Lalani et al., 1997). MT-7 binds CXCL8 (interleukin 8) (Zlotnik and Yoshie, 2000) through the C-terminal heparin binding domain and has been proposed to prevent chemokine–GAG interactions. The 35 kDa/M-T1 protein in vaccinia virus (VV), MV, cowpox virus and variola virus, designated vCKBP-2, binds CC chemokines with high affinity and inhibits their interaction with chemokine receptors and activity (Graham et al., 1997; Smith et al., 1997; Alcami et al., 1998). VCKBP-3 is encoded by the murine gammaherpesvirus 68 (MHV-68) M3 protein and has broad binding specificity for C, CC, CXC and CX3C chemokines (Parry et al., 2000; van Berkel et al., 2000). Similar to vCKBP-2, M3 inhibits chemokine binding to cellular receptors and activation of biological effects. Neither M3, identified as the first and only vCKBP from a herpesvirus, nor the poxvirus-encoded vCKBP-2 show sequence similarity to human or mouse proteins.
Genes encoding homologues of chemokines or chemokine receptors have been identified in beta- and gammaherpesviruses (Lalani et al., 2000; Murphy, 2001). In contrast, none of these genes has been found in alphaherpesviruses, with the exception of a biologically active CXCL8 homologue encoded by Marek’s disease virus (MDV), which infects domestic chickens (Parcells et al., 2001).
Members of the alphaherpesvirus subfamily have variable host range and the capacity to establish latent infections primarily, but not exclusively, in sensory ganglia (Roizman and Pellet, 2001). Alphaherpesviruses include human pathogens such as herpes simplex virus 1 (HSV-1) and HSV-2, which replicate in oral or genital mucosal tissue, and varicella–zoster virus (VZV), the causative agent of chickenpox and herpes zoster. The alphaherpesviruses equine herpesviruses 1, 3 and 4 (EHV-1, EHV-3 and EHV-4) are important pathogens of domestic horses that cause immunosuppression, rhinitis, bronchiolitis, abortions and neurological disorders. Bovine herpesvirus 1 (BHV-1) is a major pathogen of cattle associated with abortions and with respiratory and genital infections, and is related to alphaherpesviruses that cause disorders in ruminants: BHV-5, caprine herpesvirus 1 (CaHV-1), cervine herpesvirus 1 (CeHV-1) and rangiferine herpesvirus 1 (RanHV-1).
Several herpesvirus glycoproteins are present in the virion envelope and play a role in virus morphogenesis, cell attachment and tropism. Glycoprotein G (gG) encoded by alphaherpesviruses is found in the virus envelope and, in some viruses, is also secreted after proteolytic processing, but its function remains largely unknown. Alphaherpesvirus gG was first characterized in HSV-2 as a secreted 92 kDa glycoprotein (Marsden et al., 1984; Su et al., 1987). Homologues of gG were found in HSV-1 (Richman et al., 1986), EHV-1 (Drummer et al., 1998), EHV-3 (Hartley et al., 1999), EHV-4 (Crabb et al., 1992; Drummer et al., 1998), BHV-1 (Keil et al., 1996) and BHV-5 (Engelhardt and Keil, 1996), but the gG gene is absent in VZV (Gomi et al., 2002).
We report here that gG homologues encoded by EHV-1, EHV-3, BHV-1, BHV-5, RanHV-1, CapHV-1 and CerHV-1 have chemokine binding activity. Recombinant gG from EHV-1, BHV-1 and BHV-5 binds a broad range of chemokines with high affinity and inhibits their biological activity in vitro. We demonstrate that gG blocks the interaction of chemokines with both cellular receptors and GAGs, and show that the gG chemokine binding activity may also be expressed at the cell surface.
Results
Identification of soluble chemokine binding activity encoded by alphaherpesviruses
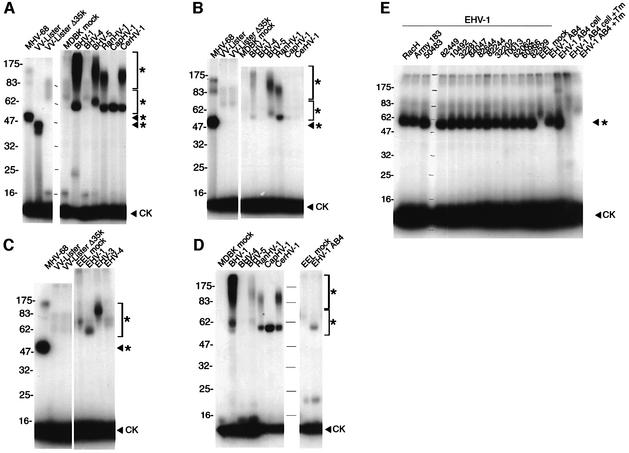
The secretion of vCKBPs from cells infected with various herpesviruses was tested by chemokine cross-linking assays with ethylene glycol-bis-succinimidyl succinate (EGS) (Figure 1A). Complexes of radiolabelled human CCL3 [macrophage inflammatory protein 1, (MIP-1α)] with a soluble vCKBP were observed for all the alphaherpesviruses tested. The gammaherpesvirus BHV-4 did not express chemokine binding activity. Binding specificity was demonstrated with the positive controls of MHV-68 (Parry et al., 2000) and VV strain Lister (Alcami et al., 1998), and the absence of a signal with mock-infected cells and a VV Lister lacking the 35 kDa vCKBP-2 (VV Lister Δ35K) (Alcami et al., 1998). The size of the ligand–vCKBP complex (54–80 kDa) suggested a vCKBP size of 46–72 kDa after subtraction of the 8 kDa monomeric CCL3. BHV-1, BHV-5, RanHV-1 and CerHV-1 also showed ligand binding to a diffusely migrating protein species of 60 kDa to >175 kDa. The size variation may reflect the degree of glycosylation or different polypeptide lengths. Weaker binding to human radiolabelled CXCL8 resulting in a ligand–vCKBP complex was observed for BHV-1, BHV-5 and RanHV-1, but not for CapHV-1 and CerHV-1, which showed CCL3 binding (Figure 1B). The formation of [125I]CXCL8–vCKBP complexes with EHV-1 strain AB4 (55–60 kDa) and EHV-3 (60–80 kDa), but not with EHV-4, was detected (Figure 1C). None of these viruses expressed detectable CCL3 binding activity in this assay (not shown). In an attempt to identify potential new interactions of viral proteins with other chemokines representing different receptor binding specificities, we performed cross-linking assays of all these viral supernatants to radio-iodinated human CCL1 (I-309), CCL2 (monocyte chemoattractant protein 1), CCL11 (eotaxin), CCL17 (thymus and activation-regulated chemokine), CCL20 (MIP-3α), CXCL1 (growth-related oncogene-α), CXCL10 (IFN-γ-inducible protein 10), CXCL21 (stromal cell-derived factor 1α) and CX3CL1 (fractalkine) using EGS, 1-ethyl-3-(3-dimethyl-aminopropyl)-carbodiimide (EDC) or bis(sulfosuccinimidyl) suberate (BS3) as a cross-linker. As shown for CXCL12 (Figure 1D), the results obtained with most of these chemokines were similar to those observed previously with CCL3 and CXCL8, with the exception of CX3CL1 and CXCL10, for which no chemokine–vCKBP interaction was detected (not shown). Medium from cultures infected with HSV-1, HSV-2 or VZV was also tested by cross-linking to all the chemokines used here, but no vCKBP was detected (not shown).
Fig. 1. Soluble chemokine binding activity produced by herpesviruses. Cross-linking of (A) [125I]CCL3, (B, C and E) [125I]CXCL8 or (D) [125I]CXCL12 with EGS to medium from cultures uninfected (mock) or infected with the indicated viruses. The field isolates of EHV-1 are indicated with an identification number (E). Cross-linking with [125I]CXCL8 was also performed to supernatants and cell extracts (cell) from cultures infected with EHV-1 strain AB4 in the absence or presence of tunicamycin (Tm) (E). Samples were analysed by SDS–PAGE and autoradiography. Molecular masses are in kilodaltons and the positions of 125I-labelled chemokine (CK) and vCKBP–chemokine complexes (asterisks) are indicated.
Expression of vCKBP by EHV-1
The chemokine binding activity was conserved throughout a collection of EHV-1 field isolates, as well as the virulent laboratory strains AB4 and Army 183 (Turtinen and Allen, 1982; Smith et al., 1992) and the apathogenic vaccine strain RacH (Osterrieder et al., 1994) (Figure 1E). CXCL8 binding activity was detected in lysates of EHV-1-infected cells by cross-linking (Figure 1E). Supernatants and cell extracts from infections performed in the presence of tunicamycin lacked chemokine binding activity, suggesting that the vCKBP is N-glycosylated and that the lack of carbohydrate resulted in aberrant expression of the vCKBP or loss of activity (Figure 1E).
Chemokine binding specificity of naturally produced EHV-1 and BHV-1 vCKBP
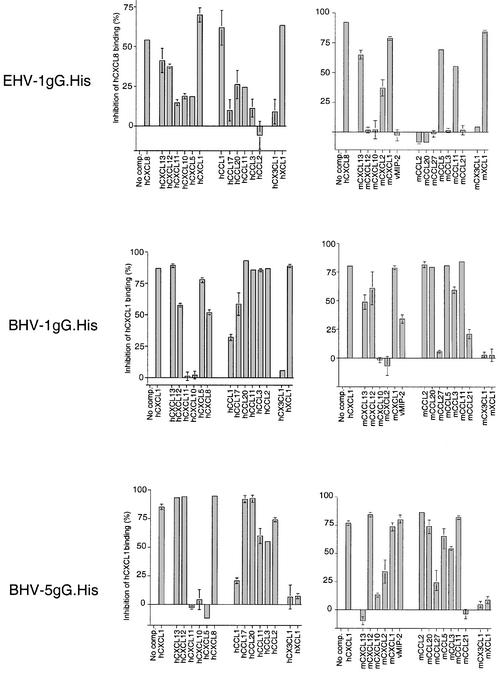
The chemokine binding specificity was also investigated in cross-linking assays in the presence of excess unlabelled chemokines. Figure 2A shows that the CXC chemokines of human or mouse origin and human XCL1 (lymphotactin) bound the EHV-1 strain AB4 vCKBP and competitively inhibited binding to [125I]CXCL8. Some inhibition was observed with human CCL3 and CCL5, but not with CCL2 or CX3CL1.
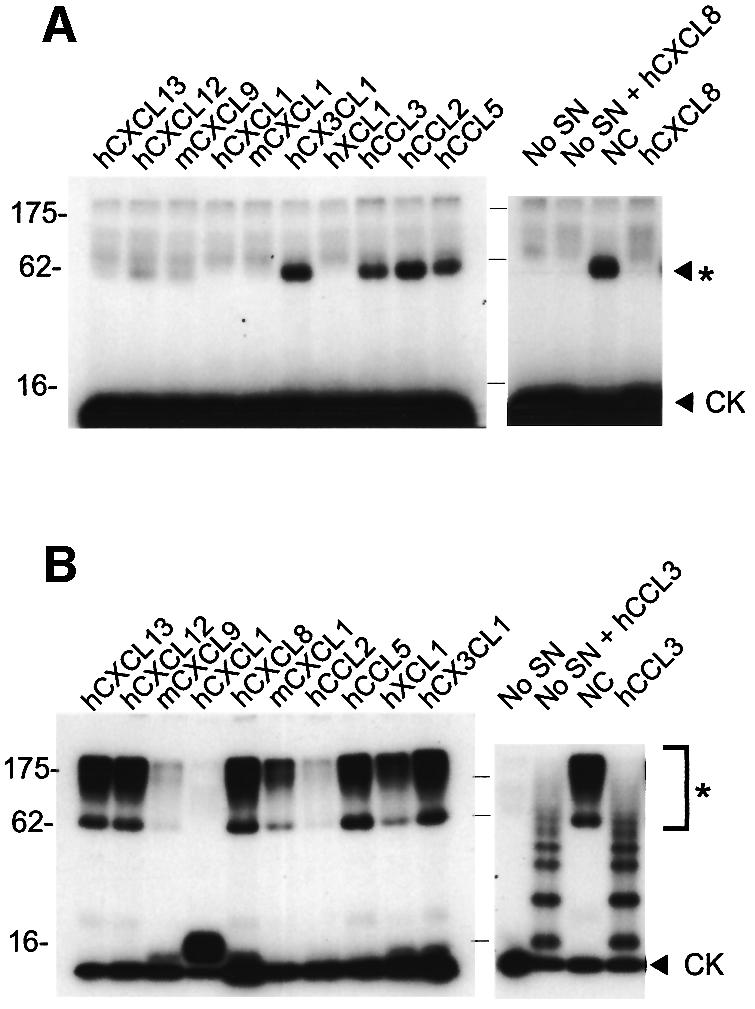
Fig. 2. Chemokine binding specificity of the vCKBP secreted from cells infected with EHV-1 or BHV-1. Cross-linking assay with EGS of (A) [125I]CXCL8 to supernatants from EHV-1 infections or (B) [125I]CCL3 to supernatants from BHV-1 infections in the absence (NC) or presence of a 2000-fold excess of the indicated unlabelled chemokines of human (h) or mouse (m) origin. Cross-linking to binding medium alone (No SN) was performed as a control. Samples were analysed by SDS–PAGE and autoradiography. Molecular masses are in kilodaltons and the positions of 125I-labelled chemokine (CK) and vCKBP-chemokine complexes (asterisks) are indicated.
A different binding pattern was observed with BHV-1 supernatants (Figure 2B). Addition of excess CXCL8 did not inhibit binding to [125I]CCL3, whereas a weak [125I]CXCL8–vCKBP complex was observed by direct cross-linking (Figure 1A), suggesting a much higher affinity for CCL3. Different degrees of inhibition, and hence of affinity, were observed with various CXC and CC chemokines. XCL1 partially inhibited CCL3 binding but no inhibition was observed with CX3CL1. The addition of unlabelled CCL3 resulted in the formation of multimers.
Identification of the secreted forms of gG as the vCKBP
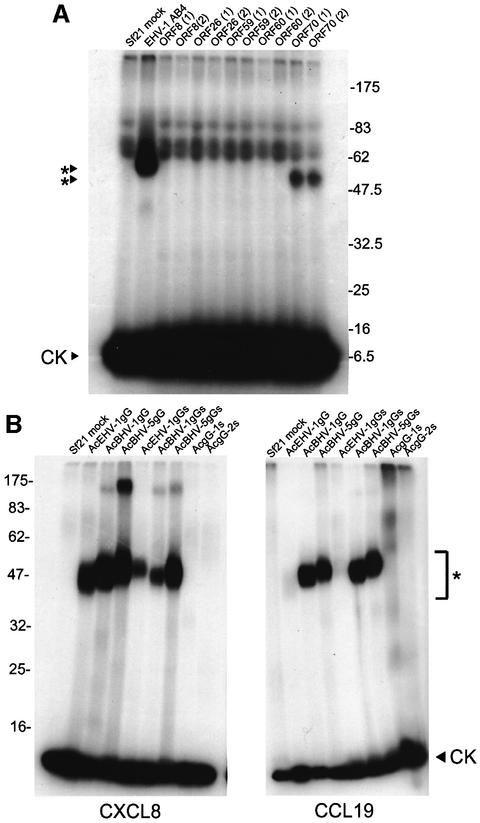
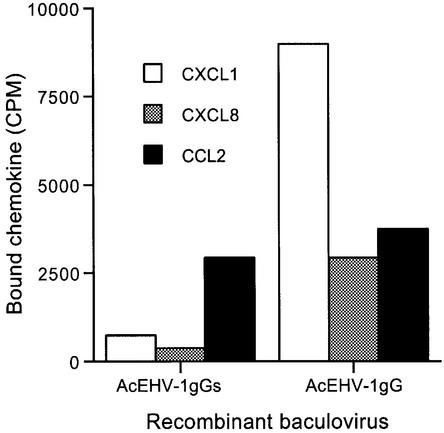
Open reading frames (ORFs) from the EHV-1 strain AB4 genome sequence (Telford et al., 1992) were selected based on the presence of a predicted signal peptide and/or a size consistent with that observed by cross-linking. EHV-1 ORF8, ORF26, ORF59, ORF60 and ORF70, which have unknown function, were expressed as full-length polypeptides in the baculovirus system. Figure 3A shows that a [125I]CXCL8–vCKBP complex of 50–55 kDa was detected in supernatants from cultures expressing ORF70 (AcEHV-1gG). The smaller size compared with that observed in natural infections (Figure 1A) is probably due to incomplete glycosylation in insect cells. EHV-1 ORF70 encodes gG, which is presumably anchored through the C-terminal transmembrane domain into the virus particle envelope and is also proteolytically cleaved and released into the medium (Crabb et al., 1992; Telford et al., 1992; Drummer et al., 1998). Insect cells can cleave the membrane form of gG to produce a secreted active protein.
Fig. 3. Identification of gG as the vCKBP encoded by alphaherpesviruses. (A) Cross-linking with EGS of [125I]CXCL8 to supernatants from insect cell cultures infected with recombinant baculoviruses expressing the indicated ORF from EHV-1 strain AB4. The baculovirus clone number is shown in parenthesis. (B) Cross-linking with EGS of [125I]CXCL8 or [125I]CCL19 to supernatants from insect cell cultures infected with the indicated recombinant baculoviruses expressing full-length or secreted versions (S) of gG encoded by EHV-1, BHV-1, BHV-5, HSV-1 and HSV-2. Samples were analysed by SDS–PAGE and autoradiography. Molecular masses are in kilodaltons and the positions of 125I-labelled chemokine (CK) and vCKBP-chemokine complexes (asterisks) are indicated.
To determine whether gG homologues from alphaherpesviruses encode chemokine binding activity, full-length and/or truncated versions, lacking the transmembrane and cytoplasmic domains and in the case of EHV-1, BHV-1 and BHV-5 fused to a C-terminal His6 tag, were expressed in the baculovirus system. Supernatants from infected cultures were tested in cross-linking assays to 125I-labelled CCL19 and CXCL8 (Figure 3B). A truncated version of EHV-1 gG was secreted and bound CXCL8 and, with lower affinity, CCL19. Full-length and truncated versions of gG encoded by both BHV-1 and BHV-5 were secreted and bound both CCL19 and CXCL8, confirming that gG from these viruses encodes the vCKBP. BHV-1 gG consists of a glycosylated form (gG; 65 kDa) and a complex glycoproteoglycan of higher molecular mass (gpgG; 90 to >240 kDa) which is modified by the addition of chondroitin sulfate (Keil et al., 1996), and this was consistent with chemokines cross-linking to a diffusely migrating species of proteins between 60 and >175 kDa (Figure 1A). These high molecular size forms were also observed after chemokine cross-linking with BHV-5, RanHV-1 and CerHV-1 samples, but not with CapHV-1. However, similar high molecular size forms were only weakly observed after cross-linking of recombinant forms of BHV-1 and BHV-5 gG to CXCL8, suggesting that the glycoproteoglycan form may be produced in very low amounts by insect cells. Expression of secreted versions of gG from HSV-1 (gG-1) and HSV-2 (gG-2) in the baculovirus system confirmed the absence of chemokine binding activity to these chemokines, consistent with the lack of activity in HSV-1- and HSV-2-infected cultures (not shown).
Chemokine binding affinity and specificity of purified recombinant gG encoded by EHV-1, BHV-1 and BHV-5
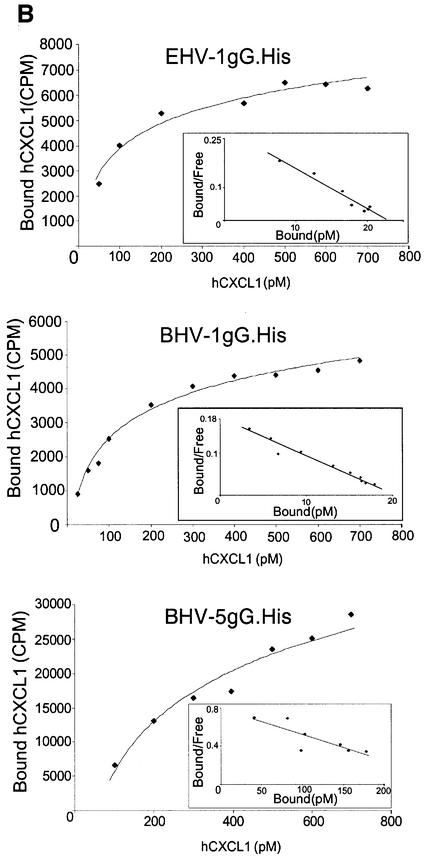
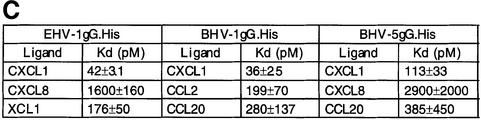
Recombinant secreted forms of gG fused to a C-terminal His6 tag and expressed in the baculovirus system were purified in nickel chelate columns (gG.His), and chemokine binding activity with human [125I]CXCL1 was demonstrated by cross-linking (Figure 4A). Chemokine binding affinity was determined with human [125I]CXCL1 using a scintillation proximity assay. Nickel chelate FlashPlates (Perkin-Elmer Life Sciences), containing a layer of scintillant in the interior of each well, were coated with purified gG.His. Increasing doses of [125I]CXCL1 were added and bound chemokine was determined in a scintillation counter. The affinity constants were determined by Scatchard analysis of the saturation curves (Figure 4B and C). Affinity constants for other chemokines were determined in competition assays of [125I]CXCL1 binding in the presence of increasing doses of unlabelled chemokines (Figure 4C). In all cases, the interaction of gG with chemokines was of high affinity.
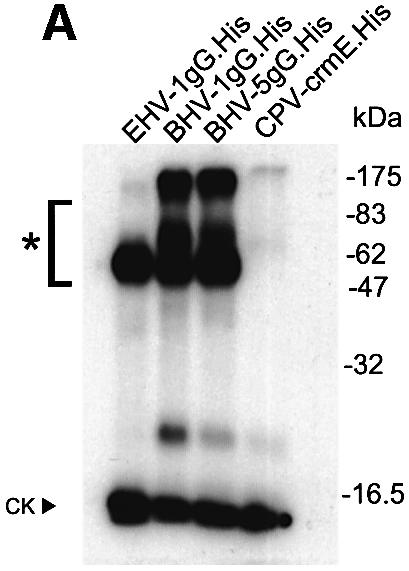
Fig. 4. Affinity constant of gG binding to chemokines. (A) Cross-linking with EGS of 1µg of purified gG.His from EHV-1, BHV-1 or BHV-5 to [125I]CXCL1. Purified His-tagged CrmE from cowpox virus (CPV-crmE.His) was a negative control. Samples were analysed by SDS–PAGE and autoradiography. Molecular masses are in kilodaltons and the positions of 125I-labelled chemokine (CK) and vCKBP-chemokine complexes (asterisks) are indicated. (B) Saturation curve and Scatchard analysis of [125I]CXCL1 binding to gG.His from EHV-1 (0.5 ng), BHV-1 (0.5 ng) and BHV-5 (5 ng). Nickel chelate FlashPlates precoated overnight with purified gG.His were incubated in triplicate with the indicated doses of [125I]CXCL1. (C) Affinity constants for the interaction of gGs with chemokines. The data for CXCL1 were derived from Scatchard analysis of the saturation curves shown in (B), and those for the other chemokines from competition experiments of binding of [125I]CXCL1 in the presence of increasing doses of unlabelled chemokine. The data were analysed using the Ligand program.
Binding of [125I]chemokines to purified gG.His in the presence of a 500-fold excess of unlabelled chemokines was determined in the FlashPlate assay. This provided a broad spectrum of binding specificity for 30 chemokines of human, mouse or viral origin (Figure 5). However, we cannot rule out the possibility that those chemokines that do not competitively inhibit the binding of [125I]chemokines may bind to a different site in gG. Purified gG.His from BHV-1 and BHV-5 bound to most human and mouse CC chemokines, and to some human and mouse CXC chemokines. In contrast, EHV-1 gG.His showed a narrower binding specificity for both human and mouse CC and CXC chemokines. EHV-1 gG.His bound to human and mouse XCL1, whereas BHV-1 gG.His bound only to human XCL1 and BHV-5 gG.His did not bind to the C chemokine. None of the recombinant proteins bound to CX3CL1. BHV-5 gG.His, and to a lesser extent BHV-1 gG.His, interacted with vMIP-2 encoded by Kaposi’s sarcoma-associated herpesvirus.
Fig. 5. Binding specificity of gG encoded by EHV-1, BHV-1 and BHV-5. Nickel chelate FlashPlates were coated with gG.His from EHV-1, BHV-1 or BHV-5. Three hundred picomolar iodinated human CXCL8 (EHV-1) or CXCL1 (BHV-1 and BHV-5) was added in the absence or presence of a 500-fold excess of unlabelled competitor CC, CXC, C or CX3C chemokines of human (h) or mouse (m) origin, or vMIP-2. Bars represent the mean percentage inhibition of binding ± SD of the triplicate sample.
The binding specificity of recombinant gG.His in the FlashPlate assay (Figure 5) was generally consistent with that obtained by cross-linking to gG secreted from EHV-1- and BHV-1-infected cells (Figure 2). The only exception was with human CXCL8, CXCL12 and CXCL13, which inhibited [125I]CXCL1 binding to recombinant BHV-1 gG.His but did not block the cross-linking of [125I]CCL3 to BHV-1-produced gG. An apparent higher affinity of BHV-1 for CC chemokines than for CXC chemokines (Figure 1A, B and D) may explain the lack of competition when using [125I]CCL3.
Inhibition of chemokine binding to cells by gG
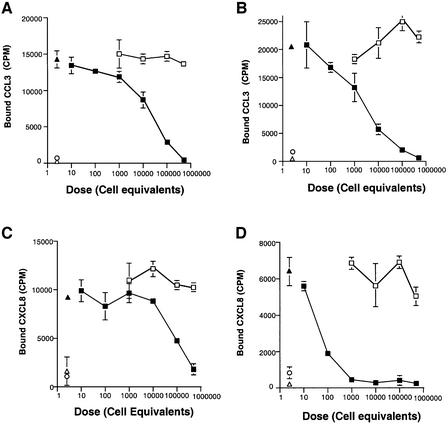
Full-length naturally cleaved recombinant gG from EHV-1 and BHV-1 expressed in the baculovirus system inhibited the specific binding of [125I]CXCL8 or [125I]CCL3 to U937 cells in a dose-dependent manner (Figure 6). This suggested that untagged versions of gG have a high affinity for chemokines, sufficient to block the binding of chemokines to high affinity cellular receptors. Purified recombinant EHV-1 gG.His also blocked the binding of CXCL8 to U937 cells (not shown). Recombinant secreted forms of HSV-1 and HSV-2 gG did not inhibit CCL3 binding to cells, confirming the absence of activity (not shown).
Fig. 6. Secreted gG from EHV-1 and BHV-1 expressed in the baculovirus system inhibits chemokine binding to cells in a dose-dependent manner. Binding assay of [125I]CCL3 and [125I]CXCL8 to U937 cells in the absence (solid triangles) or presence of increasing amounts of supernatants from Sf21 cells infected with recombinant baculovirus expressing full-length gG from either EHV-1 (B and D) or BHV-1 (A and C) (solid squares), or with control baculovirus (AcNPV) (open squares). The dose of supernatant is expressed as cell equivalents. Binding specificity was determined in the presence of a 500-fold excess of unlabelled CCL3 or CXCL8 (open circles). Purified M3 protein was used as a positive control (open triangles). Binding of chemokines is expressed as the mean ± SD of triplicate assays.
Inhibition of the biological activity of chemokines by gG
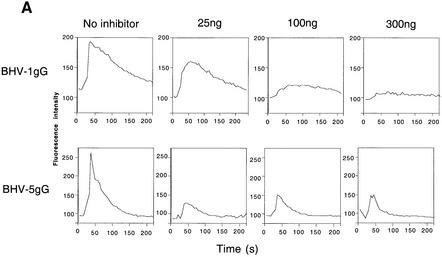
Figure 7A shows that purified BHV-1 gG.His inhibited, in a dose-dependent manner, calcium mobilization induced by human CXCL1 in U937 cells. Similarly, purified BHV-5 gG.His inhibited calcium mobilization in HeLa cells expressing CCR5 and CXCR4 in response to human CCL3 (Figure 7A).
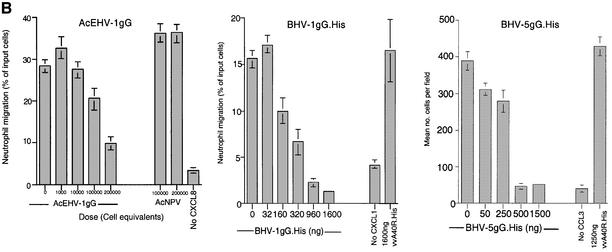
Fig. 7. Inhibition of chemokine biological activity by gG. (A) Chemokine-induced calcium flux. The indicated amount of purified BHV-1 gG.His was pre-incubated with 75 ng of CXCL1 before addition to Indo-1-loaded U937 cells. Similarly, purified BHV-5 gG. His was pre-incubated with 75 ng of CCL3 before addition to Indo-1-loaded HeLa cells transfected with CCR5 and CXCR4. Intracellular calcium mobilization was measured by FACS analysis. (B) Chemokine-induced chemotaxis in a Transwell migration assay. The top panel shows the migration of human neutrophils in response to 100 ng/ml CXCL8 in the presence of supernatants, expressed as cell equivalents, from insect cells infected with a recombinant baculovirus expressing full-length EHV-1 gG. The middle panel shows chemotaxis of human neutrophils induced by 100 ng/ml CXCL1 in the presence of increasing doses of purified BHV-1 gG.His. The bottom panel shows migration of U937 cells in response to 100 ng/ml CCL3 in the presence of purified BHV-5 gG.His. Supernatants from AcNPV-infected insect cells or an irrelevant purified His-tagged VV protein (A40R.His) were used as negative controls. Leukocytes migrating in duplicate assays are expressed as the percentage (mean ± SD) of the total cells added or as the number of cells counted (mean ± SD) in four high powered fields.
Cell migration in response to chemokine gradients in vitro, a measure of their ability to induce cell infiltration into tissues, was also inhibited by gG (Figure 7B). Neutrophil migration in response to CXCL8 or CXCL1 was specifically inhibited in a dose-dependent manner by supernatants containing recombinant EHV-1 gG or by purified BHV-1 gG.His, respectively. Similarly, purified BHV-5 gG.His specifically inhibited CCL3-induced chemotaxis in differentiated U937 cells pretreated with IFN-α.
gG can alter the heparin binding characteristics of chemokines
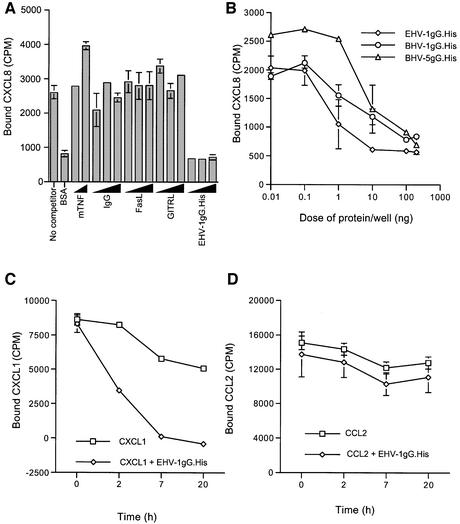
Figure 8B shows that pre-incubating purified gG.His from EHV-1, BHV-1 and BHV-5 with [125I]CXCL8 prior to addition to FlashPlates precoated with heparin–bovine serum albumin (BSA) greatly reduced binding of the chemokine to heparin. No effect was observed with several control proteins, some of which expressed a His tag (Figure 8A). Moreover, gG also disrupted pre-established chemokine–GAG interactions. FlashPlates coated with heparin–BSA were pre-incubated with [125I]CXCL1, and addition of EHV-1 gG.His efficiently detached [125I]CXCL1 from heparin–BSA over time (Figure 8C). In contrast, no effect was observed on the binding of [125I]CCL2, a chemokine not recognized by EHV-1 gG.His, to heparin–BSA (Figure 8D).
Fig. 8. Inhibition of chemokine–heparin interactions by purified gG. (A and B) FlashPlates precoated with heparin–BSA were incubated with [125I]CXCL8 in the absence or presence of purified gG from EHV-1 (EHV-1gG.His), BHV-1 (BHV-1gG.His) or BHV-5 (BHV-5gG.His), and the bound radio-activity (mean ± SD of duplicate samples) was determined. FlashPlates coated with BSA alone (BSA) were also tested to determine background binding. Increasing doses (10, 50 and 150 ng per well) of murine TNF (mTNF), human IgG, His-tagged Fas ligand (FasL) or His-tagged glucocorticoid-induced TNFR-superfamily-related protein ligand (GITRL) were also tested. (C and D) FlashPlates precoated with heparin–BSA were pre-incubated with [125I]CXCL1 or [125I]CCL2. Purified recombinant EHV-1 gG (300 ng per well) or medium was added to the wells and the bound radiolabelled chemokine (mean ± SD of triplicate samples) was determined at the indicated times. Background binding to BSA alone was subtracted.
gG encodes chemokine binding activity at the cell surface
To determine whether the membrane form of gG also binds chemokines, insect cells infected with baculoviruses expressing the full-length gG (AcEHV-1gG) or a truncated form lacking the C-terminal transmembrane domain of EHV-1 gG (AcEHV-1gGs) were tested. The ability of the surfaces of cells infected with AcEHV-1gG to bind human CXCL1 and CXCL8 was clearly increased compared with cells infected with AcEHV-1gGs (Figure 9). No difference was observed in the binding to CCL2, a chemokine not recognized by EHV-1 gG. Chemokine binding activity was observed in media from cells infected with either recombinant (Figure 3B; data not shown).
Fig. 9. Chemokine binding activity expressed at the surface of recombinant baculovirus-infected insect cells. Sf21 insect cells were infected with AcEHV-1gG or AcEHV-1gGs for 24 h and incubated for 30 min at room temperature with the indicated chemokines in suspension. The bound chemokine was determined. The binding difference between AcEHV-1gG and AcEHV-1gGs was observed three times for CXCL8 and twice for CXCL1.
Discussion
This paper describes the identification and characterization of a family of novel soluble vCKBPs (Table I). We demonstrate that secreted forms of gG encoded by several alphaherpesviruses bind chemokines with high affinity. Baculovirus-expressed gGs from EHV-1, BHV-1 and BHV-5 bind some of the CC, CXC and C chemokines, but not CX3CL1. Biological assays confirmed that gG inhibits signal transduction and migration induced by both CC and CXC chemokines. Remarkably, the alphaherpesvirus vCKBP blocks the interaction of chemokines with both specific receptors and GAGs. We also identified gG as the first vCKBP that interacts with chemokines both at the cell surface and in solution.
Table I. Comparison of vCKBPs encoded by different viruses.
| Class | Virus and protein | Chemokine binding specificity | Mechanism of action |
|---|---|---|---|
| vCKBP-1 | MV M-T7 | CC, CXC, C | Secreted, blockade of chemokine–GAG interactions |
| vCKBP-2 | MV M-T1VV 35 kDa | CC | Secreted, blockade of chemokine–receptor interactions |
| CPV CCI | |||
| vCKBP-3 | MHV-68 M3 | CC, CXC, C, CX3C | Secreted, blockade of chemokine–receptor and chemokine–GAG interactions |
| vCKBP-4 | Alphaherpesvirus isoforms of gG | CC, CXC, C | Secreted and membrane bound, blockade of chemokine–receptor and chemokine–GAG interactions |
CCI, chemokine inhibitor; GAG, glycosaminoglycan; gG, glycoprotein G; MHV-69, murine gammaherpesvirus 68; MV, myxoma virus; vCKBP, viral chemokine binding protein; VV, vaccinia virus.
Many viral molecules that modulate chemokine activity have been described in beta- and gammaherpesviruses (Lalani et al., 2000; Murphy, 2001), but a biologically active CXCL8 homologue encoded by MDV is the only such molecule found in alphaherpesviruses to date (Parcells et al., 2001). We show that expression of soluble vCKBPs is a strategy encoded by many alphaherpesviruses to block chemokine activity. vCKBPs were initially described in the poxvirus family, and the gammaherpesvirus MHV-68 M3 protein was the only one previously identified in the herpesvirus family (Parry et al., 2000; van Berkel et al., 2000).
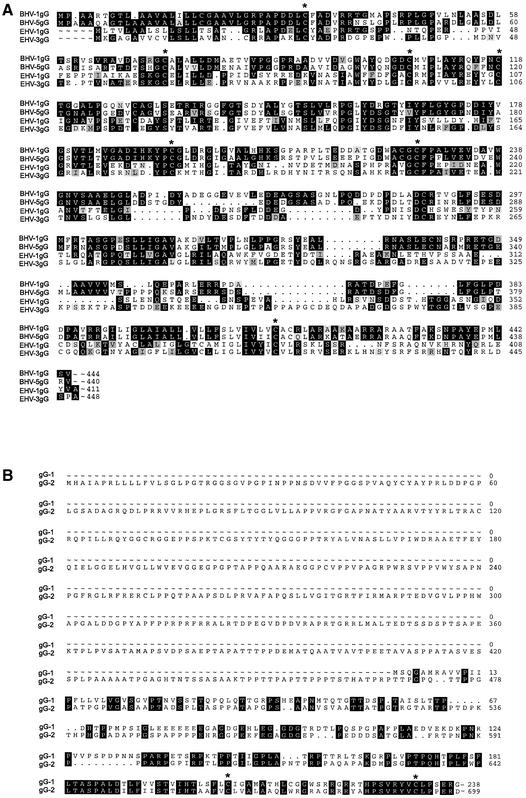
gG is unique to alphaherpesvirus, with no sequence similarity to known cellular proteins or previously identified vCKBPs (Figure 10), suggesting that gG interacts with chemokines in a different way. Expression of secreted gG provides a mechanism to sequester chemokines and to neutralize their effect by blocking chemokine–receptor interaction and activation of cell migration.
Fig. 10. Amino acid sequence similarity among members of the gG family. Sequence comparisons (A) among gGs encoded by the equine and bovine herpesviruses and (B) between gGs encoded by HSV-1 and HSV-2. Regions of identity are shown in black boxes and regions of high similarity are shown in shaded boxes. Conserved cysteine residues are marked with an asterisk. DDBJ/EMBL/GenBank Accession Nos are EHV-1 gG (p28967), BHV-1 (o39504), BHV-5 (x99755), EHV-3 (o90421), HSV-1 (p06484) and HSV-2 (p13290).
Interestingly, we found that gG blocks the interaction of chemokines with heparin. Recent experiments show that EHV-1 and BHV-1 gG interfere with the interaction of chemokines with complex GAGs at the surface of CHO-K1 cells (N.A.Bryant and A.Alcami, unpublished data). Chemokines are retained at the surface of endothelial cells by interacting with GAGs, an interaction critical for correct chemokine presentation to the passing leukocyte (Middleton et al., 1997; Cinamon et al., 2001). The chemokine domains that interact with receptors or GAGs may be different and have been well defined for some chemokines (Proudfoot et al., 2001). Thus, the alphaherpesvirus vCKBP may mask both receptor and GAG-binding sites in the chemokine, or alternatively may induce a conformational change in the chemokine. Future studies will identify the chemokine domains involved in the interaction with gG. We have not been able to detect direct binding of purified gG to heparin–Sepharose colums (N.A.Bryant and A.Alcami, unpublished data). MV M-T7 binds to the GAG binding domain of CXCL8 and has been proposed to block chemokine–GAG interactions (Lalani et al., 1997) (Table I). The poxvirus 35 kDa/M-T1 and gammaherpesvirus M3 proteins block the interaction of chemokines with their receptors (Smith et al., 1997; Alcami et al., 1998; Parry et al., 2000). This novel property of gG of blocking both receptor and GAG binding should enhance its ability to inhibit chemokine function. gG can also displace chemokine once it has bound to heparin, suggesting that it may disrupt established chemokine gradients. Recently, we have found that MHV-68 M3 also disrupts chemokine–GAG interactions (L.M.C.Webb, V.P.Smith, I.Clark-Lewis and A.Alcami, unpublished data).
The diversity of chemokine binding specificities among gGs from different viruses is probably related to host cell and tissue tropism and the requirement to block different subsets of chemokines in vivo. We have only been able to test human and mouse chemokines in these studies, but these are not the natural hosts of EHV-1, BHV-1 and BHV-5. However, species cross-reactivity exists among chemokines and their receptors, and thus the overall binding specificity of gGs for chemokine from natural hosts is likely to be similar. In contrast to previously identified vCKBPs, the finding of a family of vCKBPs with different chemokine specificities suggests that gG may provide the molecular scaffolding for designing soluble chemokine inhibitors with different specificities. The broad binding specificity of gGs, together with their ability to interfere with the interaction of chemokines with both specific receptors and GAGs, may be advantageous to inhibit chemokine-mediated inflammatory reactions. This family of vCKBPs have the potential of being used, like other virus-encoded chemokine inhibitors (Murphy, 2000), as therapeutic reagents in immune-related diseases.
A unique feature not found previously in vCKBPs is that gG is a membrane protein, anchored by a C-terminal transmembrane domain, which may be secreted after proteolytic cleavage. Secretion of gG has been demonstrated during infections with EHV-1, EHV-4, BHV-1, BHV-5 and HSV-2 (Marsden et al., 1984; Su et al., 1987; Crabb et al., 1992; Engelhardt and Keil, 1996; Keil et al., 1996; Drummer et al., 1998). The finding of chemokine binding activity secreted from cells infected with EHV-3, RanHV-1, CapHV-1 and CerHV-1 suggests that gG from these viruses is also proteolytically cleaved and secreted. Inhibition of N-linked glycosylation via tunicamycin blocked EHV-1 expression of chemokine binding activity, consistent with previous evidence that EHV-4 gG expression is greatly reduced and no detectable gG is secreted in the presence of tunicamycin (Drummer et al., 1998).
When expressed naturally, BHV-1 and BHV-5 gG is secreted as a 65 kDa polypeptide and a diffusely migrating species of 90–240 kDa (gpgG), which consists of the 65 kDa gG linked to GAGs (Engelhardt and Keil, 1996; Keil et al., 1996). Chemokines may interact with the GAG present in BHV-1 and BHV-5 gG, but they bind to gG itself since cross-linking to chemokines was observed for the 65 kDa species. The high molecular size species were identified by chemokine cross-linking in supernatants from cells infected with RanHV-1 and CerHV-1, but not with CapHV-1. The secretion of EHV-1 and EHV-4 gG as a 120 kDa disulfide-linked homodimer was reported (Drummer et al., 1998), but we have not detected complexes of [125I]chemokines with dimeric EHV-1 gG after cross-linking and SDS–PAGE in the absence of reducing agents (not shown).
Expression of gG on the surface of virus particles has been observed for EHV-1, EHV-4, HSV-1 and HSV-2 (Richman et al., 1986; Su et al., 1987; Drummer et al., 1998). We show chemokine binding activity of recombinant EHV-1 gG when expressed at the surface of insect cells. Membrane-bound EHV-1 gG expressed from infected cells may function as a decoy receptor preventing the interaction of chemokines with cellular chemokine receptors and subsequent signal transduction. It is tempting to speculate that the chemokine binding activity of gG incorporated into the EHV-1 virion envelope may mediate initial virus attachment to cell surfaces presenting chemokines, and play a role in determining cell and tissue tropism in vivo. Further studies will determine whether the chemokine binding activity of EHV-1 gG confers an advantage for virus attachment and replication.
No chemokine binding activity has yet been associated with HSV-1, HSV-2 or EHV-4 gGs, despite the latter having 72.7% amino acid identity with EHV-1 gG (Telford et al., 1998). It is not possible to predict from sequence information whether a particular gG will bind chemokines (Figure 10). We have used laboratory strains of HSV-1 and HSV-2, and gG from clinical isolates may have different properties. However, this is unlikely considering the high degree of conservation of the gG gene (Rekabdar et al., 1999; Liljeqvist et al., 2000). gG from these viruses may bind chemokines different to those included in this study (CCL1, CCL2, CCL3, CCL11, CCL17, CCL20, CXCL1, CXCL8, CXCL10, CXCL12 and CX3CL1). Membrane-bound gGs from HSV-1 and HSV-2 were not tested for chemokine binding, but it is unlikely that the secreted forms tested here will have lost the binding properties of the membrane-bound form. Alternatively, the chemokine binding activity of gG may have been lost from these viruses during evolution. It is notable that HSV-1 gG is significantly shorter that other alphaherpesvirus gGs and is not secreted into the medium of infected cell cultures (Richman et al., 1986). Moreover, the gene encoding gG is not found in VZV or MDV, suggesting that some alphaherpesviruses do not require this function for successful replication in the host. Three VZV strains, including a clinical isolate, lack the gG gene (Gomi et al., 2002) and we have been unable to amplify by PCR the gG gene from two clinical isolates (N.A.Bryant and A.Alcami, unpublished data). MDV encodes a chemokine homologue and hence subverts the chemokine signalling pathways in an alternative way.
A role for BHV-1 gG in viral infection in vivo has been reported. Deletion of the gene encoding gG from BHV-1 caused viral attenuation in calves and the mutant virus was more immunogenic (Kaashoek et al., 1998). The lack of chemokine binding activity encoded by BHV-1 gG may explain the observed phenotype in vivo. Others have suggested that BHV-1 gG facilitates viral cell-to-cell spread (Nakamichi et al., 2002). In the case of HSV-1, a gG mutant showed no phenotype in vitro and marginal attenuation in the mouse ear model (Weber et al., 1987; Balan et al., 1994), and another study implicated HSV-1 gG in virus entry through apical surfaces of polarized epithelial cells (Tran et al., 2000). Thus, HSV-1 gG may have other functions unrelated to chemokine binding. However, it should be noted that certain functions ascribed to gG from the study of such deletion mutants may need to be re-assessed in the light of evidence that disruption of the upstream gene US3, a protein kinase homologue, can occur with gG deletion mutants of pseudorabies virus, an alphaherpesvirus of swine (Demmin et al., 2001).
In conclusion, we report the identification and characterization of a novel family of viral proteins that interact with chemokines. These findings provide insights into the interaction of viruses with the chemokine network and new strategies of immune modulation.
Materials and methods
Reagents
Radio-iodinated recombinant human CCL1, CCL2, CCL3, CCL11, CCL17, CCL19, CCL20, CXCL1, CXCL8, CXCL10, CXCL12 and CX3CL1 (2200 Ci/mmol) were from Perkin-Elmer Life Sciences (Boston, MA). Recombinant human CCL2, CCL3, CXCL1, CXCL8 and CXCL12, and murine CCL11, CXCL10, CXCL13 (B-cell-attracting chemokine 1), XCL1, CX3CL1 and TNF were from R&D Systems (Minneapolis, MN). Recombinant human CCL1, CCL5 (regulated upon activation, normal T-cell expressed and secreted), CCL11, CCL17, CCL20, CXCL5 (epithelial cell-derived protein 78), CXCL10, CXCL11 (IFN-inducible T-cell-α chemoattractant), CXCL13, XCL1, CX3CL1 and vMIP-2, and murine CCL2, CCL5, CCL20, CCL21, CCL27, CXCL1, CXCL2 (MIP-2), CXCL9 (monokine induced by IFN-γ) and CXCL12 were from PeproTech (Rocky Hill, NJ). Recombinant His-tagged human GITRL and FasL were from R&D Systems. Human IgG was from Sigma.
Cells and viruses
Human neutrophils were isolated from blood samples in Lymphoprep gradients (Baly et al., 1997). The EHV-1 strains AB4, V592, RacH, Army 183 and those described in Figure 1E and EHV-3 were grown in equine embryonic lung cells. EHV-4 strain MD was grown in equine embryonic kidney cells. Primary equine cells were derived from lung or kidney of an aborted fetus via enzymatic digestion, and recovered cells were passaged in 10% fetal calf serum (FCS) MEM. MDBK were infected with BHV-1 (strain Jura), BHV-4 (strain V. test), BHV-5 (strain N569), RanHV-1 (strain CvHV-2), CapHV-1 (strain E-CH) and CerHV-1 (strain CvHV-1). HSV-1 strain SC16 and HSV-2 strain HVD were grown in Vero cells. VZV was provided by A.Davison (Institute of Virology, Glasgow, UK) and grown in human MRC-5 cells. VV strain Lister and VV Lister Δ35K were grown in BSC-1 cells (Alcami et al., 1998).
Construction of recombinant baculoviruses
The gG gene was PCR amplified from infected cell DNA using Pfu polymerase and specific oligonucleotides (Table II), and cloned into pBAC-1 (Novagen). Except for the expression of EHV-1 candidate genes, the nucleotide sequence of the genes was determined. Autographa californica nuclear polyhedrosis virus (AcNPV) recombinants were constructed as described previously (Alcami et al., 1998). Full-length and secreted forms of gG with and without a C-terminal His6 tag were produced in Spodoptera frugiperda (Sf) 21 insect cells infected with recombinant baculoviruses.
Table II. Oligonucleotides used for PCR amplification of gG from alphaherpesviruses and expression in the baculovirus system.
| Virus | DDBJ/EMBL/GenBank accession No. | His6 tag | Protein form | Primers |
|---|---|---|---|---|
| EHV-1 | M86664 | No | Full length | 5′-CGCGAATTCATGTTGACTGTCTTAGCAGC-3′ |
| (411 amino acids) | 5′-CGCAAGCTTTTAAGCAACGTACTCAAGTC-3′ | |||
| C-terminal | Secreted form | 5′-CGCGAATTCATGTTGACTGTCTTAGCAGC-3′ | ||
| (355 amino acids) | 5′-CGCAAGCTTGTGTGTACTGTCATCGCTGTT-3′ | |||
| BHV-1 | AJ004801 | No | Full length | 5′-ACCGGATCCATGCCTGCCGCCCGGACCGG-3′ |
| (444 amino acids) | 5′-TCCAAGCTTTCAGACGCTGAGCATCGGCT-3′ | |||
| C-terminal | Secreted form | 5′-ACCGGATCCATGCCTGCCGCCCGGACCGG-3′ | ||
| (391 amino acids) | 5′-CCGGCGGCCGCGCCAAAGAGCCCAAACTCCG-3′ | |||
| BHV-5 | X99755 | No | Full length | 5′-CATGGATCCATGCCCGCCGCCGCTCAAGC-3′ |
| (440 amino acids) | 5′-TCCAAGCTTCTAGACGCGGAGCATGGGCT-3′ | |||
| C-terminal | Secreted form | 5′-CATGGATCCATGCCCGCCGCCGCTCAAGC-3′ | ||
| (396 amino acids) | 5′-CTCGCGGCCGCGCGCCGCGTGGCGGGATCAG-3′ | |||
| HSV-1 | X14112 | No | Secreted form | 5′-ACGGAATTCATGTCGCAGGGCGCCATGCG-3′ |
| (181 amino acids) | 5′-CTCAAGCTTCTATCGTCCCTTTGAGGTGAGTC-3′ | |||
| HSV-2 | Z86099 | No | Secreted form | 5′-CTCGAATTCATGCAGCCATCGCTCCCAG-3′ |
| (471 amino acids) | 5′-CCTAAGCTTCTATGCGGAGGCGCTGCTGGTG-3′ |
The full-length EHV-1 AB4 gG ORF sequence was identical to the published sequence (Telford et al., 1992), but the truncated version had an amino acid substitution V208A (T623C at the nucleotide level). Full-length and secreted forms of the BHV-1 (strain Jura) (Metzler et al., 1986) gG showed an E331K substitution (G991A) when compared with the Cooper strain (DDBJ/EMBL/GenBank Accession No. AJ004801). Full-length and secreted BHV-5 (strain N569) (French, 1962) gG had five amino acid substitutions compared with the published sequence: V157L (C580G, G581C), E220D (C660G), P221A (G661C), A266G (C797G) and A392T (G1174A). The sequences of the secreted forms of gG from HSV-1 strain SC16 and HSV-2 strain HVD were identical to the published sequences from HSV-1 strain 17 and HSV-2 strain HG52, respectively (McGeoch et al., 1985; Dolan et al., 1998).
Preparation of medium for binding and biological assays
FCS-free supernatants from cultures infected with 5–10 plaque-forming units per cell were harvested 2–3 days post-infection and prepared as described previously (Alcami et al., 1998). Infected cells were cultured for 24 h with tunicamycin (5 µg/ml; Sigma) when indicated. Infectious virus in the supernatants was inactivated with 4,5′,8-trimethylpsoralen (Sigma) and exposure to UV radiation, and concentrated and dialysed against phosphate-buffered saline (PBS) (Alcami et al., 1998).
Protein purification
Recombinant gG.His fusion proteins were purified on nickel–Sepharose columns (Pharmacia) from supernatants of baculovirus-infected Hi5 insect cells grown in EX-Cell 405 (JRH Biosciences). Purified His-tagged proteins were visualized by SDS–PAGE and Coomassie Blue staining, and were quantified by protein assay (Bio-Rad).
Chemokine cross-linking assay
The binding medium was RPMI containing 20 mM HEPES pH 7.4 and 0.1% BSA. Cross-linking experiments with BS3 (Pierce), EGS (Sigma) or EDC (Sigma) to [125I]chemokines (0.4 nM) were performed as described previously (Alcami et al., 1998). The amount of medium used was equivalent to 5–7 × 104 cells. Samples were analysed by 12% SDS–PAGE. Purified His-tagged CrmE, a TNFR homologue encoded by cowpox virus (Saraiva and Alcami, 2001), was provided by M.Saraiva and used as a control in some experiments.
Chemokine binding to cells
To determine chemokine binding to U937 cells, samples were pre-incubated with 100 pM [125I]chemokine for 1 h at 4°C, and U937 cells were added and incubated for 2 h at 4°C. Purified M3 protein was provided by C.Bunce and M.Wilson (Xenova Research Ltd, Cambridge, UK). To determine the binding of chemokines to Sf21 cells, 2 × 106 cells were scraped and incubated in suspension with 200 pM [125I]chemokines for 30 min at room temperature. Bound chemokine was determined by phthalate oil centrifugation (Alcami et al., 1998).
Determination of chemokine binding specificity and affinity constants
Nickel-chelate-coated FlashPlates (Perkin-Elmer Life Sciences) were used. Purified His-tagged protein (0.5–10 ng/well) was incubated overnight in 0.1% BSA in PBS, washed and incubated at room temperature for 1–2 h with increasing doses of [125I]chemokine in the absence or presence of excess unlabelled chemokine. Bound [125I]chemokine was determined in a Packard TopCount Microplate Scintillation Counter. Non-specific binding, determined in the presence of 100-fold excess unlabelled chemokine, was subtracted and binding data were analysed by the Ligand program (Munson and Rodbard, 1980).
Inhibition of chemokine binding to heparin
Basic FlashPlates were coated with heparin–BSA (Sigma) (2.5 µg/well) in 10 mM phosphate buffer pH 7.0. The wells were washed, blocked for 2 h with phosphate buffer containing 2% BSA and washed again. Iodinated chemokine (400 pM), pre-incubated for 1 h at room temperature with purified gG or control proteins, was then added to the well. Plates were read after overnight incubation at 4°C. Time course experiments were conducted as above except that [125I]chemokine was bound to immobilized heparin–BSA overnight before the addition of purified proteins. Background binding was measured by [125I]chemokine binding to wells treated only with 2% BSA in binding buffer.
Calcium mobilization assay
U937 cells or HeLa cells transfected with CCR5 and CXCR4 (B7), provided by J.Alcami (Instituto Carlos III, Madrid, Spain), were loaded with 3 µM Indo-1/acetoxymethyl ester in 0.1% FCS RPMI for 30 min at 37°C. Cells were washed in 0.1% FCS HBSS and 105 cells were stimulated with 75 ng of CCL3 or CXCL1, alone or pre-incubated for 30 min with purified protein. Calcium mobilization was analysed on a FACScan (Becton Dickinson).
Chemotaxis assay
Twenty-four-well Transwell plates (Costar) were used. Chemokine diluted in 0.1% FCS RPMI was placed in the lower compartment and 5 × 105 primary neutrophils or 106 IFN-α-treated U937 cells (Zella et al., 1999) were placed in the upper chamber, separated by a 5 µm pore size filter. After incubating the neutrophils for 2 h and the U937 cells for 6 h at 37°C, the cells in the lower chamber were counted by microscopy. The purified His-tagged VV protein A40R.His expressed in Escherichia coli, used as a control, was provided by V.Smith (Cambridge University, Cambridge, UK).
Acknowledgments
Acknowledgements
We thank L.M.C.Webb for advice and critical reading of the manuscript, and R.Hicks for help with the calcium mobilization assay. This work was supported by the Wellcome Trust (grant no. 051087/Z/97/Z) and a Concerted Action Awards Program of the French Community of Belgium (ARC 98/03-220). N.A.B. is funded by a BBSRC Studentship, N.D.-P. is supported by a Tetra Laval Senior Research Fellowship, A.V. is a Senior Research Associate of the Fonds National Belge de la Recherche Scientifique and A.A. is a Wellcome Trust Senior Research Fellow.
References
- Alcami A. and Koszinowski,U.H. (2000) Viral mechanisms of immune evasion. Immunol. Today, 21, 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A., Symons,J.A., Collins,P.D., Williams,T.J. and Smith,G.L. (1998) Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. J. Immunol., 160, 624–633. [PubMed] [Google Scholar]
- Baggiolini M. (1998) Chemokines and leukocyte traffic. Nature, 392, 565–568. [DOI] [PubMed] [Google Scholar]
- Balan P., Davis-Poynter,N., Bell,S., Atkinson,H., Browne,H. and Minson,T. (1994) An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol., 75, 1245–1258. [DOI] [PubMed] [Google Scholar]
- Baly D., Gibson,U., Allison,D. and DeForge,L. (1997) Biological assays for C-X-C chemokines. Methods Enzymol., 287, 69–88. [DOI] [PubMed] [Google Scholar]
- Cinamon G., Shinder,V. and Alon,R. (2001) Shear forces promote lymphocyte migration across vascular endothelium bearing apical chemokines. Nat. Immunol., 2, 515–522. [DOI] [PubMed] [Google Scholar]
- Crabb B.S., Nagesha,H.S. and Studdert,M.J. (1992) Identification of equine herpesvirus 4 glycoprotein G: a type-specific, secreted glycoprotein. Virology, 190, 143–154. [DOI] [PubMed] [Google Scholar]
- Demmin G.L., Clase,A.C., Randall,J.A., Enquist,L.W. and Banfield,B.W. (2001) Insertions in the gG gene of pseudorabies virus reduce expression of the upstream Us3 protein and inhibit cell-to-cell spread of virus infection. J. Virol., 75, 10856–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan A., Jamieson,F.E., Cunningham,C., Barnett,B.C. and McGeoch,D.J. (1998) The genome sequence of herpes simplex virus type 2. J. Virol., 72, 2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummer H.E., Studdert,M.J. and Crabb,B.S. (1998) Equine herpesvirus-4 glycoprotein G is secreted as a disulphide-linked homodimer and is present as two homodimeric species in the virion. J. Gen. Virol., 79, 1205–1213. [DOI] [PubMed] [Google Scholar]
- Engelhardt T. and Keil,G.M. (1996) Identification and characterization of the bovine herpesvirus 5 US4 gene and gene products. Virology, 225, 126–135. [DOI] [PubMed] [Google Scholar]
- French E.L. (1962) Relationship between infectious bovine rhinotracheitis (IBR) virus and a virus isolated from calves with encephalitis. Aust. Vet. J., 38, 555. [Google Scholar]
- Gomi Y., Sunamachi,H., Mori,Y., Nagaike,K., Takahashi,M. and Yamanishi,K. (2002) Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol., 76, 11447–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.A. et al. (1997) The T1/35kDa family of poxvirus-secreted proteins bind chemokines and modulate leukocyte influx into virus-infected tissues. Virology, 229, 12–24. [DOI] [PubMed] [Google Scholar]
- Hartley C.A., Drummer,H.E. and Studdert,M.J. (1999) The nucleotide sequence of the glycoprotein G homologue of equine herpesvirus 3 (EHV3) indicates EHV3 is a distinct equid alphaherpesvirus. Arch. Virol., 144, 2023–2033. [DOI] [PubMed] [Google Scholar]
- Kaashoek M.J., Rijsewijk,F.A., Ruuls,R.C., Keil,G.M., Thiry,E., Pastoret,P.P. and Van Oirschot,J.T. (1998) Virulence, immunogenicity and reactivation of bovine herpesvirus 1 mutants with a deletion in the gC, gG, gI, gE, or in both the gI and gE gene. Vaccine, 16, 802–809. [DOI] [PubMed] [Google Scholar]
- Keil G.M., Engelhardt,T., Karger,A. and Enz,M. (1996) Bovine herpesvirus 1 U(s) open reading frame 4 encodes a glycoproteoglycan. J. Virol., 70, 3032–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani A.S., Graham,K., Mossman,K., Rajarathnam,K., Clark-Lewis,I., Kelvin,D. and McFadden,G. (1997) The purified myxoma virus gamma interferon receptor homolog M-T7 interacts with the heparin-binding domains of chemokines. J. Virol., 71, 4356–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani A.S., Barrett,J.W. and McFadden,G. (2000) Modulating chemokines: more lessons from viruses. Immunol. Today, 21, 100–106. [DOI] [PubMed] [Google Scholar]
- Liljeqvist J.A., Svennerholm,B. and Bergstrom,T. (2000) Conservation of type-specific B-cell epitopes of glycoprotein G in clinical herpes simplex virus type 2 isolates. J. Clin. Microbiol., 38, 4517–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden H.S., Buckmaster,A., Palfreyman,J.W., Hope,R.G. and Minson,A.C. (1984) Characterization of the 92 000-dalton glyco protein induced by herpes simplex virus type 2. J. Virol., 50, 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D.J., Dolan,A., Donald,S. and Rixon,F.J. (1985) Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol., 181, 1–13. [DOI] [PubMed] [Google Scholar]
- Metzler A.E., Schudel,A.A. and Engels,M. (1986) Bovine herpesvirus 1: molecular and antigenic characteristics of variant viruses isolated from calves with neurological disease. Arch. Virol., 87, 205–217. [DOI] [PubMed] [Google Scholar]
- Middleton J., Neil,S., Wintle,J., Clark-Lewis,I., Moore,H., Lam,C., Auer,M., Hub,E. and Rot,A. (1997) Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell, 91, 385–395. [DOI] [PubMed] [Google Scholar]
- Munson P.J. and Rodbard,D. (1980) Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem., 107, 220–239. [DOI] [PubMed] [Google Scholar]
- Murphy P.M. (2000) Viral antichemokines: from pathogenesis to drug discovery. J. Clin. Invest., 105, 1515–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy P.M. (2001) Viral exploitation and subversion of the immune system through chemokine mimicry. Nat. Immunol., 2, 116–122. [DOI] [PubMed] [Google Scholar]
- Nakamichi K., Matsumoto,Y. and Otsuka,H. (2002) Bovine herpesvirus 1 glycoprotein G is necessary for maintaining cell-to-cell junctional adherence among infected cells. Virology, 294, 22–30. [DOI] [PubMed] [Google Scholar]
- Osterrieder N., Hubert,P.H., Brandmuller,C. and Kaaden,O.R. (1994) A touchdown PCR for the differentiation of equine herpesvirus type 1 (EHV-1) field strains from the modified live vaccine strain RacH. J. Virol. Methods, 50, 129–136. [DOI] [PubMed] [Google Scholar]
- Parcells M.S. et al. (2001) Marek’s disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol., 75, 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry C.M., Simas,J.P., Smith,V.P., Stewart,C.A., Minson,A.C., Efstathiou,S. and Alcami,A. (2000) A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med., 191, 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot A.E. et al. (2001) The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem., 276, 10620–10626. [DOI] [PubMed] [Google Scholar]
- Rekabdar E., Tunback,P., Liljeqvist,J.A. and Bergstrom,T. (1999) Variability of the glycoprotein G gene in clinical isolates of herpes simplex virus type 1. Clin. Diagn. Lab. Immunol., 6, 826–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D.D., Buckmaster,A., Bell,S., Hodgman,C. and Minson,A.C. (1986) Identification of a new glycoprotein of herpes simplex virus type 1 and genetic mapping of the gene that codes for it. J. Virol., 57, 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. and Pellet,P.E. (2001) The family herpesviridae: an introduction. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology, Vol. 2. Lippincott-Williams and Wilkins, Philadelphia, PA, pp. 2381–2397.
- Saraiva M. and Alcami,A. (2001) CrmE, a novel soluble tumor necrosis factor receptor encoded by poxviruses. J. Virol., 75, 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.A. et al. (1997) Poxvirus genomes encode a secreted, soluble protein that preferentially inhibits β chemokine activity yet lacks sequence homology to known chemokine receptors. Virology, 236, 316–327. [DOI] [PubMed] [Google Scholar]
- Smith K.C., Whitwell,K.E., Binns,M.M., Dolby,C.A., Hannant,D. and Mumford,J.A. (1992) Abortion of virologically negative foetuses following experimental challenge of pregnant pony mares with equid herpesvirus 1. Equine Vet. J., 24, 256–259. [DOI] [PubMed] [Google Scholar]
- Su H.K., Eberle,R. and Courtney,R.J. (1987) Processing of the herpes simplex virus type 2 glycoprotein gG-2 results in secretion of a 34 000-Mr cleavage product. J. Virol., 61, 1735–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford E.A., Watson,M.S., McBride,K. and Davison,A.J. (1992) The DNA sequence of equine herpesvirus-1. Virology, 189, 304–316. [DOI] [PubMed] [Google Scholar]
- Telford E.A., Watson,M.S., Perry,J., Cullinane,A.A. and Davison,A.J. (1998) The DNA sequence of equine herpesvirus-4. J. Gen. Virol., 79, 1197–1203. [DOI] [PubMed] [Google Scholar]
- Tortorella D., Gewurz,B.E., Furman,M.H., Schust,D.J. and Ploegh,H.L. (2000) Viral subversion of the immune system. Annu. Rev. Immunol., 18, 861–926. [DOI] [PubMed] [Google Scholar]
- Tran L.C., Kissner,J.M., Westerman,L.E. and Sears,A.E. (2000) A herpes simplex virus 1 recombinant lacking the glycoprotein G coding sequences is defective in entry through apical surfaces of polarized epithelial cells in culture and in vivo. Proc. Natl Acad. Sci. USA, 97, 1818–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtinen L.W. and Allen,G.P. (1982) Identification of the envelope surface glycoproteins of equine herpesvirus type 1. J. Gen. Virol., 63, 481–485. [DOI] [PubMed] [Google Scholar]
- van Berkel V., Barrett,J., Tiffany,H.L., Fremont,D.H., Murphy,P.M., McFadden,G., Speck,S.H. and Virgin,H.I. (2000) Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J. Virol., 74, 6741–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P.C., Levine,M. and Glorioso,J.C. (1987) Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science, 236, 576–579. [DOI] [PubMed] [Google Scholar]
- Zella D., Barabitskaja,O., Casareto,L., Romerio,F., Secchiero,P., Reitz,M.S.,Jr, Gallo,R.C. and Weichold,F.F. (1999) Recombinant IFN-α (2b) increases the expression of apoptosis receptor CD95 and chemokine receptors CCR1 and CCR3 in monocytoid cells. J. Immunol., 163, 3169–3175. [PubMed] [Google Scholar]
- Zlotnik A. and Yoshie,O. (2000) Chemokines: a new classification system and their role in immunity. Immunity, 12, 121–127. [DOI] [PubMed] [Google Scholar]