Abstract
The solution structure of the 6F11F22F2 fragment from the gelatin-binding region of fibronectin has been determined (Protein Data Bank entry codes 1e88 and 1e8b). The structure reveals an extensive hydrophobic interface between the non-contiguous 6F1 and 2F2 modules. The buried surface area between 6F1 and 2F2 (∼870 Å2) is the largest intermodule interface seen in fibronectin to date. The dissection of 6F11F22F2 into the 6F11F2 pair and 2F2 results in near-complete loss of gelatin-binding activity. The hairpin topology of 6F11F22F2 may facilitate intramolecular contact between the matrix assembly regions flanking the gelatin-binding domain. This is the first high-resolution study to reveal a compact, globular arrangement of modules in fibronectin. This arrangement is not consistent with the view that fibronectin is simply a linear ‘string of beads’.
Keywords: assembly/collagen/dissection/extracellular matrix/fibronectin
Introduction
The extracellular matrix glycoprotein fibronectin is a large, multifunctional molecule involved in adhesion and migration events in a range of important physiological processes such as embryogenesis, wound healing, haemostasis and thrombosis (Hynes, 1990). As a soluble dimer in plasma, it is involved in blood coagulation through its affinity for fibrin and platelets. As an insoluble network in the extracellular matrix, it interacts with cell surface receptors and with other matrix components such as collagens and proteoglycans, thus assisting cell migration and the maintenance of tissue integrity (Hynes, 1990).
The interaction between fibronectin and collagen in the extracellular matrix is well documented (Hynes, 1990) but poorly understood at the molecular level. The two proteins are co-distributed in tissues, as shown by immunofluorescence and immunocytochemical studies. Addition of extraneous fibronectin also promotes the attachment of fibroblastic cells to collagen substrates in vitro. Further more, fibronectin is observed in a regularly distributed array along collagen fibres synthesized in culture. Knowledge of the molecular basis of fibronectin’s interaction with collagen would provide a better understanding of the structure and function of the extracellular matrix.
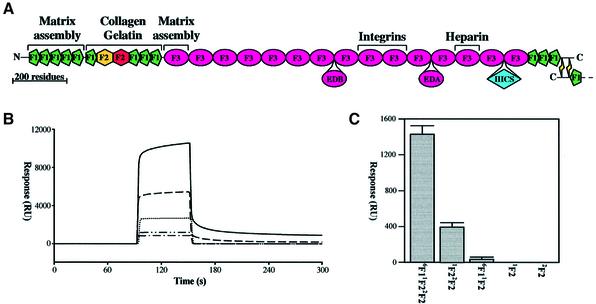
The only viable approach to correlating the structure and function of the large, flexible fibronectin monomer at the atomic level is to dissect it into manageable units (Campbell and Downing, 1998). Fortunately, like many other proteins of the extracellular matrix, fibronectin is a mosaic protein consisting of repeating sequence elements or ‘modules’ that are capable of folding independently (Bork et al., 1996). Its primary sequence is composed almost entirely of three types of module (F1, F2 and F3), which are organized into functional domains (Figure 1A). These domains may be isolated in the form of proteolytic fragments that retain affinity for various ligands. Consequently, many of the ligand-binding sites have been mapped to specific regions of the fibronectin polypeptide. The collagen-binding domain can be isolated as a 42 kDa proteolytic fragment that retains affinity for heat-denatured collagen (gelatin) (Hynes, 1990). This domain has the module composition 6F11F22F27F18F19F1, where nFX represents the nth type X module in the native protein. Further dissection of this gelatin-binding domain by proteolysis produces three non-overlapping module pairs (6F11F2, 2F27F1 and 8F19F1) each of which retains some degree of gelatin-binding activity (Ingham et al., 1989; Litvinovich et al., 1991). This suggests that the gelatin-binding site (or sites) spans multiple modules in the domain.
Fig. 1. Binding of fibronectin fragments to immobilized collagen α1(I) chains. (A) The mosaic structure of a fibronectin monomer is shown, with the positions of the alternatively spliced regions EDB, EDA and IIICS depicted below. The major binding sites for cells and for other matrix components are labelled. (B) Assaying the binding of fibronectin fragments to immobilized α1 chains by surface plasmon resonance (SPR). A 20 µl aliquot of 300 µM samples of 6F11F22F2 (—), 1F22F2 (– – –), 6F11F2 (........), 1F2 (– - – -) and 2F2 (– - - –) was injected over the same flow cell of immobilized collagen α1(I) chains at a flow rate of 20 µl/min. The surface was regenerated with 50 mM HCl after each injection and the individual sensorgrams overlaid with the BIAevaluation 3.0 software (Biacore). (C) Response (RU) 25 s after the end of the injection of the fibronectin fragments as a comparison of collagen α1(I) chain binding ability. Significant binding was only detected in this assay for 6F11F22F2 and 1F22F2. Values represent the mean of triplicate samples, with standard deviation of replicate samples shown as error bars.
Attempts to localize the gelatin-binding site further by recombinant expression in Escherichia coli have yielded conflicting results. Analysis of recombinant fragments produced as β-galactosidase fusion proteins showed that both the 1F2 module and the 1F22F2 module pair could bind immobilized gelatin (Banyai et al., 1990). In an earlier study, however, the 6F11F22F2 module construct only bound to gelatin if the 14 N-terminal residues of 7F1 were present at the C-terminus of the triplet fragment (Owens and Baralle, 1986). A third study identified the smallest recombinant fragment capable of binding to immobilized gelatin as 6F11F22F27F1 (Skorstengaard et al., 1994). The occurrence of F2 modules in other gelatin-binding proteins, such as the matrix metalloproteinases (MMP) 2 and 9, provides evidence for their involvement in gelatin binding by fibronectin (Collier et al., 1988; Wilhelm et al., 1989). Furthermore, recombinant expression of F2 modules from these MMPs has produced fragments with high affinity for gelatin (Banyai and Patthy, 1991; Collier et al., 1992; Banyai et al., 1994), whereas recombinant MMP2 lacking F2 modules was devoid of gelatin-binding activity (Murphy et al., 1994; Allan et al., 1995).
Here we describe the gelatin-binding properties and solution structure of the 6F11F22F2 fragment from fibro nectin (Protein Data Bank entry codes 1e88 and 1e8b). Dissection of this fragment into the 6F11F2 module pair and the individual 2F2 module results in a drastic reduction in gelatin affinity, suggesting that module– module interactions are essential for optimal binding activity. A comparison of backbone amide chemical shifts of 6F11F22F2 with 6F11F2 and 1F22F2 revealed long-range interactions between the non-contiguous 6F1 and 2F2 modules. In the solution structure of 6F11F22F2, the 6F1 and 2F2 modules interact via an extensive hydrophobic interface whose buried surface area is the largest intermodule contact yet seen in fibronectin. The central 1F2 module shows no non-covalent interactions with either 6F1 or 2F2, implying that the 6F1–1F2 interface observed previously (Bocquier et al., 1999) is disrupted in the presence of additional modules. The hairpin topology of 6F11F22F2 may facilitate intramolecular contact between the flanking 1F12F13F14F15F1 and 1F3 fragments, an interaction that is believed to modulate fibronectin fibrillogenesis in the extracellular matrix. Its conformation may also account for the previously noted disruptions in the otherwise uniform strand-like images seen in electron micrographs of fibronectin at high ionic strength. This is the first high-resolution study to reveal a compact, globular arrangement of modules in fibronectin.
Results and discussion
Enhanced gelatin-binding activity in the 6F11F22F2 fragment
The isolated 1F2 and 2F2 modules, the 6F11F2 and 1F22F2 module pairs and the 6F11F22F2 module triplet were produced as described in Materials and methods. During their purification, the isolated 1F2 and 2F2 modules and the 6F11F2 module pair bound weakly to the gelatin affinity column and were separated from the non-binding contaminants by isocratic elution; the 1F22F2 module pair and the 6F11F22F2 module triplet bound more tightly and were eluted with a urea gradient.
The binding of 1F2, 2F2, 6F11F2, 1F22F2 and 6F11F22F2 to immobilized collagen α1(I) chains was analysed in greater detail using surface plasmon resonance (SPR) (Figure 1B and C). For the 6F11F22F2 binding to the α1(I) polypeptide chains, an acceptable fit (χ2 <2) could be obtained with a model that assumed surface heterogeneity, a common observation of immobilization through amine side chains. This treatment resolved the data into two components, with the major component exhibiting association (kon) and dissociation (koff) rates of 115 ± 12 M–1s–1 and 0.0036 ± 0.001 s–1, respectively, and an equilibrium dissociation constant of 31 ± 6 µM (Kd = koff/kon). No significant changes in the 6F11F22F2 binding were observed under acidic conditions (pH 4.5). The 1F22F2 bound with much lower affinity, with kon and koff values of 94 ± 21 M–1s–1 and 0.012 ± 0.001 s–1, respectively, which is equivalent to an equilibrium dissociation constant of 131 ± 23 µM. The binding of 1F2, 2F2 and 6F11F2 to immobilized α1(I) chains was too weak to quantify the kinetics with this assay.
The very low affinity of the 1F2 and 2F2 modules and of the 6F11F2 module pair for α1(I) chains demonstrates that the gelatin-binding site of 6F11F22F2 does not reside entirely within any single module (Figure 1C). The moderate affinity of the 1F22F2 module pair may result from cooperativity between weak, independent binding sites on each module since there is no defined interface between the modules and they tumble independently of each other (Smith et al., 2000). The enhanced binding activity of 6F11F22F2 must therefore involve the formation of a composite binding site involving 1F2 and/or 2F2 and the 6F1 module, or cooperativity between independent, weak gelatin-binding sites on separate modules. The structural basis for this affinity was investigated by nuclear magnetic resonance (NMR) spectroscopy.
6F1 and 2F2 interact in the 6F11F22F2 fragment
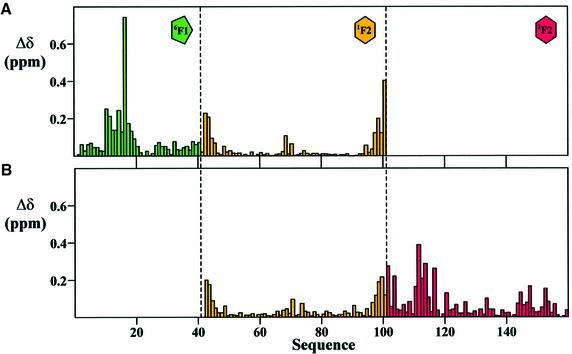
Preliminary information regarding the nature and site of any intermodule interaction in a mosaic protein can be derived from a comparison of the NMR chemical shifts of overlapping fragments under identical solution conditions. We compared the backbone amide resonances (NH and HN) of the 6F11F22F2 fragment with the overlapping 6F11F2 and 1F22F2 module pairs (Figure 2). Removal of the 2F2 module from 6F11F22F2 results in shift perturbations at the C-terminus of the 1F2 module (Figure 2A), as expected from the change in charge. However, many major shift changes are seen in 6F1, in the region of Val10–Lys20. Similarly, removal of 6F1 from 6F11F22F2 results in shift perturbations at the N-terminus of the 1F2 module, but the largest shift changes are seen in the 2F2 module, in the region of Ser111–His117 (Figure 2B). Therefore, the 6F1 and 2F2 modules must interact in 6F11F22F2 despite being separated in sequence by the 1F2 module.
Fig. 2. Amide chemical shift perturbation upon dissection of 6F11F22F2. (A and B) Combined chemical shift perturbation (Δδ) of the HN and NH backbone resonances, upon (A) removal of the 2F2 module from 6F11F22F2, and (B) removal of the 6F1 module from 6F11F22F2. In each case, Δδ = {|ΔδHN| + (|ΔδNH|/5)}/2, where ΔδHN and ΔδNH are the amide proton and amide nitrogen chemical shift differences, respectively.
Solution structure of 6F11F22F2
The solution structure of the 6F11F22F2 fragment could, in theory, be modelled by combining the NMR restraints derived from the independent studies on the overlapping 6F11F2 and 1F22F2 module pairs (Bocquier et al., 1999; Smith et al., 2000). However, because of the extensive amide chemical shift changes in the 6F1 and 2F2 modules, and thus the possibility of significant structural alterations, the solution structure was determined ab initio using only restraints derived from NMR experiments on 6F11F22F2 itself. Of the 100 structures calculated, 20 were selected on the basis of their good agreement with the experimental restraints and their minimal deviations from ideal covalent geometry (Table I).
Table I. Experimental restraints and structural statistics.
| R.m.s. deviations from experimental dataa | |
| all 2024 NOE restraints | 0.009 ± 0.001 Å |
| 515 intraresidue NOEs {i = j} | 0.008 ± 0.002 Å |
| 598 sequential NOEs {|i–j| = 1} | 0.007 ± 0.002 Å |
| 231 short-range NOEs {1 < |i–j| < 5} | 0.014 ± 0.003 Å |
| 539 long-range NOEs {|i–j| > 4} | 0.009 ± 0.002 Å |
| 41 6F1–2F2 intermodule NOEs | 0.006 ± 0.003 Å |
| 100 ambiguous NOEs | 0.011 ± 0.004 Å |
| 106 hydrogen bond restraints | 0.010 ± 0.002 Å |
| 69 dihedral φ angles | 0.066 ± 0.036° |
| R.m.s. deviations from ideal covalent geometry | |
| bonds | 0.0013 ± 0.0001 Å |
| angles | 0.245 ± 0.004° |
| impropers | 0.123 ± 0.014° |
| Ramachandran analysisb | |
| residues in favoured regions | 55.0% |
| residues in additional allowed regions | 39.9% |
| residues in generously allowed regions | 4.4% |
| residues in disallowed regions | 0.7% |
| Coordinate precision: secondary structure backbone, all heavy atomsc | |
| 6F1(1F22F2) | 0.46 ± 0.17 Å, 1.21 ± 0.82 Å |
| (6F1)1F2(2F2) | 0.33 ± 0.09 Å, 0.99 ± 0.88 Å |
| (6F11F2)2F2 | 0.37 ± 0.06 Å, 0.98 ± 0.69 Å |
| 6F1(1F2)2F2 | 0.53 ± 0.14 Å, 1.15 ± 0.73 Å |
| (6F1)1F2(2F2) | 11.7 ± 3.48 Å, 10.2 ± 3.59 Å |
aNone of the 20 accepted structures showed distance restraint violations of >0.3 Å or dihedral restraint violations of >2°. No distance or dihedral restraints were consistently violated by >0.1 Å or 1°, respectively.
bProlines, glycines and terminal residues are excluded.
cCoordinate r.m.s. deviations were calculated following best-fit superposition over the secondary structure elements of the underlined module(s). Residues from modules in parentheses were excluded from the calculation.
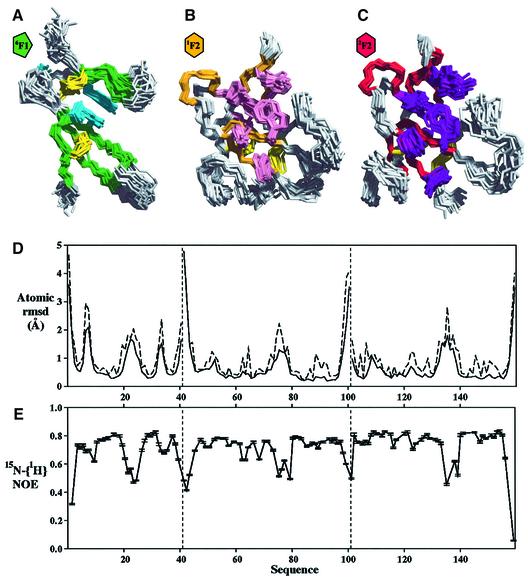
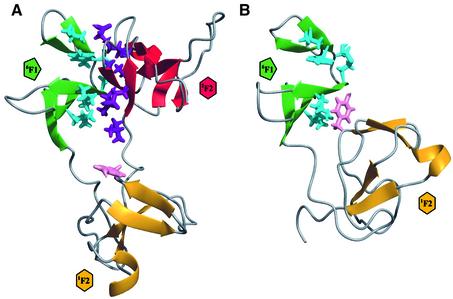
The tertiary structures of the individual modules in 6F11F22F2 are very similar to those in previous studies of the 6F11F2 module pair (Bocquier et al., 1999) and the isolated 2F2 module (Sticht et al., 1998). The back bone heavy atom (NH, Cα, C) root mean square (r.m.s.) deviations over secondary structure elements are 0.98 Å for 6F1, 0.65 Å for 1F2 and 1.21 Å for 2F2. The 6F1 module (Figure 3A) comprises a short N-terminal double-stranded antiparallel β-sheet (AB) that folds over a triple-stranded β-sheet (CDE). The β-sheets are linked by two, conserved disulfide bridges in a 1–3 and 2–4 pattern connecting strands A–D and D–E, respectively. The module core is composed primarily of two highly conserved aromatic residues Tyr12 and Trp18, and a hydrophobic residue, Val36.
Fig. 3. Structural definition of the individual modules in 6F11F22F2. (A–C) Individual module overlays for the ensemble of 20 lowest energy structures of 6F11F22F2. Each module has been overlaid onto the lowest energy structure by best-fit superposition over the backbone heavy atoms of its secondary structure elements. The secondary structure is coloured green in 6F1, gold in 1F2 and red in 2F2. The disulfide bridges are shown in yellow, and the side chains of non-polar residues that are invariant or highly conserved between modules are shown in cyan for 6F1, pink for 1F2 and purple for 2F2. (D) Average atomic r.m.s. deviation between the 20 accepted structures for the backbone heavy atoms (solid line) and all heavy atoms (dotted line). The vertical dashed lines mark the exon boundaries between the modules. (E) 15N-{1H}-NOE for the 6F11F22F2 fragment. Lower 15N-{1H}-NOE values indicate regions with increased backbone flexibility.
The 1F2 module (Figure 3B) comprises two double-stranded antiparallel β-sheets (AB and CD), oriented approximately perpendicular to each other, with a single α-helical turn located between strands C and D. The cleft between the β-sheets is occupied by side chains of invariant hydrophobic and aromatic residues, which make up the module core. On the opposite face of the second β-sheet, two disulfide bonds link the invariant cysteines, with connectivities 1–3 and 2–4. The topology of 2F2 (Figure 3C) is very similar to that of 1F2, but includes the additional A′A′′ β-sheet preceding the first cysteine residue.
In general, the individual 6F1, 1F2 and 2F2 modules in 6F11F22F2 are well defined (Figure 3A–C) with low backbone heavy atom r.m.s. deviations over their secondary structure elements (Table I). The low 15N-{1H}-NOE for the C–D loop in 6F1, and the B–C loops in 1F2 and 2F2, each of which have backbone r.m.s. deviations >1.0 Å, shows that the poorer definition of these regions arises from backbone flexibility rather than a lack of experimental data (Figure 3D and E).
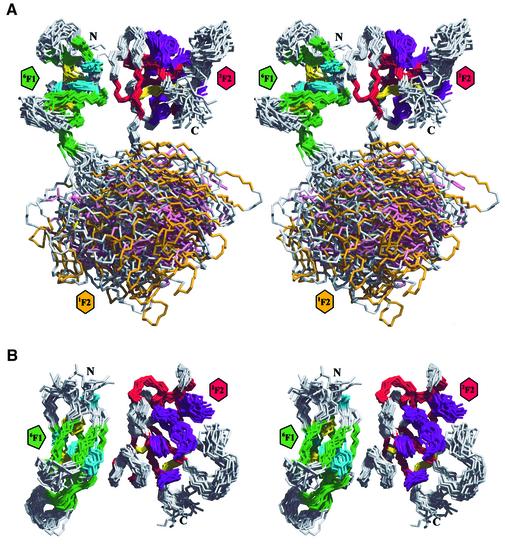
The list of long-range NOEs used in the final round of 6F11F22F2 structure calculations included an extensive array of intermodule restraints between nine residues in 6F1 and eight in 2F2 (Table I). This resulted in a precise definition of the relative locations and orientations of the 6F1 and 2F2 modules in 6F11F22F2 (Figure 4; Table I). The 6F1–2F2 interface is formed by the docking of the external edges of β-strands C of 6F1 and A′ of 2F2, and by the annealing of two extensive hydrophobic surfaces involving Val10, Tyr12, Met16, Leu19 and Leu28 from 6F1, and Leu103, Ala114, Leu115 and Thr145 from 2F2 (Figure 5A). These non-polar residues account for 53% of the total buried surface area between the two modules. There are no titratable groups involved in the remaining 47% that could disrupt the hairpin topology within the pH range 4.5–7.4.
Fig. 4. Solution structure of 6F11F22F2. (A) A stereoview of the ensemble of the 20 lowest energy structures of 6F11F22F2 is shown. The structures were superimposed on the backbone heavy atoms of the secondary structure elements of the 6F1 and 2F2 modules of the lowest energy structure. The colour scheme for the protein backbone and side chain residues is the same as in Figure 3A–C. (B) Orthogonal stereoview of the 6F11F22F2 ensemble generated by a 90° rotation about the horizontal. For clarity, the disordered 1F2 module has been omitted.
Fig. 5. Module reorganization upon dissection of 6F11F22F2. Ribbon diagrams of the minimized average structures of (A) 6F11F22F2 and (B) 6F11F2. The colour scheme for the secondary structure elements is as in Figure 33A–C. Side chains for which 6F1–2F2 intermodule NOEs were observed (V10, Y12, S13, M16, L19 and L28 of 6F1, and L103, Q105, S111, N112, A114, L115, T145 and K153 for 2F2) are shown in cyan for 6F1 and purple for 2F2. Removal of the 2F2 module allows the side chain of Y68 (pink) in 1F2 to interact with L19 and L28 in 6F1.
No long- or short-range NOEs were observed, either between the 6F1 and 1F2 modules, or between the 1F2 and 2F2 modules. The 6F1–1F2 and 1F2–2F2 linker sequences are flexible, as indicated by their low 15N-{1H}-NOE values (Figure 3E). Hence, the relative location and orientation of 1F2 are somewhat ill defined (Figure 4A; Table I) since the only positional constraints on this central module are the covalent tethering of its N- and C-termini, and steric hindrance from 6F1 and 2F2.
The gelatin-binding site(s) of 6F11F22F2
In each F2 module, five of the core aromatic residues (Tyr62, Trp81, Tyr88, Tyr94 and Phe96 in 1F2; Tyr122, Trp141, Tyr148, Phe154 and Phe156 in 2F2) form an extensive, solvent-exposed hydrophobic surface (Figure 3B and C). A pocket in each of these surfaces is thought to provide a binding site for non-polar residues in type I collagen (Pickford et al., 1997). This is supported by the binding of the collagen-like peptide (Pro-Pro-Gly)6 to the 2F2 module from MMP2, which produced backbone HN and NH chemical shift perturbations in the equivalent residues (Briknarová et al., 1999). Furthermore, in the crystal structure of pro-MMP2, a phenylalanine side chain in the inhibitory propeptide occupies the equivalent hydrophobic pocket in the 3F2 module, preventing it from binding its gelatin substrate (Morgunova et al., 1999).
In all structures of the 6F11F22F2 ensemble, solvent access to the putative binding sites on the F2 modules is unhindered by the 6F1–2F2 interface. In the average 6F11F22F2 structure (Figure 5A), the binding sites are oriented in opposite directions. This configuration of F2 modules observed here is reminiscent of the pro-MMP2 crystal structure where the three F2 modules are also oriented with their binding sites facing outwards (Morgunova et al., 1999). Recombinant 1F22F23F2 from MMP2 has been shown to be capable of binding multiple collagen triple helices (Steffensen et al., 1995), suggesting that F2 modules form separate binding surfaces that intercalate between molecules in a collagen fibril. The flexibility of the 1F2 module with respect to 6F1 and 2F2 may permit a variety of collagen-binding conformations. It has also been proposed that the observed pliability of the 9F310F3 module pair may allow it to accommodate some variation in the integrin structure to which it binds (Copié et al., 1998).
The structural basis for the 4-fold increase in affinity for α(I) chains from 1F22F2 to 6F11F22F2 is not known, but may arise from an extension of the 2F2 binding site onto 6F1. The 6F1 module docks onto 2F2 alongside its gelatin-binding site in an orientation that may allow solvent-exposed residues in the AB sheet of 6F1 to contribute to binding (Figure 4B). Alternatively, the binding enhancement may arise from stabilization of the 2F2 module due to the 6F1–2F2 interface. The backbone HN protons of residues within the AB sheet of 6F1 and the A′A′′ sheet of 2F2 undergo much slower solvent exchange in 6F11F22F2 than observed previously for 6F11F2 (Bocquier et al., 1999) and 2F2 (Sticht et al., 1998). Stabilization of the 2F2 module was observed previously in differential scanning calorimetric studies on the 6F11F22F27F1 proteolytic fragment, although this was originally attributed to an interaction between 1F2 and 2F2 (Litvinovich et al., 1991).
The contributions of 7F1, 8F1 and 9F1 to collagen binding have yet to be determined. The only structural information available for these domains is the isolated 7F1 (Baron et al., 1990), but there is no information on its interactions with adjacent modules. Given the hairpin structure observed in 6F11F22F2, we believe that the extension of the 6F11F22F2 nucleus with these modules will provide a better examination of the collagen-binding domain function.
Module reorganization upon dissection
The two techniques capable of providing high-resolution structural information on binding surfaces, namely X-ray diffraction and NMR, are both limited in the size of mosaic proteins that can be studied (Campbell and Downing, 1998). In the case of X-ray diffraction, the limitation arises from the difficulty in crystallizing a protein that is inherently flexible, whilst for NMR the molecular weight is the limiting factor. Hence, for both techniques, large mosaic proteins must frequently be dissected into fragments that are amenable to analysis. However, a comparison of the solution structures of 6F11F2 and 6F11F22F2 illustrates a potential complication associated with this dissection strategy (Figure 5).
The F1 and F2 modules in 6F11F2 were found to interact via a small hydrophobic interface of ∼340 Å2, involving the side chains of Leu19 and Leu28 from 6F1, and Tyr68 from 1F2 (Bocquier et al., 1999). This interaction resulted in a significant upfield shift in the NH resonance of Ser69 relative to the isolated 1F2 module (Hashimoto et al., 2000). However, this 6F1–1F2 interface is inconsistent with the solution structure of 6F11F22F2 presented here. The upfield shift of the Ser69 NH resonance is reversed in 6F11F22F2, consistent with a break up of the weak 6F1–1F2 interface (Figure 2A). None of the 18 weak intermodule NOEs previously observed between 6F1 and 1F2 is apparent in the spectra of 6F11F22F2; manual incorporation of these restraints into the structure calculations of 6F11F22F2 also resulted in structures with 20–25% higher potential energy. Therefore, this contact between the modules must have arisen from a module reorganization; removal of the 2F2 module relieves the covalent and steric constraints on the 1F2 module, allowing a hydrophobic collapse of non-polar residues that are distant and/or buried in the intact protein (Figure 5). Such rearrangements are a well-known phenomenon in intracellular proteins, for example, the rearrangement of SH2 and SH3 domains in Src family tyrosine kinases (Sicheri et al., 1997).
A globular domain in a fibrillar protein
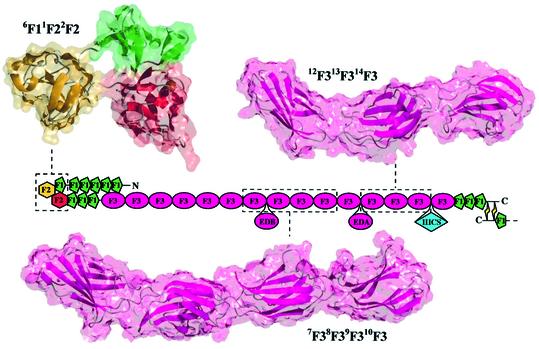
The 6F1–2F2 interface gives the 6F11F22F2 fragment a compact, hairpin topology (with average dimensions 15 × 19 × 32 Å). This is in sharp contrast to the extended, near-linear arrangement of F3 modules in the crystal structures of the cell-binding 7F38F39F310F3 (Leahy et al., 1996) and heparin-binding 12F313F314F3 (Sharma et al., 1999) fragments of fibronectin (Figure 6). The global topology of 6F11F22F2 is in agreement with previous calorimetric studies, which suggested that the gelatin-binding domain has a relatively compact structure (Litvinovich et al., 1991).
Fig. 6. Global topologies of multimodule fibronectin fragments. Solvent-accessible surfaces have been superimposed over ribbon diagrams for the minimized average structure of 6F11F22F2, and the crystal structures of 7F38F39F310F3 (Leahy et al., 1996) and 12F313F314F3 (Sharma et al., 1999). The fragment structures are mapped onto the mosaic illustration of fibronectin, which has been folded to account for the hairpin structure of 6F11F22F2.
Numerous biophysical and biochemical studies have shown that intact fibronectin undergoes a substantial change in structure from a compact conformation at low ionic strength to a more extended conformation in high salt (Engel et al., 1981; Erickson et al., 1981; Erickson and Carrell, 1983; Rocco et al., 1983; Lai et al., 1993). However, even at high ionic strength, numerous kinks are seen in electron micrographs of the molecule (Engel et al., 1981; Erickson et al., 1981; Erickson and Carrell, 1983; Rocco et al., 1983; Lai et al., 1993; Johnson et al., 1999). The resistance of these bends in the protein to high salt concentrations suggests that the interactions involved are predominantly non-polar in nature. Thus, the solution structure of 6F11F22F2 presented here, with its hairpin conformation and extensive hydrophobic interface between the 6F1 and 2F2 modules, is one possible explanation for the kinks observed towards the N-terminal ends of the fibronectin dimer (Figure 6).
Many of the models for the compact structure of fibronectin at low ionic strength involve a folding over of the 1F12F13F14F15F1 fragment allowing it to form interdomain interactions with the F3 modules in the protein (Williams et al., 1982; Homandberg and Erickson, 1986; Rocco et al., 1987; Ingham et al., 1988; Khan et al., 1990). Of particular interest is the potential contact between the 1F12F13F14F15F1 and 1F3 fragments (Figure 1A), an intramolecular interaction that is believed to suppress fibronectin fibrillogenesis (Aguirre et al., 1994; Schwarzbauer and Sechler, 1999). While this contact would be unlikely to take place if the gelatin-binding domain had a more extended organization, akin to those of the cell-binding and heparin-binding fragments (Figure 6), the hairpin conformation of 6F11F22F2 in the gelatin-binding domain may facilitate intramolecular contact between the flanking 1F12F13F14F15F1 and 1F3 fragments. Interestingly, the addition of collagen or the cyanogen bromide fragment CB7 of the α1(I) chain results in a reorganization and increased accumulation of fibronectin fibrils at the surface of collagen-deficient MOV-13 cells (Dzamba et al., 1993). Thus, collagen binding may induce a conformational change that disengages the inhibitory 1F12F13F14F15F1–1F3 intramolecular interaction, thus facilitating matrix assembly; this is also supported by the observed partial unfolding of plasma fibronectin upon binding of the α1(I)-CB7 fragment (Williams et al., 1982).
It has been proposed that stretching of the fibronectin molecule by the cell might regulate matrix function by exposing new binding sites and affecting cell adhesion (Schwarzbauer and Sechler, 1999). Applied tension could extend the molecule through a breakdown of the above interdomain interactions, the local disruption of intermodule interfaces or even complete unfolding of F3 modules (Erickson, 1994; Oberhauser et al., 1998; Ohashi et al., 1999). For example, the disruption of the 9F3–10F3 interface due to an extension of the intermodule linker resulted in a reduction of integrin-mediated cell adhesion and intracellular signalling (Grant et al., 1997). Thus, it could be argued that stretching might also provide a means for actively modulating the affinity of fibronectin for collagen by disrupting the 6F1–2F2 intermodule interface that enhances binding. However, the buried surface area between 6F1 and 2F2 in the 6F11F22F2 structure ensemble is on average 868 Å2 (±48 Å2), much greater than the 340 Å2 buried between 9F3 and 10F3 (Leahy et al., 1996). Therefore, the smaller and more flexible 9F3–10F3 interface (Copié et al., 1998) is likely to deform more easily in response to stress, with the result that the cell retracts from the fibronectin matrix before it can apply the necessary tension to disrupt the fibronectin–collagen interaction.
Materials and methods
Preparation of recombinant fibronectin modules
Fragments of the collagen-binding domain of human fibronectin were prepared by recombinant expression from the methylotrophic yeast Pichia pastoris. Expression and purification of the isolated 1F2 and 2F2 modules and the 6F11F2 and 1F22F2 module pairs have been described previously (Pickford et al., 1997; Sticht et al., 1998; Bocquier et al., 1999; Smith et al., 2000). The P.pastoris clone expressing the 6F11F22F2 triplet (corresponding to residues 274–433 of mature human fibronectin) was produced in analogous fashion to that described previously for the 1F2 module (Pickford et al., 1997). Expression of unlabelled and uniformly 15N-labelled ([u-15N]) proteins was carried out in a 1 l fermentor (Electrolab Ltd, Tewkesbury, UK) following the detailed protocol for the 4F15F1 module pair (Bright et al., 1999). Each fragment was purified by a combination of cation exchange chromatography on SP-Sepharose Fast Flow (Amersham Pharmacia Biotech), affinity chromatography on gelatin–Sepharose 4B (Amersham Pharmacia Biotech) and reverse phase high-performance liquid chromatography (HPLC) on a C8 column (Rainin). Prior to the last purification step, those fragments containing the 2F2 module were treated with Endo Hf (New England Biolabs) to trim the high mannose sugar attached to residue Asn25 back to a single N-acetylglucosamine (GlcNAc) (Sticht et al., 1998). The identity and purity of each fragment were confirmed by electrospray mass spectrometry and N-terminal sequence analysis.
Surface plasmon resonance
The collagen α1(I) chain was purified from human placental type I collagen (Sigma) by size-exclusion chromatography through Sephacryl-S400HR (Amersham Pharmacia Biotech) followed by cation exchange chromatography at 42°C on a Mono-S HR5/5 column (Amersham Pharmacia Biotech). SPR experiments were performed on a BIAcore 2000 instrument (Biacore AB, Uppsala, Sweden). Purified collagen α1(I) chains were immobilized on the dextran matrix of a CM5 sensorchip (Biacore AB) in 10 mM acetate buffer pH 4.0, and covalently bound using the amine coupling method as described in the BIAapplications handbook (Biacore AB). A control flow cell was also created by derivatizing the surface for amine coupling in the absence of protein. For comparative binding of the fibronectin fragments, a collagen α1(I) immobilization level of ∼7000 resonance units (RU) was used. In order to avoid mass transport limitation, this immobilization level was reduced to ∼2000 RU for the kinetic analysis of fibronectin fragment binding. The same sensorchip surface was used in each set of experiments. Binding experiments were carried out at 25°C in HBS-EP running buffer [10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) Surfactant P20] at a flow rate of 20 µl/min. A regeneration step of 30 s exposure to 50 mM HCl was applied after each injection to return to the baseline. To compare the binding of the fibronectin fragments, 20 µl of 300 µM samples of 6F11F22F2, 1F22F2, 6F11F2, 1F2 and 2F2 were injected over immobilized collagen α1(I) chains, and the amount of protein bound to the sensorchip was monitored by the change in RU. For the kinetic analyses, triplicates of 20 µl of five dilutions (25–200 µM) of the fibronectin fragments 1F22F2 and 6F11F22F2 were injected over the flow cells. The sensorgram data were analysed using global fitting procedures in the BIAevaluation 3.0 program (Karlsson and Falt, 1997). The association (kon) and dissociation (koff) rates were evaluated by analysing appropriate components of the sensorgram curve.
NMR spectroscopy
All NMR experiments were acquired at 25°C on a spectrometer built in-house at the Oxford Centre for Molecular Sciences incorporating an Oxford Instruments magnet (750.1 MHz for 1H) and a GE/Omega computer. Experiments were recorded in a phase-sensitive manner using the States/TPPI method for quadrature detection in the indirectly detected dimensions. In all heteronuclear experiments, 1H–15N decoupling was achieved using a GARP pulse-train with a 1.7 kHz decoupling bandwidth. Samples for amide chemical shift comparison were prepared by dissolving either [u-15N]6F11F2, [u-15N]1F22F2 or [u-15N]6F11F22F2 to a final concentration of 1.0 mM in 90% H2O/10% D2O containing 1 mM 1,4-dioxane, and adjusting the pH to 4.5 (meter uncorrected for deuterium). For each sample, a one-dimensional 1H spectrum and a two-dimensional gradient-enhanced [1H–15N]-HSQC spectrum (Kay et al., 1992) were acquired. Samples for 6F11F22F2 assignment and structure determination were prepared by dissolving [u-15N]6F11F22F2 to a final concentration of 2.0 mM in either 90% H2O/10% D2O or 99.9% D2O, adding 1,4-dioxane to 1 mM, and adjusting the pH to 4.5. The following spectra were recorded in H2O: a two-dimensional [1H–15N]-HMQC-J (Kay and Bax, 1990), a three-dimensional gradient-enhanced [1H–15N]-TOCSY-HSQC with 46 ms mixing time (Marion et al., 1989) and a three-dimensional gradient-enhanced [1H–15N]-NOESY-HSQC with 60 ms mixing time (Marion et al., 1989). The following spectra were recorded in D2O: a two-dimensional [1H–1H]-DQF-COSY (Rance et al., 1983), a two-dimensional [1H–1H]-NOESY with 60 ms mixing time (Kumar et al., 1980) and a two-dimensional [1H–1H]-TOCSY with 46 ms mixing time (Davis and Bax, 1985). For measuring the 15N-{1H}-NOE, two experiments were recorded, either with (NOE) or without (NONOE) 1H saturation, during the recycle delay (Kay et al., 1989). Slowly exchanging amide protons were identified by lyophilizing [u-15N]6F11F22F2 from H2O, dissolving in D2O and recording multiple gradient-enhanced [1H–15N]-HSQC spectra at two-hourly intervals.
Data processing and analysis
Data processing was performed using the FELIX 2.3 software package (Biosym Technologies Inc.). Homonuclear DQF-COSY, TOCSY and NOESY experiments were processed as described previously (Pickford et al., 1997). The three-dimensional gradient-enhanced [1H–15N]-TOCSY-HSQC and [1H–15N]-NOESY-HSQC data sets were processed using a Lorentzian–Gaussian multiplication in t3, linear prediction and apodization in t2 using a 70° phase-shifted squared sine-bell window function, and a Kaiser function for apodization in t1. Proton chemical shifts were referenced relative to the internal standard 1,4-dioxane at 3.743 p.p.m., with indirect referencing in the 15N dimension using a 15N/1H frequency ratio of 0.101329118 (Wishart et al., 1995). The program NMRView v3.0.b1 (Merck and Co., Inc.) was used for spectral assignment and the derivation of structural restraints. Complete assignment of the backbone NH, HN and Hα resonances, and of most side chain proton and nitrogen resonances of 6F11F22F2 at 25°C and pH 4.5 was achieved using standard homonuclear and heteronuclear techniques. The assignment process was assisted by previous studies on the 6F11F2 module pair (Bocquier et al., 1999) and the isolated 2F2 module (Sticht et al., 1998) under the same conditions. Backbone NH and HN resonances of the 1F22F2 module pair, which had previously been assigned at pH 6.0 (Smith et al., 2000), were reassigned at pH 4.5. An iterative procedure was used in the assignment of NOEs: those that could not be assigned unambiguously were included as ambiguous restraints during initial structure calculations (Nilges, 1995) followed, where possible, by resolution of the ambiguity by inspection of preliminary structures. NOEs were calibrated using interproton distances in regions of regular secondary structure, and converted into three distance restraint categories (‘strong’, ‘medium’ and ‘weak’) with upper distance limits of 2.8, 3.5 and 5.0 Å, respectively. Hydrogen bond restraints were introduced in the final round of the calculation if three criteria were met: slow solvent exchange of the HN proton, an HN–O distance <2.3 Å and an O–HN–NH angle >120° in at least 70% of the unrestrained structures. For each hydrogen bond, two distance restraints were introduced into the calculation (dHN–O = 1.7–2.3 Å and dN–O = 2.4–3.3 Å). Backbone φ torsion angle restraints were derived by measuring 3JHN–Hα spin–spin coupling constants from the [1H–15N]-HMQC-J spectra using spectral simulations (Redfield et al., 1991). For those residues with 3JHN–Hα <6 Hz or 3JHN–Hα >8 Hz, estimates of φ angles were obtained using a modified Karplus equation (Pardi et al., 1984) and included as restraints in the structure calculations with an error of ±30°.
Structure calculations and analysis
Structure calculations were performed using an ab initio simulated annealing protocol within the program CNS v0.9 (Brünger et al., 1998). The ‘parallhdg.pro’ forcefield (version 5.1) was used to describe the covalent and non-bonded interactions for the polypeptide (Linge and Nilges, 1999). Parameters for the N-linked GlcNAc on Asn25 of the 2F2 module were derived as previously described (Sticht et al., 1998). The non-bonded energy was calculated using a purely repulsive function with a final value of the van der Waals radii scaled by a factor of 0.75 (Linge and Nilges, 1999). A total of 100 structures were calculated using a simulated annealing profile similar to that described previously (Sticht et al., 1998). It comprised four stages: a high temperature conformational search phase in cartesian space (50 ps at 2000 K with a 2 fs time step), two cooling phases (2000 to 1000 K in 25 ps, and 1000 to 100 K in 25 ps, each with a 1 fs time step) and a final minimization phase. The final values for the force constants were Kbond = 1000 kcal/mol/Å2, Kangl = 500 kcal/mol/rad2, Kimpr = 500 kcal/mol/rad2, Kvdw = 4 kcal/mol, Knoe = 50 kcal/mol/Å2 and Kcdih = 200 kcal/mol/rad2. The structures were refined using an additional cycle of simulated annealing similar to the second cooling phase above, followed by extensive restrained energy minimization. The floating assignment of prochiral groups was achieved using a novel procedure named SOPHIE (for ‘spinning of prochiral hydrogens’). Throughout the conformational search and cooling phases, the diastereospecifically unassigned groups were allowed to rotate freely about the bond connecting their pseudoatom and prochiral centre. Then, during the final minimization phase, each group was eased into either the pro-R or pro-S position by enforcing the correct bond angles at the prochiral centre. The stereochemical quality of the structures was assessed using the program PROCHECK_NMR (Laskowski et al., 1993). Buried surface areas were calculated in CNS using a probe radius of 1.4 Å. Atomic r.m.s. deviations were calculated following best-fit superposition of each accepted structure onto the secondary structure backbone heavy atoms of the lowest energy structure. The average structure was calculated by superimposing over the backbone heavy atoms (N, Cα, C) of the secondary structure elements of each module. Geometric strain was removed from this average structure by extensive restrained energy minimization in CNS (Brünger et al., 1998). Molecular models were generated with the programs MOLMOL (Koradi et al., 1996) and POV-Ray (http://www.povray.org).
Accession codes
The list of 1H and 15N resonance assignments of 6F11F22F2 at pH 4.5 and 25°C has been deposited at the BioMagResBank with the accession number 4830. The coordinates of the 6F11F22F2 NMR structure ensemble and the minimized average structure have been deposited in the Brookhaven Protein Data Bank with the ID codes 1e88 and 1e8b, respectively.
Acknowledgments
Acknowledgements
We thank Tony Willis, Robin Aplin, Nick Soffe and Jörn Werner for help and advice concerning N-terminal sequence analysis, electrospray mass spectrometry, NMR spectroscopy and protein dynamics. This is a contribution from the Oxford Centre for Molecular Sciences, which is supported by the BBSRC, EPSRC and MRC. A.R.P. and D.S. thank the Wellcome Trust for financial support. S.P.S. is a recipient of a Burroughs Wellcome Fund Hitchings–Elion Fellowship.
References
- Aguirre K.M., McCormick,R.J. and Schwarzbauer,J.E. (1994) Fibronectin self-association is mediated by complementary sites within the amino-terminal one-third of the molecule. J. Biol. Chem., 269, 27863–27868. [PubMed] [Google Scholar]
- Allan J.A., Docherty,A.J.P., Barker,P.J., Huskisson,N.S., Reynolds,J.J. and Murphy,G. (1995) Binding of gelatinases A and B to type-I collagen and other matrix components. Biochem. J., 309, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyai L. and Patthy,L. (1991) Evidence for the involvement of type II domains in collagen binding by 72 kDa type IV procollagenase. FEBS Lett., 282, 23–25. [DOI] [PubMed] [Google Scholar]
- Banyai L., Trexler,M., Koncz,S., Gyenes,M., Sipos,G. and Patthy,L. (1990) The collagen-binding site of type-II units of bovine seminal fluid protein PDC-109 and fibronectin. Eur. J. Biochem., 193, 801–806. [DOI] [PubMed] [Google Scholar]
- Banyai L., Tordai,H. and Patthy,L. (1994) The gelatin-binding site of human type IV collagenase (gelatinase A). Biochem. J., 298, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M., Norman,D.G., Willis,A. and Campbell,I.D. (1990) Structure of the fibronectin type 1 module. Nature, 345, 642–646. [DOI] [PubMed] [Google Scholar]
- Bocquier A.A., Potts,J.R., Pickford,A.R. and Campbell,I.D. (1999) Solution structure of a pair of modules from the gelatin-binding domain of fibronectin. Structure, 7, 1451–1460. [DOI] [PubMed] [Google Scholar]
- Bork P., Downing,A.K., Kieffer,B. and Campbell,I.D. (1996) Structure and distribution of modules in extracellular proteins. Q. Rev. Biophys., 29, 119–167. [DOI] [PubMed] [Google Scholar]
- Bright J.R., Pickford,A.R., Potts,J.R. and Campbell,I.D. (1999) Preparation of isotopically labelled recombinant fragments of fibronectin for functional and structural study by heteronuclear magnetic resonance spectroscopy. Methods Mol. Biol., 139, 59–69. [DOI] [PubMed] [Google Scholar]
- Briknarová K., Grishaev,A., Banyai,L., Tordai,H., Patthy,L. and Llinas,M. (1999) The second type II module from human matrix metalloproteinase 2: structure, function and dynamics. Structure, 7, 1235–1245. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system (version 0.9): a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Campbell I.D. and Downing,A.K. (1998) NMR of modular proteins. Nature Struct. Biol. NMR Suppl., 496–499. [DOI] [PubMed] [Google Scholar]
- Collier I.E. et al. (1988) H-ras oncogene-transformed human bronchial epithelial cells (TBE-1) secrete a single metalloproteinase capable of degrading basement membrane collagen. J. Biol. Chem., 263, 6579–6587. [PubMed] [Google Scholar]
- Collier I.E., Krasnov,P.A., Strongin,A.Y., Birkedal-Hansen,H. and Goldberg,G.I. (1992) Alanine scanning mutagenesis and functional analysis of the fibronectin-like collagen-binding domain from human 92 kDa type IV collagenase. J. Biol. Chem., 267, 6776–6781. [PubMed] [Google Scholar]
- Copié V., Tomita,Y., Akiyama,S.K., Aota,S.-I., Yamada,K.M., Venable,R.M., Pastor,R.W., Krueger,S. and Torchia,D.A. (1998) Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure. J. Mol. Biol., 277, 663–682. [DOI] [PubMed] [Google Scholar]
- Davis D.G. and Bax,A. (1985) Assignment of complex 1H NMR spectra via two-dimensional homonuclear Hartmann–Hahn spectroscopy. J. Am. Chem. Soc., 107, 2820–2821. [Google Scholar]
- Dzamba B.J., Wu,H., Jaenisch,R. and Peters,D.M. (1993) Fibronectin binding site in type I collagen regulates fibronectin fibril formation. J. Cell Biol., 121, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J., Odermatt,E., Engel,A., Madri,J.A., Furthmayr,H., Rohde,H. and Timpl,R. (1981) Shapes, domain organizations and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix. J. Mol. Biol., 150, 97–120. [DOI] [PubMed] [Google Scholar]
- Erickson H.P. (1994) Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Natl Acad. Sci. USA, 91, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H.P. and Carrell,N.A. (1983) Fibronectin in extended and compact conformations. Electron microscopy and sedimentation analysis. J. Biol. Chem., 258, 14539–14544. [PubMed] [Google Scholar]
- Erickson H.P., Carrell,N. and McDonagh,J. (1981) Fibronectin molecule visualized in electron microscopy: a long, thin, flexible strand. J. Cell Biol., 91, 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R.P., Spitzfaden,C., Altroff,H., Campbell,I.D. and Mardon,H.J. (1997) Structural requirements for biological activity of the ninth and tenth FIII domains of human fibronectin. J. Biol. Chem., 272, 6159–6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Smith,S.P., Pickford,A.R., Bocquier,A.A., Campbell,I.D. and Werner,J.M. (2000) The relative orientation of the fibronectin 6F11F2 module pair: a 15N NMR relaxation study. J. Biomol. NMR, 17, 203–214. [DOI] [PubMed] [Google Scholar]
- Homandberg G.A. and Erickson,J.W. (1986) Model of fibronectin tertiary structure based on studies of interactions between fragments. Biochemistry, 25, 6917–6925. [DOI] [PubMed] [Google Scholar]
- Hynes R.O. (1990) Fibronectins. Springer-Verlag, Berlin, Germany.
- Ingham K.C., Brew,S.A. and Isaacs,B.S. (1988) Interaction of fibronectin and its gelatin-binding domains with fluorescent labelled chains of type I collagen. J. Biol. Chem., 263, 4624–4628. [PubMed] [Google Scholar]
- Ingham K.C., Brew,S.A. and Migliorini,M.M. (1989) Further localization of the gelatin-binding determinants within fibronectin. J. Biol. Chem., 264, 16977–16980. [PubMed] [Google Scholar]
- Johnson K.J., Sage,H., Briscoe,G. and Erickson,H.P. (1999) The compact conformation of fibronectin is determined by intra molecular ionic interactions. J. Biol. Chem., 274, 15473–15479. [DOI] [PubMed] [Google Scholar]
- Karlsson R. and Falt,A. (1997) Experimental design for kinetic analysis of protein–protein interactions with surface plasmon resonance biosensors. J. Immunol. Methods, 200, 121–133. [DOI] [PubMed] [Google Scholar]
- Kay L.E. and Bax,A. (1990) New methods for the measurement of NH–CαH coupling constants in 15N-labelled proteins. J. Magn. Reson., 86, 110–126. [Google Scholar]
- Kay L.E., Torchia,D.A. and Bax,A. (1989) Backbone dynamics as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry, 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Kiefer,R. and Saarinen,T. (1992) Pure absorption gradient-enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc., 114, 10663–10665. [Google Scholar]
- Khan M.Y., Medow,M.S. and Newman,S.A. (1990) Unfolding of fibronectin and its domains. Biochem. J., 270, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Kumar A., Ernst,R.R. and Wüthrich,K. (1980) A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for elucidation of complete proton–proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun., 95, 1–6. [DOI] [PubMed] [Google Scholar]
- Lai C.S., Wolff,C.E., Novello,D., Griffone,L., Cuniberti,C., Molina,F. and Rocco,M. (1993) Solution structure of human plasma fibronectin under different solvent conditions: fluorescence energy transfer, circular dichroism and light-scattering studies. J. Mol. Biol., 230, 625–640. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Leahy D.J., Aukhil,I. and Erickson,H.P. (1996) 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell, 84, 155–164. [DOI] [PubMed] [Google Scholar]
- Linge J.P. and Nilges,M. (1999) Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR, 13, 51–59. [DOI] [PubMed] [Google Scholar]
- Litvinovich S.V., Strickland,D.K., Medved,L.V. and Ingham,K.C. (1991) Domain structure and interactions of the type I and type II modules in the gelatin-binding region of fibronectin. All six modules are independently folded. J. Mol. Biol., 217, 563–575. [DOI] [PubMed] [Google Scholar]
- Marion D., Driscoll,P.C., Kay,L.E., Wingfield,P.T., Bax,A., Gronenborn,A.M. and Clore,G.M. (1989) Overcoming the overlap problem in the assignment of 1H-NMR spectra of larger proteins by use of 3-dimensional 1H–15N Hartman Hahn multiple quantum coherence and nuclear Overhauser multiple quantum coherence spectroscopy. Biochemistry, 28, 6150–6156. [DOI] [PubMed] [Google Scholar]
- Morgunova E., Tuuttila,A., Bergmann,U., Isupov,M., Lindqvist,Y., Schneider,G. and Tryggvason,K. (1999) Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed. Science, 284, 1667–1670. [DOI] [PubMed] [Google Scholar]
- Murphy G., Nguyen,Q., Cockett,M.I., Atkinson,S.J., Allan,J.A., Knight,C.G., Willenbrock,F. and Docherty,A.J. (1994) Assessment of the role of the fibronectin-like domain of gelatinase A by analysis of a deletion mutant. J. Biol. Chem., 269, 6632–6636. [PubMed] [Google Scholar]
- Nilges M. (1995) Calculation of protein structures with ambiguous distance restraints: automated assignment of ambiguous NOE crosspeaks and disulphide connectivities. J. Mol. Biol., 245, 645–660. [DOI] [PubMed] [Google Scholar]
- Oberhauser A.F., Marszalek,P.E., Erickson,H.P. and Fernandez,J.M. (1998) The molecular elasticity of the extracellular matrix protein tenascin. Nature, 393, 181–185. [DOI] [PubMed] [Google Scholar]
- Ohashi T., Kiehart,D.P. and Erickson,H.P. (1999) Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin–green fluorescent protein. Proc. Natl Acad. Sci. USA, 96, 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R.J. and Baralle,F.E. (1986) Mapping the collagen-binding site of human fibronectin by expression in Escherichia coli. EMBO J., 5, 2825–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi A., Billeter,M. and Wüthrich,K. (1984) Angular dependence of the amide proton–Cα proton coupling constants, 3JHNα, in a globular protein. J. Mol. Biol., 180, 741–751. [DOI] [PubMed] [Google Scholar]
- Pickford A.R., Potts,J.R., Bright,J.R., Phan,I. and Campbell,I.D. (1997) Solution structure of a type 2 module from fibronectin: implications for the structure and function of the gelatin-binding domain. Structure, 5, 359–370. [DOI] [PubMed] [Google Scholar]
- Rance M., Sørensen,O.W., Bodenhausen,G., Wagner,G., Ernst,R.R. and Wüthrich,K. (1983) Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun., 177, 479–485. [DOI] [PubMed] [Google Scholar]
- Redfield C., Smith,L.J., Boyd,J., Lawrence,G.M., Edwards,R.G., Smith,R.A. and Dobson,C.M. (1991) Secondary structure and topology of human interleukin 4 in solution. Biochemistry, 30, 11029–11035. [DOI] [PubMed] [Google Scholar]
- Rocco M., Carson,M., Hantgan,R., McDonagh,J. and Hermans,J. (1983) Dependence of the shape of the plasma fibronectin molecule on solvent composition. Ionic strength and glycerol content. J. Biol. Chem., 258, 14545–14549. [PubMed] [Google Scholar]
- Rocco M., Infusini,E., Daga,M.G., Gogioso,L. and Cuniberti,C. (1987) Models of fibronectin. EMBO J., 6, 2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J.E. and Sechler,J.L. (1999) Fibronectin fibrillogenesis: a paradigm for extracellular matrix assembly. Curr. Opin. Cell Biol., 11, 622–627. [DOI] [PubMed] [Google Scholar]
- Sharma A., Askari,J.A., Humphries,M.J., Jones,E.Y. and Stuart,D.I. (1999) Crystal structure of a heparin- and integrin-binding segment of human fibronectin. EMBO J., 18, 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheri F., Moarefi,I. and Kuriyan,J. (1997) Crystal structure of the Src family tyrosine kinase Hck. Nature, 385, 602–609. [DOI] [PubMed] [Google Scholar]
- Skorstengaard K., Holtet,T.L., Etzerodt,M. and Thorgersen,H.C. (1994) Collagen-binding recombinant fibronectin fragments containing type II domains. FEBS Lett., 343, 47–50. [DOI] [PubMed] [Google Scholar]
- Smith S.P., Hashimoto,Y., Pickford,A.R., Campbell,I.D. and Werner,J.M. (2000) Interface characterization of the type II module pair from fibronectin. Biochemistry, 39, 8374–8381. [DOI] [PubMed] [Google Scholar]
- Steffensen B.J., Wallon,U.M. and Overall,C.M. (1995) Extracellular matrix binding properties of recombinant fibronectin type II-like modules of human 72-kDa gelatinase/type IV collagenase. High affinity binding to native type I collagen but not native type IV collagen. J. Biol. Chem., 270, 11555–11566. [DOI] [PubMed] [Google Scholar]
- Sticht H., Pickford,A.R., Potts,J.R. and Campbell,I.D. (1998) Solution structure of the glycosylated second type 2 module of fibronectin. J. Mol. Biol., 276, 177–187. [DOI] [PubMed] [Google Scholar]
- Wilhelm S.M., Collier,I.E., Marmer,B.L., Eisen,A.Z., Grant,G.A. and Goldberg,G.I. (1989) SV40-transformed human lung fibroblasts secrete a 92 kDa type IV collagenase which is identical to that secreted by normal human macrophages. J. Biol. Chem., 264, 17213–17221. [PubMed] [Google Scholar]
- Williams E.C., Janmey,P.A., Ferry,J.D. and Mosher,D.F. (1982) Con formational states of fibronectin. J. Biol. Chem., 257, 14973–14978. [PubMed] [Google Scholar]
- Wishart D.S., Bigam,C.G., Yao,J., Abildgaard,F., Dyson,H.J., Oldfield,E., Markley,J.L. and Sykes,B.D. (1995) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR, 6, 135–140. [DOI] [PubMed] [Google Scholar]